Abstract
Protease nexin-1 (PN1) is a specific and extremely efficient inhibitor of thrombin. However, unlike other thrombin inhibitors belonging to the serpin family, PN1 is not synthesized in the liver and does not circulate in the blood. Rather, PN1 is expressed by multiple cell types, including macrophages, smooth muscle cells, and platelets, and it is on the surface of these cells, bound to glycosaminoglycans, that PN1 inhibits the signaling functions of thrombin. PN1 sets the threshold for thrombin-induced platelet activation and has been implicated in atherosclerosis. However, in spite of the emerging importance of PN1 in thrombosis and atherosclerosis, little is know about how it associates to cells and how it inhibits thrombin at rates that surpass the diffusion limit. To address these issues, we determined the crystal structures of PN1 in complex with heparin, and in complex with catalytically inert thrombin. The crystal structures suggest a unique 2-step mechanism of thrombin recognition involving rapid electrostatics-driven association to form an initial glycosaminoglycan-bridged complex, followed by a large conformational rearrangement to form the productive Michaelis complex.
Introduction
Protease nexin-1 (PN1, also known as glia-derived nexin and SERPINE2) is a member of the serine protease inhibitor (serpin) family.1,2 The mechanism by which serpins inhibit proteases involves a major conformational change concomitant with a distortion of the target protease.3 This mechanism is irreversible with a high degree of complexity and was naturally selected over other inhibitor families to regulate the tightly controlled processes in human such as hemostasis and fibrinolysis.4 Most of the proteases involved in blood coagulation are regulated by serpins.5 Thrombin is the central enzyme in blood coagulation and is of vital importance to human life.6 All endogenous inhibitors of thrombin are serpins, namely, antithrombin (AT), for inhibiting the majority of thrombin generated during blood coagulation; heparin cofactor II (HCII) that inhibits thrombin in a dermatan sulfate–dependent manner in the arterial vasculature; protein C inhibitor (PCI) that inhibits thrombin in complex with thrombomodulin; and PN1 that inhibits thrombin at the vessel wall and platelet surface. Despite the conserved protein fold and inhibition mechanism, these serpins have evolved radically different regulatory mechanisms with regard to thrombin recognition.5,7 AT circulates at a high concentration in a low-activity state, and it is of principal importance for inhibiting luminal thrombin. In the microvasculature, AT associates with heparan sulfate with high affinity to increase its rate of thrombin inhibition. HCII also circulates at a high concentration but in an inert state until it associates loosely to dermatan sulfate. PCI circulates at a low concentration in an active state, but it is a poor inhibitor of thrombin in the absence of cofactors such as thrombomodulin. PN1, in contrast, is found exclusively bound to glycosaminoglycans (GAGs) on various cell surfaces, and it is not found in the circulation in its free (apo) form.2 PN1 is only free of GAGs in vitro, after purification from fibroblasts cells or from recombinant systems. In its apo state, it is the fastest thrombin inhibitor of all the serpins (1 × 106M−1s−1), and in the presence of GAGs (such as heparin) its rate of inhibition (∼ 109M−1s−1) exceeds the diffusion limit for a normal bimolecular reaction.8 The extremely high rate of inhibition ensures that any thrombin that reaches the surface of a cell on which PN1 resides will be inhibited before thrombin can effect any other activity, until, that is, the PN1 is exhausted. PN1 therefore has been proposed to play a protective role against the proliferative effects of thrombin on smooth muscle cells9 and to set the “threshold” for platelet activation by thrombin.10 Recent studies have demonstrated an important antithrombotic effect of platelet PN1 in vitro and in vivo.11
We have reported previously on the structural basis of thrombin recognition by serpins AT,12 HCII,13 and PCI.14 Here, we complete the structural study of thrombin recognition by serpins by solving the crystal structures of the PN1-heparin complex and of PN1 in complex with S195A thrombin. We show that native PN1 has a typical serpin fold with a highly positively charged helix D harboring the heparin-binding residues and an additional basic surface on and around the reactive center loop (RCL) that facilitates the formation of the initial encounter complex with thrombin. Two distinct complexes were found in the crystal of PN1 and thrombin: one complex in which the heparin-binding sites of the 2 proteins are aligned, but there is a nonproductive engagement of the RCL in thrombin's active site; and another complex that is a productive Michaelis complex but is unable to be bridged by heparin. We also observed that the formation of the productive Michaelis complex was associated with a disruption of the structure of thrombin, in particular, the unfolding of the heparin-binding exosite II. We propose a 2-step mechanism of thrombin recognition by PN1 where an initial heparin-bridged complex is rapidly formed because of electrostatic attraction, without the full engagement of the RCL, followed by a rotation of thrombin relative to PN1 to dock the RCL and fully engage recognition exosites. Formation of the second complex results in the dissociation of heparin from thrombin before the translocation of thrombin to form the final inhibitory complex. The final PN1-thrombin complex remains bound to the cell surface through the persistently tight interaction between PN1 and GAGs to promote internalization and cell signaling.
Methods
Protein production and purification
Catalytically inactive S195A thrombin was the kind gift of Charles T. Esmon (Oklahoma Medical Research Foundation) and was produced as described previously.12 For generating protease nexin-1, full-length cDNA of the β-isoform of human PN1, starting at serine 20 (NM_006216, after signal peptide), was cloned into pET19b between NdeI and XhoI site (XhoI site in the PN1 gene was mutated out without changing the amino acid sequence before cloning). The insert was confirmed by DNA sequencing. Bacteria strain Rossetta DE3 pLysS was used for protein expression. In brief, cells were grown at 37°C until mid-log phase followed by isopropyl β-d-thiogalactoside induction at 25°C overnight. Cells were harvested the next day and resuspended in Tris-buffered saline with Complete protease inhibitor (EDTA-free; Roche). After sonication, cell pellets were separated by centrifugation, and the supernatant was loaded onto a 5-mL nickel-nitrilotriacetic acid column (GE Healthcare). Subsequent heparin-Sepharose and S-75 gel filtration chromatography resulted in PN1 of greater than 99% purity.
Crystallization, data collection, and refinement
The PN1-heparin complex was formed by mixing recombinant PN1 at 7.8 mg/mL with 2.5-fold molar excess of 10mer heparin (Iduron) in a buffer containing 20mM Tris-HCl, pH 7.4, 100mM NaCl, 5mM EDTA, and 1mM DTT. Diffraction quality crystals were obtained from hanging drop plates with a 1:1 ratio of protein and 25mM sodium acetate buffer, pH 4.8, 13% polyethylene glycol (PEG)–3350, and 200mM ammonium sulfate. Crystals were cryoprotected in 25mM sodium acetate buffer, pH 4.8, 13% PEG-3350, 0.2M ammonium sulfate, and 10μM 10mer heparin, with 25% glycerol. The PN1-thrombin complex was formed by mixing PN1 and S195A thrombin in a 1:1 molar ratio at low concentration, followed by dialysis into 20mM Tris-HCl pH 7.4, and 50mM NaCl. The complex was then concentrated to 4 mg/mL before setting up crystallization trials. Crystals appeared within a week in 140mM calcium acetate and 13% PEG-3350 and were cryoprotected in 140mM calcium acetate, 20% PEG-3350, and 20% glycerol. Data were collected at 100K at the Diamond Light Source station I02 (Diamond Light Source) from a single crystal for each complex. Data were processed using Mosflm, Scala, and Truncate,15 and structures were solved by molecular replacement using Phaser.16 Iterative rebuilding and refinement were conducted using Coot17 and Refmac.18 Figures were produced using Pymol,19 and electrostatic potentials were calculated using PDB2PQR20 server and APBS software.21 Crystallographic data for the 2 structures are given in Table 1, and the coordinates and structure factors have been deposited in the Protein Databank under accession codes 4DY0 and 4DY7.
Crystals, data processing, refinement, and models
| . | PN1–10mer (4DY0) . | PN1-Thr (4DY7) . |
|---|---|---|
| Crystals | ||
| Space group | P41212 | C2 |
| Cell dimensions, Å | a = b = 140.65 c = 93.55 α = β = γ = 90° | a = 191.96 b = 86.67 c = 101.89 β = 94.44° |
| Solvent content, % | 46.8 | 59.8 |
| Data processing statistics | ||
| Wavelength, Å | 0.9795 | 0.9796 |
| Resolution, Å | 68.14-2.35 (2.48-2.35) | 51.37-2.80 (2.95-2.80) |
| Total reflections | 279 231 | 117 303 |
| Unique reflections | 39 468 | 39 308 |
| < I/σ(I) > | 11.1 (2.5) | 6.4 (1.6) |
| Completeness (%) | 100 (100) | 95.3 (96.7) |
| Multiplicity | 7.1 (6.9) | 3.0 (2.9) |
| Rpim* | 0.051 (0.335) | 0.073 (0.425) |
| Model | ||
| No. of protein/other atoms | 5 633/317 | 9 859/126 |
| Average B-factor, Å2 | 26.3 | 42.7 |
| Refinement statistics | ||
| Reflections in working/free set | 35 419/1 976 | 35 364/1 956 |
| R-factor/R-free | 18.17/22.63 | 22.28/26.79 |
| Root-mean-square deviation of bonds (Å)/angles (°) from ideality | 0.004/0.83 | 0.002/0.56 |
| Ramachandran plot† | ||
| Favored, % | 98.1 | 91.5 |
| Outlier, % | 0.1 | 0.7 |
| . | PN1–10mer (4DY0) . | PN1-Thr (4DY7) . |
|---|---|---|
| Crystals | ||
| Space group | P41212 | C2 |
| Cell dimensions, Å | a = b = 140.65 c = 93.55 α = β = γ = 90° | a = 191.96 b = 86.67 c = 101.89 β = 94.44° |
| Solvent content, % | 46.8 | 59.8 |
| Data processing statistics | ||
| Wavelength, Å | 0.9795 | 0.9796 |
| Resolution, Å | 68.14-2.35 (2.48-2.35) | 51.37-2.80 (2.95-2.80) |
| Total reflections | 279 231 | 117 303 |
| Unique reflections | 39 468 | 39 308 |
| < I/σ(I) > | 11.1 (2.5) | 6.4 (1.6) |
| Completeness (%) | 100 (100) | 95.3 (96.7) |
| Multiplicity | 7.1 (6.9) | 3.0 (2.9) |
| Rpim* | 0.051 (0.335) | 0.073 (0.425) |
| Model | ||
| No. of protein/other atoms | 5 633/317 | 9 859/126 |
| Average B-factor, Å2 | 26.3 | 42.7 |
| Refinement statistics | ||
| Reflections in working/free set | 35 419/1 976 | 35 364/1 956 |
| R-factor/R-free | 18.17/22.63 | 22.28/26.79 |
| Root-mean-square deviation of bonds (Å)/angles (°) from ideality | 0.004/0.83 | 0.002/0.56 |
| Ramachandran plot† | ||
| Favored, % | 98.1 | 91.5 |
| Outlier, % | 0.1 | 0.7 |
Precision-indicating merging R factor value.
Calculated using MolProbity.42
Results
Overall structure of heparin-bound PN1
Two copies of PN1 were found in the asymmetric unit (supplemental Figure 1A-B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The overall fold is identical for the 2 monomers, with a Cα RMSD of 0.88 Å, for 361 residues. However, the structures diverged significantly from one another in the conformation of 4 segments: 97 to 102, comprising the loop between helix E and strand 3A; 120 to 161, comprising strand 1A, helix F, and the loop following helix F; 309 to 314, comprising the loop preceding strand 5A; and 341 to 349, corresponding to the C-terminal portion of the RCL (supplemental Figure 1B-C). The 120-161 region seems to shift as a single rigid body, whereas the other changes are plastic. All of the apparent conformational differences can be accounted for by differences in crystal contacts for molecules A and B and reflect regions of inherent mobility in the serpin structure. Monomer B was chosen as the representative native structure of PN1 because it was the most fully traced in electron density (370 vs 362 of the 379 residues) and because it matched more closely the structures of PN1 bound to thrombin. The model is complete, with the exception of the inherently flexible N terminus (residues 1-3) and the N-terminal portion of the RCL (335-340, corresponding to P12-P7, using the traditional substrate nomenclature22 ).
The structure of native PN1 is unsurprisingly similar to that of the archetypal serpin α1-antitrypsin23 (α1AT), composed of the same secondary structural elements (9 helices and 3 β-sheets) arranged in a similar manner to make the typical serpin fold (Figure 1A). However, as reflected in a Cα RMSD of 2.45 Å, PN1 deviates from the structure of α1AT (1QLP) significantly in certain features, in particular, in the orientation of the helices (supplemental Figure 2A). PN1 is most similar in sequence to PAI-1 (41% identical), and the native structures of these 2 serpins correspond very well (supplemental Figure 2B), with a Cα RMSD of 1.04 Å (1DB224 ). The only major structural differences were in the helix F loop (position of stabilizing mutations in the PAI-1 structure) and the helix E-s1A feature (the region of PAI-1 that interacts with the somatomedin B domain of vitronectin25 ).
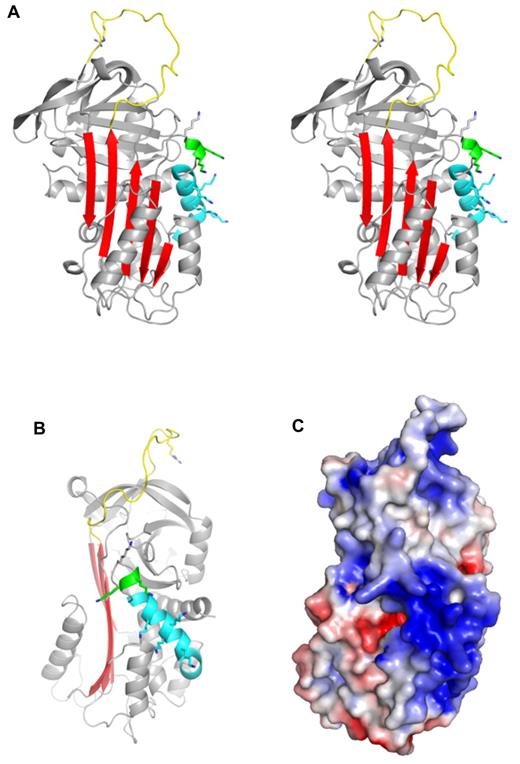
Crystal structure of native PN1. (A) A stereo diagram representation of PN1 (monomer B) is shown in the classic orientation, with the RCL (yellow) on top and β-sheet A (red) facing. The missing N-terminal portion of the RCL is included for clarity, and the P1 arginine side chain is shown as sticks. Helix D is colored cyan, and the 310 helix C-terminal to helix D is green. Putative heparin-binding lysine side chains are shown to illustrate their distribution. (B) Native PN1 is colored as in panel A but oriented so that the heparin-binding site (helix D) is facing. (C) The surface electrostatic representation of PN1 is shown in the orientation of panel B (blue for positive and red for negative).
