Abstract
Abstract 1265
Acquired severe aplastic anemia (SAA) is a human disease characterized by severe pancytopenia due to marked reduction in the numbers of hematopoietic progenitors and stem cells. Immunosuppressive treatment with antithymoglobulin and cyclosporine results in improvement in blood counts in above 2/3 of patients. The most serious long-term complication in SAA is progression to myelodysplasia (MDS) and acute myeloid leukemia, usually associated with the cytogenetic abnormality monosomy 7. Clonal evolution usually requires a hematopoietic stem cell transplant and it is the major cause of morbidity and mortality.
Decreased average telomere length in leukocytes on presentation has been associated with increase risk of progression to MDS (Scheinberg et. al., 2010 JAMA, vol. 304(12):1358–64). Single telomere length assay (STELA) uses single molecule polymerase chain reaction (PCR) to amplify chromosome-specific telomeres based on the specificity of their subtelomeric region (Baird et. al., 2003 Nature Genetics, vol. 33(2):203–7). We have utilized STELA for identify SAA patients at risk of progression to MDS prior to development of the cytogenetic abnormality.
Peripheral blood samples were obtained at diagnosis and sequentially after immunosuppressive treatment from SAA patients who fulfilled entry criteria for protocols at the National Institutes of Health. Chromosome-specific PCR amplification of telomere using primers specific for Xp/Yp subtelomeric regions was performed. A non-radioactive, digoxin-labeled telomeric probe was used for Southern blotting to identify discrete bands, allowing quantification of very short telomeres. Results were confirmed using chromosome-specific primers for 2p, 11q, 12q, and 17p.
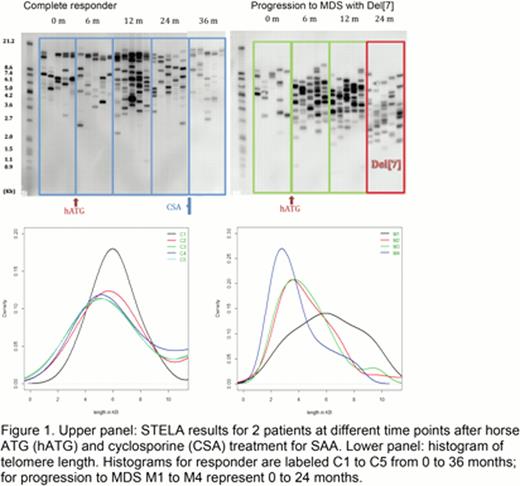
Initially, multiple serial samples from five patients who had progressed to monosomy 7 MDS were analyzed by STELA of DNA extracted from peripheral blood leukocytes at the diagnosis of SAA and again at progression to MDS. All five patients had increase in very short telomeres (<3kb) at the time of progression to MDS. Average telomere length obtained by STELA was consistent with the telomere length measured by qPCR and Southern blotting. Serial samples were next analyzed using both patients that had progressed to MDS as well as SAA patients who had been treated with the same regimen and were sustained complete responders. Very short telomeres were present in a higher proportion in all SAA patients progressing to MDS when compared with complete responders, as shown in Figure 1.
This novel, non-radioactive method of quantifying chromosome-specific telomere length is capable of identifying very short telomeres in SAA patients progressing to MDS with monosomy 7. Previous studies have associated average short telomere length with chromosomal instability and progression to malignant phenotype. Identifying very short telomere in a subpopulation of circulating cells by STELA regardless of the average telomere length by qPCR is a more sensitive method. Chromosome-specific primers and analysis of end-to-end fusion of different chromosomes will allow for better characterization of the chromosomal instability leading to malignant transformation in these patients. Allogeneic stem cell transplant is the definitive treatment for MDS with monosomy 7 but requires time for identification of a suitable donor. Identification of critically short telomere before development of the cytogenetic abnormality will allow for timely management of patients at risk of clonal evolution. Therapeutic telomerase upregulation by androgens might reduce very short telomeres and potentially decrease the transformation risk.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal