Abstract
Peripheral T-cell lymphoma (PTCL) is a rare, heterogeneous type of non-Hodgkin lymphoma (NHL) that, in general, is associated with a poor clinical outcome. Therefore, a current major challenge is the discovery of new prognostic tools for this disease. In the present study, a cohort of 122 patients with PTCL was collected from a multicentric T-cell lymphoma consortium (TENOMIC). We analyzed the expression of 80 small nucleolar RNAs (snoRNAs) using high-throughput quantitative PCR. We demonstrate that snoRNA expression analysis may be useful in both the diagnosis of some subtypes of PTCL and the prognostication of both PTCL-not otherwise specified (PTCL-NOS; n = 26) and angio-immunoblastic T-cell lymphoma (AITL; n = 46) patients treated with chemotherapy. Like miRNAs, snoRNAs are globally down-regulated in tumor cells compared with their normal counterparts. In the present study, the snoRNA signature was robust enough to differentiate anaplastic large cell lymphoma (n = 32) from other PTCLs. For PTCL-NOS and AITL, we obtained 2 distinct prognostic signatures with a reduced set of 3 genes. Of particular interest was the prognostic value of HBII-239 snoRNA, which was significantly over-expressed in cases of AITL and PTCL-NOS that had favorable outcomes. Our results suggest that snoRNA expression profiles may have a diagnostic and prognostic significance for PTCL, offering new tools for patient care and follow-up.
Introduction
Peripheral T-cell lymphoma (PTCL) is a heterogeneous type of non-Hodgkin lymphoma. The 2008 World Health Organization (WHO) classification recognizes the most common tumors within the PTCL group to be PTCL-not otherwise specified (PTCL-NOS), angio-immunoblastic T-cell lymphoma (AITL), and anaplastic large cell lymphoma (ALCL) with or without the anaplastic lymphoma kinase gene rearrangement (ALK+ ALCL or ALK− ALCL, respectively).1 To help in the diagnosis of PTCL subtypes, several gene-expression profiles have been produced that are based on coding gene analysis.2-9 However, certain subtypes remain difficult to accurately classify; for example, it is difficult to differentiate ALK−ALCL from CD30+ PTCL-NOS and AITL from PTCL-NOS expressing follicular Th-cell (TFH) markers. In addition, difficulties remain in obtaining reliable biologic tools that can predict the clinical behavior of PTCL subtypes, particularly after CHOP (cyclophosphamide, hydroxydaunorubicin, vincristine, and prednisone/prednisolone) therapy (the most accepted frontline treatment). Several scoring systems are available for prognostication, including: the International Prognostic Index (IPI), the IPI for PTCL (PIT), the modified PIT (mPIT), and the International PTCL Project Score (IPTCLP), but all have a major weakness in identifying poor- and very-poor-prognosis patients, making them somewhat meaningless.10 In fact, only 20% of PTCL patients can be cured with the CHOP regimen, among which 50% have a low IPI score.11 Overall, patients diagnosed with PTCL receive a rather grim prognosis.
Recently, a molecular signature was published that refined the classification of PTCL and helped to decipher some of the molecular pathways at play in the different subtypes of PTCL.2 These gene-expression profiles have also afforded prognostic markers, but only for AITL in which a cell of origin was proposed (the TFH cell).2 In-depth analysis of PTCL has brought to light new potential therapeutic targets. Several clinical studies are ongoing, with moderate activity so far as single agent. The most challenging issue with genetic signatures is the prognostication and distinction between PTCL-NOS and ALK− ALCL.12
In oncology, a large body of data are available for miRNA expression patterns,13,14 whereas the other classes of small noncoding RNAs have drawn less attention. Among these, the 60- to 300-nucleotide small nucleolar RNAs (snoRNAs) participate in diverse biologic processes, most importantly ribosomal RNA maturation (by classic snoRNAs).15,16 Despite this, the function of many snoRNAs, referred to as orphan snoRNAs, remains unknown.15 snoRNA expression is mostly a result of disbranching from a spliced lariat of ribosomal or housekeeping gene introns and has long been thought to be invariant.17-19 Nevertheless, recent studies have shed light on new molecular functions of snoRNAs and have demonstrated new expression patterns that could be associated with pathologic features.20-28 In the present study, we demonstrate using high-throughput quantitative PCR methods that snoRNA expression analysis is helpful in the context of PTCL, both in terms of diagnosis and prognostication in patients treated with conventional chemotherapy.
Methods
Patients and healthy donors
PTCL samples (n = 122) were collected from the tissue bank of the multicentric T-cell lymphoma consortium (TENOMIC; Tables 1 and 2). For each patient, a consensus diagnosis was made by a panel of expert hemopathologists (some of the patients participated in a GELA-sponsored clinical trial5 ; the others benefited from a national review protocol for T-cell lymphomas). Fresh and thawed samples were obtained with patients' informed consent and stored at the TENOMIC collection. According to French law, the TENOMIC collection is registered by the Ministry of Higher Education and Research and a transfer agreement was obtained after approval by the ethics committee of the Comité Consultatifs de Protection des Personnes se prêtant à des Recherches Biomédicales (CCPPRB). Clinical and biologic annotation of the samples was declared to the National Committee on Data Processing and Liberties Comité National Informatique et Libertés. This study was conducted in accordance with the Declaration of Helsinki.
Main characteristics of PTCL patients
| Non-ALCL patients . | . | . | . |
|---|---|---|---|
| Characteristic . | PTCL nos (n = 26) . | AITL (n = 46) . | Total cohort (N = 78) . |
| Median age at diagnosis, y | 74 | 65 | 66 |
| Male: female ratio | 14:12; 1,2 | 28:18; 1,55 | 47:31; 1,52 |
| Stage | |||
| I | 2/26 | 1/46 | 3/78 |
| II | 1/26 | 1/46 | 2/78 |
| III | 7/26 | 11/46 | 18/78 |
| IV | 16/26 | 32/46 | 55/78 |
| NC | 0/26 | 1/46 | 1/78 |
| Elevated serum LDH, % (n/N) | 73,1 | 76,7 | 75 |
| IPI score | |||
| 0 | 3 | 1 | 4 |
| 1 | 2 | 1 | 3 |
| 2 | 3 | 7 | 12 |
| 3 | 8 | 8 | 18 |
| 4 | 5 | 15 | 22 |
| 5 | 3 | 9 | 13 |
| NC | 2 | 5 | 7 |
| OS of total cohort, % (n/N) | |||
| 1 y | 58,2 | ||
| 3 y | 40,5 | ||
| PFS of total cohort, % (n/N) | |||
| 1 y | 43 | ||
| 3 y | 24 | ||
| Patients receiving chemotherapy, % (n/N) | 98,7 | ||
| Non-ALCL patients . | . | . | . |
|---|---|---|---|
| Characteristic . | PTCL nos (n = 26) . | AITL (n = 46) . | Total cohort (N = 78) . |
| Median age at diagnosis, y | 74 | 65 | 66 |
| Male: female ratio | 14:12; 1,2 | 28:18; 1,55 | 47:31; 1,52 |
| Stage | |||
| I | 2/26 | 1/46 | 3/78 |
| II | 1/26 | 1/46 | 2/78 |
| III | 7/26 | 11/46 | 18/78 |
| IV | 16/26 | 32/46 | 55/78 |
| NC | 0/26 | 1/46 | 1/78 |
| Elevated serum LDH, % (n/N) | 73,1 | 76,7 | 75 |
| IPI score | |||
| 0 | 3 | 1 | 4 |
| 1 | 2 | 1 | 3 |
| 2 | 3 | 7 | 12 |
| 3 | 8 | 8 | 18 |
| 4 | 5 | 15 | 22 |
| 5 | 3 | 9 | 13 |
| NC | 2 | 5 | 7 |
| OS of total cohort, % (n/N) | |||
| 1 y | 58,2 | ||
| 3 y | 40,5 | ||
| PFS of total cohort, % (n/N) | |||
| 1 y | 43 | ||
| 3 y | 24 | ||
| Patients receiving chemotherapy, % (n/N) | 98,7 | ||
Total cohort of non-ALCL patients included 6 rare diagnostics (HSTL, NK/T, and EATL).
NC indicates not communicated; and LDH, lactate dehydrogenase.
Main characteristics of ALCL patients
| ALCL patients (n = 32) . | |
|---|---|
| Characteristic | |
| Median age at diagnosis, y | 13 |
| Male: female ratio | 21:11; 1,7 |
| ALK translocation | |
| + | 22 |
| − | 10 |
| Ann Arbor stage | |
| I | 5 |
| II | 6 |
| III | 5 |
| IV | 6 |
| NC | 10 |
| Elevated serum LDH, % (n/N) | 62 |
| IPI score | |
| 0 | 4 |
| 1 | 5 |
| 2 | 4 |
| 3 | 6 |
| 4 | 0 |
| 5 | 0 |
| NC | 13 |
| OS of total cohort, % (n/N) | |
| 1 y | 83,3 |
| 3 y | 62,5 |
| Complete remission, % (n/N) | 91,3 |
| ALCL patients (n = 32) . | |
|---|---|
| Characteristic | |
| Median age at diagnosis, y | 13 |
| Male: female ratio | 21:11; 1,7 |
| ALK translocation | |
| + | 22 |
| − | 10 |
| Ann Arbor stage | |
| I | 5 |
| II | 6 |
| III | 5 |
| IV | 6 |
| NC | 10 |
| Elevated serum LDH, % (n/N) | 62 |
| IPI score | |
| 0 | 4 |
| 1 | 5 |
| 2 | 4 |
| 3 | 6 |
| 4 | 0 |
| 5 | 0 |
| NC | 13 |
| OS of total cohort, % (n/N) | |
| 1 y | 83,3 |
| 3 y | 62,5 |
| Complete remission, % (n/N) | 91,3 |
T lymphocytes from healthy donors (n = 35) were collected from blood samples (provided by the Etablissement Français du Sang) or lymph nodes. Cells were sorted by CD3+ (n = 10), CD4+, or CD8+ selection on columns (Miltenyi Biotec) and cell purity was determined by flow cytometry (only purities more than 94% were retained). HLA-DR+ and HLA-DR− were obtained from lymph node–dissociated cells by magnetic bead depletion of CD16+, CD14+, CD19+, and CD8+ cells and HLA-DR magnetic sorting. TFH cells and non-TFH cells were obtained from lymph node–dissociated cells by magnetic bead depletion of CD16+, CD14+, CD19+, and CD8+ cells, followed by flow cytometric cell sorting using the CD4, CXCR5, and ICOS29 markers.
Quantitative PCR method
Total RNA from patient samples and CD3+-sorted T lymphocytes were extracted using the TRIzol method; RNA integrity was evaluated using an Agilent Nano Chip (Agilent 2100 Bioanalyzer). Only samples with an RIN (RNA integrity number) more than 7.8 were used in this study. All samples were reverse transcribed using the Superscript II reverse transcription kit (Invitrogen) according to the manufacturer's protocol. The Fluidigm high-throughput quantitative PCR method (Biomark) was used as described previously.30 Each primer used for this study had been previously tested and the amplification efficiency was more than 85%. Relative RNA quantity was determined by the 2−ΔCt method.
Hsa-miRNA-768-5p and Hsa-miRNA-768-3p expression was quantified first by TaqMan reverse transcription (Life Technologies), then by TaqMan PCR quantification on an Applied Biosystems 7300 thermocycler according to the manufacturer's protocol. The random selection of cases was carried out using Research Randomizer (http://www.randomizer.org/).
The specificity of the miRNA-768-3p TaqMan miRNA assay (Life Technologies) and HBII-239 snoRNA primers was verified with experiments of in vitro transcription (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Building of the snoRNA chip
Eighty small noncoding RNAs (supplemental Table 1) were selected on the basis that they were: (1) orphan snoRNAs, many of which are encoded by multiple, tandemly arranged intronic variant genes (eg, SNORD112-114 snoRNAs31 ); (2) already known as miRNA precursors (eg, HBII-239 snoRNA21,32 ); (3) previously linked to a medical condition (eg, SNORD116 snoRNAs33 ); and (4) predicted to guide chemical modification of ribosomal RNA using the snoRNABase dataase (http://www-snorna.biotoul.fr/). We also tested U8 and U3 snoRNAs because they are under the control of their own promoter. U2, U4, and U5 small nuclear RNAs corresponding to 3 spliceosomal machinery components were tested in parallel. Among the 5 housekeeping genes used on the snoRNA chips, S14 and 5s-rRNA displayed the lowest coefficient of variation (supplemental Figure 2). We selected 5s-rRNA because of its similarity with snoRNAs (it is a noncoding RNA of similar length) and because this gene is widely used for noncoding RNA normalization.
Statistical considerations
Dendrograms were generated by dChip 2010.01 software using correlation determination and the centroid method. A filter of gene expression was applied to all samples with a cutoff of approximately 0.25. Comparison of gene expression in each group of samples was done using a fold change cutoff above 1.3 with P < .05. Specific gene expression comparison between each assigned group was then performed using an unpaired t test.
Progression-free survival (PFS) was calculated as time from diagnosis to first occurrence of progression, death, secondary malignancy, or time of last patient contact if no event occurred. Overall survival (OS) time was calculated from diagnosis time until death or time of last contact if the patient was alive. The Kaplan-Meier method was used to generate survival curves, and curves were compared using a log-rank test (MedCalc Version 12.3 software).
miRNA-768-3p effects on FEPD cells
A total of 50 000 synchronized cells (after serum starvation) from the ALK− ALCL cell line (FEPD; DSMZ) were transfected for 4 days using lipofectamine RNAi-max (Invitrogen) with a 30nM concentration of miRNA-768 mimic (customized by Life Technology) or a miRNA-negative control (Eurogentec). Cell growth was evaluated using an MTS proliferation assay (Promega) according to the manufacturer's protocol. The impact of miRNA-768-3p on the cell cycle was evaluated by the same transfection protocol using mutated miRNA-768-3p in the seed region as an additional control (miRNA-768-3pM, also customized by Life Technology).
Cell-cycle analysis
Cells were washed with PBS and fixed in cold 70% ethanol for 20 minutes, washed twice with PBS containing 0.1% BSA and once with PBS, and then labeled using propidium iodide staining for 30 minutes (Invitrogen). Cell-cycle distribution was evaluated by fluorescence analysis on a FACScan flow cytometer (BD Bioscience). Cell doublets were excluded and 20 000 events per condition were analyzed. Cyclin A expression analysis was by Western blot using anti–β-actin (MAB150L) and anti–cyclin A (Sc-239), according to the manufacturer's recommendations.
Results
Patient characteristics in PTCL
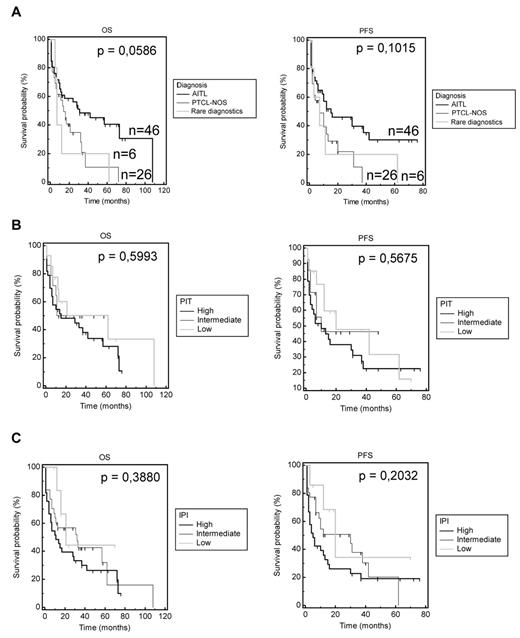
The distinct PTCL entities (n = 78 non-ALCL and n = 32 ALCL) and their respective clinical characteristics are summarized in Tables 1 and 2. Of the 90 non-ALCL patients enrolled in this study, full clinical records were available for 78 patients, 46 of which were diagnosed with AITL, 26 with PTCL-NOS, and 6 with a rare entity including enteropathy-associated T-cell lymphoma, hepatosplenic T-cell lymphoma, or extranodal NK/T-cell lymphoma. The median age of patients in the PTCL-NOS/AITL group was 67 years. Although a slightly better outcome was observed for AITL patients, the 3-year-OS and PFS were not significantly different between AITL and PTCL-NOS patients (Figure 1A). Prognostic indices were not significant and did not differentiate better prognosis patients from others (patients had intermediate-high and high-risk IPI/ PIT scores of 74% and 65.5%, respectively; Figure 1B-C). Initial therapy approaches varied widely, but 79% of patients received an active CHOP- or cytarabine-based regimen, 11.8% a combination of oral cyclophosphamide and steroids (for AITL), 2.6% a combination of oral fludarabine and cyclophosphamide, and 6.6% a palliative oral monotherapy (steroids and chlorambucil). Only 4 of 78 patients received upfront intensification with autologous (n = 3) or allogeneic (n = 1) BM transplantation. Response rates for the non-ALCL patients included 44% complete responses (CRs), 18.2% partial responses, 7.8% with stable disease, and 30% with progressive disease. For the PTCL-NOS and AITL subgroups, these figures were 38.5% CR, 15.4% partial response, 11.5% stable disease, 34.6% progressive disease, and 50%, 17.4%, 4.3%, 28.3%, respectively (P > .05), in accordance with previous data published on CHOP efficacy in PTCL.5,11,34
OS and PFS analysis according to diagnostic and common prognostic predictive scores (IPI and PIT). (A) OS and PFS of patients for each diagnosis. (B) IPI score was divided into 3 categories: low for IPI 0-1, intermediate for IPI 2-3, and high for IPI 4-5. OS and PFS were analyzed by Kaplan-Meier curves. (C) The PIT score was also divided into 3 categories: low for PIT 0, intermediate for PIT 1-2, and high for PIT 3-4.
OS and PFS analysis according to diagnostic and common prognostic predictive scores (IPI and PIT). (A) OS and PFS of patients for each diagnosis. (B) IPI score was divided into 3 categories: low for IPI 0-1, intermediate for IPI 2-3, and high for IPI 4-5. OS and PFS were analyzed by Kaplan-Meier curves. (C) The PIT score was also divided into 3 categories: low for PIT 0, intermediate for PIT 1-2, and high for PIT 3-4.
ALCL cases were composed of 22 patients with the ALK+ ALCL and 10 patients with ALK− ALCL (Tables 1 and 2). The median age was of 13 years. ALK+ cases had a good clinical outcome (1-year OS of 94%) and ALK− cases had a poorer outcome (1-year OS of 57%). Most patients (91% of cases) who were submitted to ALCL99-based treatment35 achieved CRs.
snoRNA expression profiles in the diagnosis of PTCL
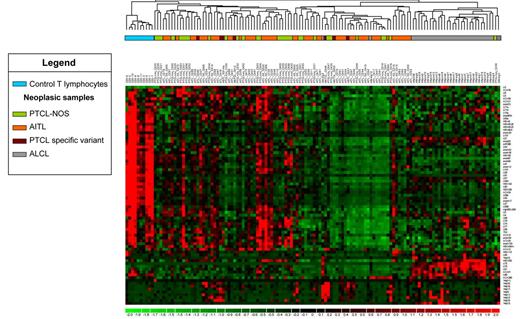
Using unsupervised hierarchical clustering, we observed that a significant global down-regulation of snoRNA expression was seen in neoplastic cells compared with nonneoplastic CD3+-sorted T cells from the peripheral blood (ranging from a 15%-96% decrease in expression; Figure 2 and supplemental Table 2). However, we noticed a significant overexpression of the snoRNAs U75, U76, and snRNA U2 in PTCL samples (supplemental Table 2). We included 25 additional controls corresponding to CD4+-sorted cells (HLA-DR+ or HLA-DR−; TFH or non-TFH) from the peripheral blood and lymph nodes and CD8+-sorted cells from the peripheral blood (supplemental Figure 3). Some variations in snoRNA expression levels were observed between these 2 sets of controls. Nevertheless, irrespective of the origin of the cells or the method of selection, we observed a significant down-regulation of snoRNA expression in neoplastic samples in 63% of the snoRNAs tested (supplemental Figure 3). In particular, there was a clear delineation between AITL, a tumor of TFH origin, and normal TFH cells from healthy donors.
snoRNA expression–based unsupervised hierarchical clustering of PTCL and normal T cells. High-throughput quantitative PCR was carried out on 80 snoRNAs in 90 non-ALCL and 32 ALCL patients. Each column represents a case and each row the expression level of a gene. Gene-expression levels are depicted according to the color scale shown. Correlation analysis and the centroid method were applied.
snoRNA expression–based unsupervised hierarchical clustering of PTCL and normal T cells. High-throughput quantitative PCR was carried out on 80 snoRNAs in 90 non-ALCL and 32 ALCL patients. Each column represents a case and each row the expression level of a gene. Gene-expression levels are depicted according to the color scale shown. Correlation analysis and the centroid method were applied.
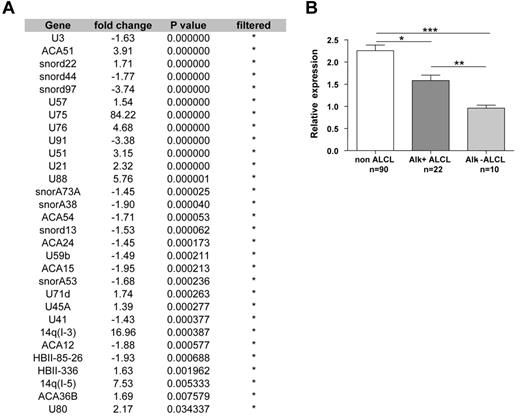
Among PTCL entities, ALCL had a specific snoRNA profile. A significant differential expression of 30 snoRNAs was obtained when ALCL and non-ALCL patients were compared (Figure 3). snoRNA profile comparison between ALK− ALCL and the other PTCL subtypes confirmed a significant differential expression of 21 of the 30 snoRNAs described in supplemental Table 3. From these genes, snoRNA U75 (belonging to the GAS5 snoRNA cluster) was the most powerful classifier in distinguishing ALCL from other PTCLs (displaying the highest fold change in expression; Figure 3 and supplemental Table 2). Within the “core” group of ALCL, unsupervised clustering was not able to distinguish ALK− from ALK+ patients. However, a supervised comparison identified snoRNA U3 as the single discriminant marker that distinguished ALK+ from ALK− ALCL samples (Figure 3). Unsupervised snoRNA clustering did not allow us to distinguish AITL from the other subgroups of PTCL (ie, PTCL-NOS, enteropathy-associated T-cell lymphoma, hepatosplenic T-cell lymphoma, and extranodal NK/T-cell lymphoma; Figure 2). Furthermore, a supervised comparison of snoRNA expression between PTCL-NOS and AITL failed to identify a specific profile. These results show that snoRNA expression profiles discriminate ALCL from other PTCL entities and that snoRNA U3 is statistically overexpressed in ALK+ compared with ALK− ALCL.
ALCL displays a specific snoRNA profile. (A) Supervised snoRNA expression analysis demonstrated a set of 30 snoRNAs differentially expressed in ALCL patients compared with other PTCL subtypes. *Filtered genes according to selected criteria (fold change > 1.3 and P < .05). (B) U3 snoRNA expression levels discriminate ALK+ from ALK− ALCL patients. *P < .05; **P < .01; ***P < .001.
ALCL displays a specific snoRNA profile. (A) Supervised snoRNA expression analysis demonstrated a set of 30 snoRNAs differentially expressed in ALCL patients compared with other PTCL subtypes. *Filtered genes according to selected criteria (fold change > 1.3 and P < .05). (B) U3 snoRNA expression levels discriminate ALK+ from ALK− ALCL patients. *P < .05; **P < .01; ***P < .001.
Prognostic value of snoRNA expression in non-ALCL PTCL
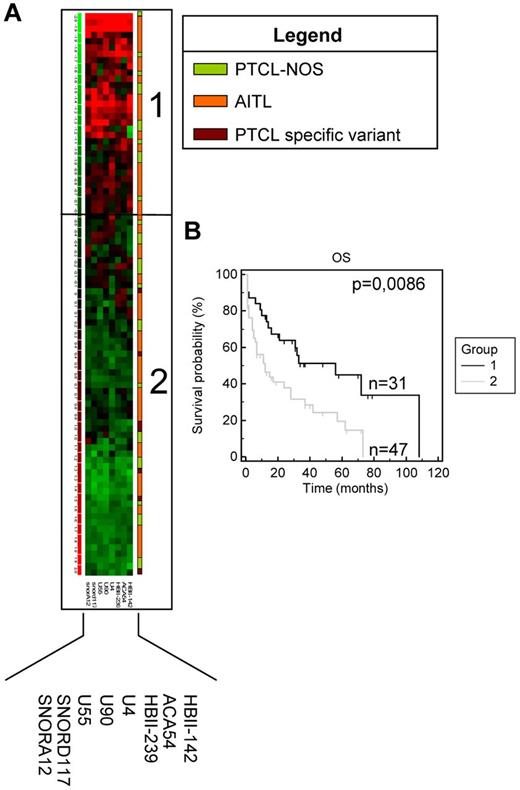
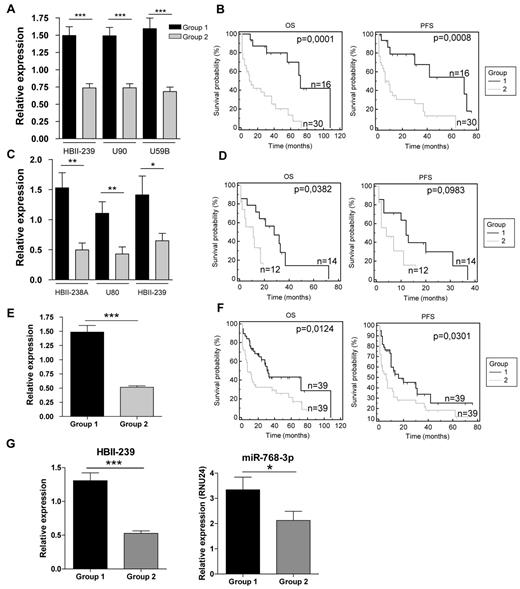
Although the snoRNA expression profiles of AITL and the other PTCL subtypes appeared very similar, we observed that AITL/PTCL-NOS patients were subdivided into 3 groups on nonsupervised clustering (Figure 2 and supplemental Table 4). Eight snoRNAs were differentially expressed between the 3 groups (each with a fold change greater than 1.5; supplemental Table 4). We also analyzed the clinical features of the 78 non-ALCL patients. Interestingly, we observed that OS was significantly prolonged in the group overexpressing the 8 snoRNA genes (Figure 4). Regarding PTCL prognosis, we performed 2 separate supervised studies on PTCL-NOS and AITL with a cutoff for OS analyses of 1 or 2 years, respectively (based on the median survival of 13 and 26 months, respectively). Supervised comparison of snoRNA expression at 2-year-OS in AITL confirmed the significant overexpression of HBII-239 (P = .0162), U59B (P = .0086), and U90 (P = .0077) in patients with long-term survival (Figure 5A). Using Kaplan-Meier analysis, both PFS and OS were significantly improved in AITL patients overexpressing these 3 snoRNA genes (Figure 5B). Interestingly, overall response rates were not significantly different between the 2 prognostic groups (supplemental Figure 4).
OS analysis based on differential expression of 8 snoRNAs in non-ALCL PTCL patients. (A) Heat map showing snoRNA expression in non-ALCL PTCL patients. Each column represents the expression level of a gene and each row a patient (diagnosis of each patient is depicted according to the color legend shown). Gene-expression levels are depicted according to the color scale shown. (B) OS comparison for patients overexpressing these snoRNAs (group 1) and other patients (group 2).
OS analysis based on differential expression of 8 snoRNAs in non-ALCL PTCL patients. (A) Heat map showing snoRNA expression in non-ALCL PTCL patients. Each column represents the expression level of a gene and each row a patient (diagnosis of each patient is depicted according to the color legend shown). Gene-expression levels are depicted according to the color scale shown. (B) OS comparison for patients overexpressing these snoRNAs (group 1) and other patients (group 2).
Prognostic impact of snoRNA signatures in non-ALCL PTCL subtypes. (A) HBII-239, U90, and U59b relative expression in AITL-overexpressing patients (group 1) and other patients (group 2) compared with the median expression in all AITL patients. (B) OS and PFS Kaplan-Meier analysis of group 2. (C) Relative expression of HBII-438A, U80, and HBII-239 in PTCL-NOS–overexpressing patients (group 1) and other patients (group 2) compared with the median expression in all PTCL-NOS samples. (D) OS and PFS Kaplan-Meier analysis of group 2. (E) HBII-239 relative expression in all non-ALCL PTCL patients compared with the median expression. (F) OS and PFS Kaplan-Meier analysis in HBII-239–overexpressing patients (group 1) and other patients (group 2). (G) HBII-239 and HBII-239–processed miRNA (has-miRNA-768-3p) relative expression in 37 randomly selected non-ALCL PTCL patients. *P < .05; **P < .01; ***P < .001.
Prognostic impact of snoRNA signatures in non-ALCL PTCL subtypes. (A) HBII-239, U90, and U59b relative expression in AITL-overexpressing patients (group 1) and other patients (group 2) compared with the median expression in all AITL patients. (B) OS and PFS Kaplan-Meier analysis of group 2. (C) Relative expression of HBII-438A, U80, and HBII-239 in PTCL-NOS–overexpressing patients (group 1) and other patients (group 2) compared with the median expression in all PTCL-NOS samples. (D) OS and PFS Kaplan-Meier analysis of group 2. (E) HBII-239 relative expression in all non-ALCL PTCL patients compared with the median expression. (F) OS and PFS Kaplan-Meier analysis in HBII-239–overexpressing patients (group 1) and other patients (group 2). (G) HBII-239 and HBII-239–processed miRNA (has-miRNA-768-3p) relative expression in 37 randomly selected non-ALCL PTCL patients. *P < .05; **P < .01; ***P < .001.
A similar approach was applied to PTCL-NOS. First, a signature of 11 snoRNAs [HBII-239, U90, U80, ACA54, HBII-99, HBII-438A, HBII-85-6, SNORA12, SNORD22, 14q(I-3) and U50B], predictive of OS by univariate analysis, was found and narrowed down to the 3 most powerful predictors by increasing the stringency of snoRNA selection (HBII-239 [P = .02], HBII-438A [P = .003], and U80 [P = .044]; Figure 5C). Again, Kaplan-Meier curves showed significantly different OS rates based on the level of snoRNA expression (Figure 5D). Nevertheless, the 3 snoRNAs had no predictive impact on PFS. Conversely, CR rates after CHOP were not significantly higher in the good-prognosis group (supplemental Figure 3).
The potential impact of the amount of malignant cells among PTCL samples (assessed morphologically) on case distribution among prognostic groups was taken into account. Except for 12 cases in which the malignant cell percentage was 50%-75%, infiltration of PTCL samples was 75%-100%. Interestingly, the percentage of tumor cells in tissue samples had no impact on the segregation of patients in the different prognostic subgroups. Indeed, the 12 patients with 50%-75% of tumor cells did not segregate together in the same snoRNA-related prognostic group. Among these 12 patients, 11 were AITL and 1 was PTCL-NOS. The snoRNA signature in AITL classified 7 of 11 of these samples in the bad-prognosis group and 4 of 11 in the good-prognosis group. For the last patient belonging to the PTCL-NOS group, the snoRNA signature classified it in the good-prognosis group. Therefore, the number of malignant cells was not significantly different between snoRNA-related good- and bad-prognosis groups.