Crystal structure of native PN1. (A) A stereo diagram representation of PN1 (monomer B) is shown in the classic orientation, with the RCL (yellow) on top and β-sheet A (red) facing. The missing N-terminal portion of the RCL is included for clarity, and the P1 arginine side chain is shown as sticks. Helix D is colored cyan, and the 310 helix C-terminal to helix D is green. Putative heparin-binding lysine side chains are shown to illustrate their distribution. (B) Native PN1 is colored as in panel A but oriented so that the heparin-binding site (helix D) is facing. (C) The surface electrostatic representation of PN1 is shown in the orientation of panel B (blue for positive and red for negative).
Heparin binding
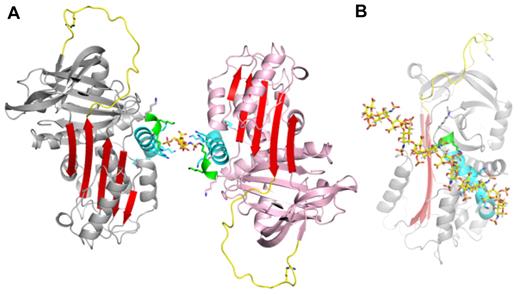
PN1 is a highly basic protein, with high positive charge densities along helix D and on the “top” of the molecule, on and adjacent to the RCL (Figure 1C). The heparin-binding site of PN1 has been shown by mutagenesis to comprise several lysine residues decorating the length of helix D.26 Our structure of PN1 reveals the positions of the lysine residues, and the unusual feature of a 310 helix at the C-terminal end of helix D, stabilized by 5 H-bonds made by the side chain of asparagine 85 (Figure 1A-B and supplemental Figure 3). The lysine side chains could not be completely built into electron density, presumably because they are sampling multiple conformations, but have been added in Figure 1 to illustrate their positions and their contributions to the electrostatics of the helix D region. Heparin is likely to be bound along the basic site of helix D, in a manner similar to what has been observed for AT binding to heparin.27 However, in spite of the high affinity of PN1 for heparin28 and the fact that crystals do not form in the absence of heparin, we could only observe weak density for a single disaccharide unit (supplemental Figure 4). The reason for this is likely to be the juxtaposition of the heparin-binding regions of the 2 monomers within the crystal. Indeed, we chose the composition of our asymmetric unit (supplemental Figure 1A) based on the intimate nature of the dimer contact (1817 Å2). However, an alternative asymmetric unit can be selected that juxtaposes the 2 heparin-binding sites with no apparent intermolecular crystallographic contact (Figure 2A). The lack of intermolecular contact is consistent with the charge repulsion between these regions (helix D), and one would expect that in the absence of bound heparin such a repulsive interface would preclude crystallization. The symmetry of the sulfation pattern of heparin allows it to bind the 2 monomers in an essentially identical manner if it were to lie along the 2-fold axis (supplemental Figure 5A-B). This binding mode corresponds well with the disaccharide unit placed into weak electron density and may indeed be the favored binding mode within the crystal lattice. However, there is good reason to believe that this mode represents only one of several heparin orientations available to PN1. Indeed, the observed or proposed orientation of heparin shown in Figure 2B would be unlikely to support bridging of thrombin (consistent with the structures described in the next section; supplemental Figure 5B). In addition, the lack of electron density for the sulfate moieties of heparin and for the lysine side chains of helix D suggest multiple heparin-binding modes of similar affinity for PN1. Moreover, the extended crystal packing is permissive of free rotation of the 10mer heparin between the heparin-binding sites at the dimer interface.
Observed heparin-binding mode of PN1. (A) A single heparin disaccharide unit (yellow sticks) was built into electron density between 2 loosely associated monomers. This alternate asymmetric unit is “glued” together by heparin that is juxtaposed between the 2 highly basic heparin-binding sites (structural elements of both molecules are colored as in Figure 1, with remaining regions in gray or pink). (B) An extension of the disaccharide to a decasaccharide along the 2-fold axis nicely satisfies the charged groups along helix D (oriented as in Figure 1B).
Observed heparin-binding mode of PN1. (A) A single heparin disaccharide unit (yellow sticks) was built into electron density between 2 loosely associated monomers. This alternate asymmetric unit is “glued” together by heparin that is juxtaposed between the 2 highly basic heparin-binding sites (structural elements of both molecules are colored as in Figure 1, with remaining regions in gray or pink). (B) An extension of the disaccharide to a decasaccharide along the 2-fold axis nicely satisfies the charged groups along helix D (oriented as in Figure 1B).
Two distinct PN1-thrombin complexes
In addition to the structure described in the preceding section, we also solved the crystal structure of PN1 bound to S195A thrombin to determine the molecular basis of recognition. We found 2 complexes in the asymmetric unit with distinct orientations of thrombin on PN1 (Figure 3). In the first complex, thrombin is oriented toward the front of PN1, aligning thrombin's heparin-binding site (exosite II, indicated) with helix D of PN1 (Figure 3A). In the second complex, thrombin is rotated ∼ 130° relative to its position in complex 1, so that exosite II is no longer in alignment with helix D of PN1 (Figure 3B). Indeed, the position of exosite II in the second complex would preclude simultaneous binding of heparin to both PN1 and thrombin (supplemental Figure 6). Both complexes engage the RCL in the active site cleft, including the insertion of the P1 arginine into the S1 pocket, but only complex 2 engages the RCL in a manner that would be permissive of proteolytic cleavage of the P1-P1′ bond (Figure 4). Thus, we surprisingly observe 2 complexes that are each at once competent and deficient—complex 1 is easily bridged by a linear heparin molecule but has failed to productively engage thrombin's active site; whereas complex 2 has correctly engaged the RCL of PN1 into the catalytic site of thrombin but is unable to be bridged by heparin.
Stereo views of the crystal structures of two separate encounter complexes between PN1 and thrombin. (A) Complex 1 is shown with PN1 colored as described in Figure 1 and thrombin colored magenta. The P1 arginine and P1′ serine side chains are shown as sticks, and the heparin-binding residues of thrombin's exosite II are depicted as blue balls. In complex 1 the heparin-binding sites of PN1 and thrombin are in close proximity and aligned. (B) Complex 2 is shown, colored as in panel A. Thrombin has rotated by 130° so that its heparin-binding site is now oriented toward the back, away from the heparin-binding helix of PN1 (cyan).
Stereo views of the crystal structures of two separate encounter complexes between PN1 and thrombin. (A) Complex 1 is shown with PN1 colored as described in Figure 1 and thrombin colored magenta. The P1 arginine and P1′ serine side chains are shown as sticks, and the heparin-binding residues of thrombin's exosite II are depicted as blue balls. In complex 1 the heparin-binding sites of PN1 and thrombin are in close proximity and aligned. (B) Complex 2 is shown, colored as in panel A. Thrombin has rotated by 130° so that its heparin-binding site is now oriented toward the back, away from the heparin-binding helix of PN1 (cyan).
Close-up stereo views of the P5-P3′ region of the RCL of PN1 in the active site of thrombin for the 2 complexes. (A) Thrombin is shown in ribbon diagram (gray) in the standard orientation, with the side chains of the catalytic triad shown as sticks and the oxyanion hole represented by blue balls. The RCL of complex 1 is in cyan and that of complex 2 is in yellow, and both are in stick representation from P5 (left) to P3′ (right). In complex 1, the main chain oxygen atom of P1 arginine is not hydrogen bonded to the oxyanion hole. (B) A surface view of thrombin (colored according to electrostatics) illustrates how the RCL of the first complex exits the active site of thrombin at P1′, and how the side chain of Glu192 (indicated) serves to bifurcate the active site into a productive engagement channel (occupied by the RCL of PN1 in complex 2, yellow), and a nonproductive channel (occupied by the RCL of PN1 in complex 1, cyan).
Close-up stereo views of the P5-P3′ region of the RCL of PN1 in the active site of thrombin for the 2 complexes. (A) Thrombin is shown in ribbon diagram (gray) in the standard orientation, with the side chains of the catalytic triad shown as sticks and the oxyanion hole represented by blue balls. The RCL of complex 1 is in cyan and that of complex 2 is in yellow, and both are in stick representation from P5 (left) to P3′ (right). In complex 1, the main chain oxygen atom of P1 arginine is not hydrogen bonded to the oxyanion hole. (B) A surface view of thrombin (colored according to electrostatics) illustrates how the RCL of the first complex exits the active site of thrombin at P1′, and how the side chain of Glu192 (indicated) serves to bifurcate the active site into a productive engagement channel (occupied by the RCL of PN1 in complex 2, yellow), and a nonproductive channel (occupied by the RCL of PN1 in complex 1, cyan).
Active site interactions
For serpin-protease Michaelis complexes it is generally considered that contacts involving P4-P3′ constitute active site interactions, and those that involve other regions are defined as exosite interactions.29 The active site interactions for the 2 PN1-thrombin complexes are clearly defined by electron density (supplemental Figure 7), but they are unusual in some aspects. In complex 1 (Figure 3A), the RCL of PN1 has adopted a helical conformation from residues 338 to 344 (P9-P3). This conformation is not seen in either of the 2 copies of native PN1 and is therefore likely to have been induced by its interaction with thrombin. This conformation allows the P4 leucine side chain to interact with the S4 pocket on thrombin; however, no other RCL-active site interactions are normal. As mentioned in the preceding section, complex 1 has engaged the active site but in a nonproductive manner, meaning that the P1 residue is partially buried in the S1 pocket but does not dock in the manner observed for other thrombin substrates or inhibitors (Figure 4). Importantly, the P1′ serine residue is not in its canonical position, instead running below the protruding glutamic acid 192 side chain (Figure 4B) and away from the oxyanion hole (Figure 4A). The RCL of complex 2 engages the P1-P1′ bond in the canonical, productive manner, but little else about the active site interaction is normal. In particular, the P4 side chain is seen pointing away from thrombin, and the S4 pocket seems to interact instead with the P3 residue, although no density for the P3 isoleucine side chain is observed. In contrast, the P′ region makes extensive contacts up to the P5′ residue.
Exosite interactions
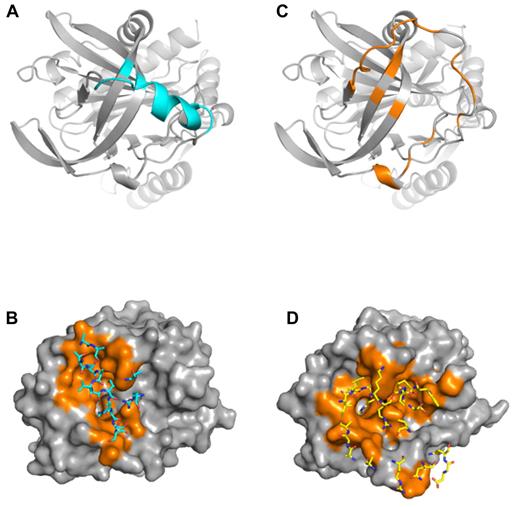
Despite the similar amount of total buried surface area for the 2 complexes (1535 Å2 for complex 1 and 2017 Å2 for complex 2), the exosite interfaces differ greatly. Of the 767 Å2 buried on PN1 in complex 1, 67% (518 Å2) comes from the P4-P3′ region of the RCL, and, by definition, the remainder (249 Å2) is contributed by exosite contacts. However, two-thirds of this “exosite” surface actually derives from the N-terminal portion of the RCL (P9-P6), and the only residues outside the RCL that contact thrombin are Ser205, Val206, and Arg269, from strands 3C and 2C, respectively (Figure 5A). The 2 principal contacts are Val206 that interacts with the side chain of Trp60d of thrombin and Arg269 that makes a double salt bridge to Glu192 and a hydrophobic contact with Trp60d. These 2 residues constitute the exosite of PN1 in the nonproductive complex and amount to a contact surface of only 79 Å2 (Figure 5A-B). In contrast, thrombin in complex 2 buries 1074 Å2 of PN1, of which 640 Å2 are in the P4-P3′ region of the RCL, 131 Å2 are in the extended N-terminal part of the RCL, and 304 Å2 are bona fide exosite contacts. These include the 177 to 182 portion of PN1 that precedes strand 4C, and residues from strands 1, 2, and 3 of β-sheet C (Figure 5C-D). The principal exosite contact buries 72 Å2 of Arg269, through the formation of 4 salt bridges to Glu192 and a side-chain H-bond with asparagine 143. Two additional H-bonds are formed between Glu39 of thrombin and glycine 271 and tryptophan 351 of PN1.
Contact regions for PN1 and thrombin in complexes 1 and 2. (A) Ribbon diagram of the top of PN1 of complex 1 is colored cyan to illustrate regions in contact with thrombin (residues with atoms within 5 Å). The principal contacts are the RCL, with only 3 residues comprising the exosite. (B) Surface representation of thrombin (gray, with contact residues in orange) illustrates the extent of interaction with PN1 in complex 1. Contacting PN1 residues are shown as sticks and colored cyan. (C) PN1 from complex 2 is shown in the same orientation as in panel A, colored orange to indicate the extensive interface with thrombin. (D) Surface of thrombin interacting with PN1 in complex 2 is shown, colored as in panel B, but with PN1 residues shown as yellow sticks.
Contact regions for PN1 and thrombin in complexes 1 and 2. (A) Ribbon diagram of the top of PN1 of complex 1 is colored cyan to illustrate regions in contact with thrombin (residues with atoms within 5 Å). The principal contacts are the RCL, with only 3 residues comprising the exosite. (B) Surface representation of thrombin (gray, with contact residues in orange) illustrates the extent of interaction with PN1 in complex 1. Contacting PN1 residues are shown as sticks and colored cyan. (C) PN1 from complex 2 is shown in the same orientation as in panel A, colored orange to indicate the extensive interface with thrombin. (D) Surface of thrombin interacting with PN1 in complex 2 is shown, colored as in panel B, but with PN1 residues shown as yellow sticks.
Conformational disorder in exosite II
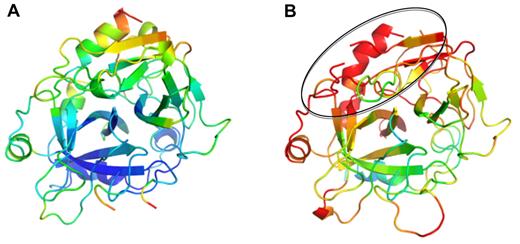
Although the 2 thrombins in complex with PN1 are similar in overall conformation (Cα RMSD of 0.75 Å for 246 residues), thrombin in the second complex is remarkably disordered. This is manifested by poor electron density and high B-factors. We were unable to model thrombin residues 93, 125 to 126, and 243 to 247 into electron density in complex 2, and the average Cα B-factors for thrombin in complex 1 is 40.1 Å2, compared with 64.2 Å2 in complex 2 (Figure 6). The missing residues and the areas of highest B-factor correspond to thrombin's exosite II. Exosite II is the heparin-binding site, and it has been shown by mutagenesis and structural studies to include histidine 91, Arg93, Arg101, Arg126, Arg233, Lys236, and Lys240.30,31 Of these, Arg93 and Arg126 cannot be modeled in electron density, and the average Cα B-factor for the remaining heparin-binding residues is 88 Å2 (compared with 50.6 Å2 for the corresponding residues in complex 1). It would thus be expected that thrombin in complex 2 would bind to exosite II ligands, including GAGs, with greatly reduced affinity, similar to what has been described for final serpin-thrombin complexes.32
Thrombin from complexes with PN1 colored according to B-factors. Ribbon diagram of thrombin is shown in the standard orientation, colored in rainbow according to B-factor (from blue to red, corresponding to B-factors from 20 Å2 to 80 Å2). Thrombin from complex 1 (A) is ordered and mostly blue; however, thrombin from complex 2 (B) is highly disordered (red), particularly in the exosite II region (indicated by oval).
Thrombin from complexes with PN1 colored according to B-factors. Ribbon diagram of thrombin is shown in the standard orientation, colored in rainbow according to B-factor (from blue to red, corresponding to B-factors from 20 Å2 to 80 Å2). Thrombin from complex 1 (A) is ordered and mostly blue; however, thrombin from complex 2 (B) is highly disordered (red), particularly in the exosite II region (indicated by oval).
Discussion
In this study, we set out to determine how PN1 recognizes heparin-like GAGs and its target protease thrombin by solving crystal structures of the complexes. These 2 interactions govern PN1 activity by localizing thrombin to cell surfaces and by conferring exquisite specificity for thrombin. Our structures reveal a normal serpin architecture, but with unusual features particular to the requirement of PN1 to bind to cell surface GAGs during each stage of its life cycle.
We were unable to obtain crystals of native PN1 in the absence of heparin. This is perhaps unsurprising because of the highly basic nature of PN1 (theoretical pI is 9.4 and net charge of +10, for the β-isoform without glycosylation).33 However, in the presence of a 10-monosaccharide unit heparin chain PN1 crystallized readily, and its structure was solved to 2.35 Å. Although the crystals were grown in the presence of heparin, we conclude that the resulting structure represents that of physiologically relevant native PN1, because in biology PN1 is always tightly bound to GAGs on cell surfaces.
The lack of electron density for heparin and for the lysine side chains along helix D suggests at least 2 heparin-binding modes. Heparin is a helical rodlike molecule because of its extreme negative charge density, and although it is not strictly symmetrical, the placement of its charges is nearly equivalent on its 2 faces.34 This means that heparin can bind most proteins with similar affinities independently of direction (nonreducing end to reducing end) and of register. Because of this, heparin is often found “sandwiched” between 2 molecules, mediating dimerization,31 and is notoriously difficult to model into electron density. However, it is normally possible to observe important sulfate moieties and interacting side chains. Here, we find 2 heparin-binding helices juxtaposed with nothing to limit the rotation of heparin along its long and short axes. The fact that we observed no strong density for the sulfate groups of heparin suggests that it is not binding in a prescribed manner and is likely to sample multiple orientations relative to helix D. Placement of heparin along the 2-fold axis, as shown in supplemental Figure 5A, nicely aligns the D helices and the lysine side chains with heparin but in an orientation incapable of bridging thrombin (Figure 2B and supplemental Figure 5B). It is thus clear that heparin can rotate to some extent across the face of helix D, while maintaining high affinity, at least within the crystal lattice. A 41° rotation from the placement of heparin along the 2-fold crystallographic axis would be sufficient to bridge PN1 to thrombin in complex 1, as shown in supplemental Figure 5B. The pivot point corresponds nicely to the weak density observed for the disaccharide in the middle of the D-helix.
Previous crystal structures of serpin-protease Michaelis complexes contained either a single complex or 2 identical complexes in the asymmetric unit. This makes interpretation easy, because the resulting structure is assumed to represent the recognition complex in solution. In the low-resolution structure of the AT-Xa Michaelis complex,35 2 identical copies were found in the asymmetric unit, supporting the conclusion that a single set of interactions defines serpin-protease Michaelis complexes. In a counter example, two very different structures of the AT-thrombin complex have been described,12,36 indicating that there may be some plasticity in the protein-protein contacts. In a single crystal of PN1 and S195A thrombin we observe 2 very different and unusual complexes: 1 complex is nonproductive because of the misalignment of the P1′ residue, but permits bridging by a linear heparin chain; the other complex is poised for proteolytic attack of the P1-P1′ bond, but the position and conformation of thrombin would not permit heparin bridging. We are thus faced with the conundrum of deciding which of the 2 complexes is relevant to the recognition of thrombin by PN1 and thus which explains the exceedingly fast rate of thrombin inhibition by PN1 in the presence of GAGs. However, because both properties are necessary (heparin bridging and proper P1-P1′ engagement), it is possible that they each represent a separate state on the pathway of inhibition. Because PN1 is resident on cell surface GAGs, it is reasonable to conclude that GAGs will always participate in approximating PN1 and thrombin. Because of the rodlike nature of heparin, formation of the initial bridged complex requires the 2 heparin-binding sites to be aligned. Additional electrostatic complementarity will orient thrombin for docking to the “top” of PN1 to aid in rapid formation and stabilization of the bridged complex. Accordingly, the region surrounding and including the RCL of PN1 presents a highly basic surface (Figure 1C) to the highly negatively charged active site of thrombin (Figure 4B). Thus, the driving force for formation of the bridged complex will be electrostatics, initiated by the simultaneous binding of thrombin to the GAG on which PN1 is already resident, followed linear diffusion along the GAG to engage the P1 arginine and Arg269 with the negatively charged active site cleft of thrombin, including the S1 pocket and Glu192. Once this has occurred, thrombin can rotate to fully engage the P1 arginine residue and Arg269, the P′ side of the RCL, and further exosite residues, to form the productive Michaelis complex. Although we do not know which of the 2 observed complexes is more stable, we can safely conclude that both are likely to be populated in solution for 2 reasons: (1) we observe both in the crystal structure, and crystal contacts stabilize a solution conformation rather than induce a novel conformation37 ; and (2) final inhibition cannot occur without a bridged complex and a productive Michaelis complex. It is possible that in the presence of heparin the bridged complex (complex 1) is more stable, because heparin can be thought of as an additional mutual exosite contact. In that case, the productive Michaelis complex might form slowly and be transiently populated. Such a scenario is still consistent with a rapid rate of thrombin inhibition, because the active site of thrombin will be blocked in the 2 docking complexes that precede final complex formation. It has been shown that serpins, including PN1, act as sticky substrates where the rate of formation of the initial docking (or Michaelis) complex is indistinguishable from the rate of formation of the final complex.38 Thus, the encounter complex between PN1 and thrombin would be as inhibitory as the final complex.
We have recently shown that thrombin in its final complex with serpins is functionally deficient with respect to exosite II binding and that the observed loss of affinity for heparin and other exosite II ligands was because of the conformational disordering of the protease.32 Surprisingly, the thrombin molecule in the second complex with PN1 was poorly ordered, with several residues lacking density and with highly elevated B-factors. Although the effect was global, disorder was particularly evident in the regions that make up exosite II. It is possible to argue that the observed disorder is due to poor crystal packing or some component of the crystallization solution, or perhaps an inadvertent destabilizing mutation. However, we have solved several structures using this thrombin preparation, including 3 serpin Michaelis complexes, and none showed any conformational disruption. Indeed, we have an internal control in complex 1 that demonstrates a normal stable fold for this thrombin preparation under these specific conditions. The disordering of thrombin in complex 2 is even more surprising, because occupancy of the active site by substrate or inhibitor is know to stabilize and order thrombin,39 but here an opposite effect is observed. Furthermore, our recent nuclear magnetic resonance study39 showed that exosite II was insensitive to ligation and was found to be stably formed even in the apo state. It is therefore necessary to conclude that something has happened to thrombin in complex 2 to destabilize thrombin and its exosite II and that the structural perturbation is caused by the productive engagement of PN1. Such a mechanism makes functional sense, because disengagement of heparin from thrombin should precede translocation of thrombin during formation of the final complex, particularly because PN1 retains high affinity for heparin throughout the process. However, although there is some precedence for allosteric linkage between the active site and exosites of thrombin,40,41 we cannot discern from the structure the cause of the apparent allosteric disordering of thrombin by PN1.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Medical Research Council (J.A.H.) and was carried out with the support of the Diamond Light Source.
Authorship
Contribution: W.L. set up PN1 expression and purification methods, produced and crystallized the PN1-heparin and PN1-thrombin complexes, collected and processed the data, solved, refined, and analyzed the structures, and wrote the paper; and J.A.H. processed the data, solved, refined, and analyzed the structures, wrote the paper, and directed the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for W.L. is Department of Medicine, University of Cambridge, Cambridge, United Kingdom.
Correspondence: Wei Li, Department of Medicine, University of Cambridge, Box 157, Addenbrooke's Hospital, Hills Road, Cambridge, United Kingdom CB2 0QQ; e-mail: wl225@cam.ac.uk; or James A. Huntington, Department of Haematology, Cambridge Institute for Medical Research, Wellcome Trust/MRC Building, Hills Road, University of Cambridge, Cambridge, United Kingdom CB2 0XY; e-mail: jah52@cam.ac.uk.