These data indicate that signatures, even with limited numbers of snoRNAs, are strong predictors of OS (for AITL and PTCL-NOS) and PFS (for AITL) independently of their IPI or PIT scores.
HBII-239 and HBII-239–processed miRNA-768: 2 powerful prognostic tools
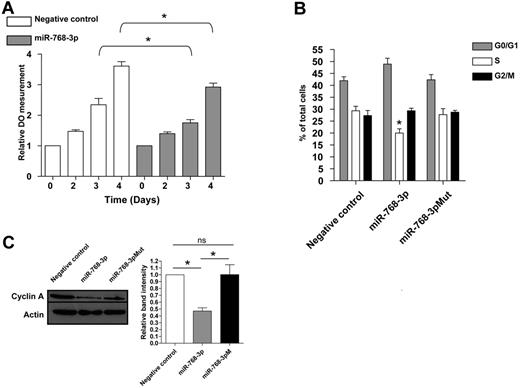
HBII-239 appears to be one of the most important snoRNAs, because its overexpression predicts good prognosis in AITL and PTCL-NOS in both unsupervised and supervised analyses. Therefore, we investigated its impact as a single marker on survival/outcome. Of the entire cohort of 78 patients, HBII-239 overexpression was statistically associated with prolonged PFS and OS (Figure 5E-F). The HBII-239 sequence has been shown previously to overlap with the hsa-miRNA-768 precursor sequence, but the latter miRNA was removed from the miRNA base because it was considered to be a snoRNA. We decided to evaluate whether the group of PTCL patients overexpressing HBII-239 showed a simultaneous overexpression of hsa-miRNA-768. miR-768-5p and miR-768-3p TaqMan miRNA assays on 37 randomly selected patients demonstrated a significant overexpression of this miRNA in the HBII-239–overexpressing group (a good prognostic group), and confirmed that only the 3p strand was processed into mature miRNA (Figure 5G). Therefore, the effect of the HBII-239–processed miRNA-768 on lymphoma T cells was tested. To address this issue, we used the FEPD cell line for in vitro experiments because of the lack of other PTCL-NOS or AITL cell lines. We first checked miRNA-768-3p basal expression in FEPD cells and found it to be significantly weaker than that of patient samples (not shown). FEPD cells were transfected with either an miRNA-768-3p miRNA mimic or an miRNA− control. We confirmed transfection efficiency by flow cytometry using a negative control labeled with a Cy3 dye and showed that the expression of miRNA-768-3p increased (median change approximately 15-fold after 2 days; supplemental Figure 5). Cell growth was monitored using an MTS proliferation assay, and a significant decrease was noticed 3 days after miRNA-768-3p transfection (Figure 6A). To further characterize this decrease in cell growth, the cell-cycle distribution of synchronized cells was analyzed after transfection of a negative control miRNA, miRNA-768-3p, and an miRNA-768-3p for which the seed region was mutated (miRNA-768-3pM). The percentage of cells in the S phase was significantly decreased in cells transfected with miRNA-768-3p (Figure 6B). To confirm this observation, cyclin A (a cyclin involved in S-phase progression and regularly used as marker of cell growth) expression was analyzed under these 3 conditions. A significant decrease in cyclin A protein expression was observed 3 days after miRNA-768-3p transfection compared with negative controls (Figure 6C). These results corroborate the notion that high miRNA-768-3p expression is associated with a decrease in cell growth that may affect prognosis.
miRNA-768-3p negatively affects PTCL growth. (A) Cell growth was monitored for 4 days in miRNA-768-3p- or miRNA negative control–transfected FEPD cells using a MTS proliferation assay. (B) Cell-cycle distribution in miRNA-768-3p-, miRNA-768-3pM-, or miRNA− control-transfected FEPD cells. (C) Cyclin A expression was analyzed by Western blot in the 3 conditions described in panel B. A paired t test was used to analyze the statistical significance of the results. *P < .05; **P < .01; ***P < .001.
miRNA-768-3p negatively affects PTCL growth. (A) Cell growth was monitored for 4 days in miRNA-768-3p- or miRNA negative control–transfected FEPD cells using a MTS proliferation assay. (B) Cell-cycle distribution in miRNA-768-3p-, miRNA-768-3pM-, or miRNA− control-transfected FEPD cells. (C) Cyclin A expression was analyzed by Western blot in the 3 conditions described in panel B. A paired t test was used to analyze the statistical significance of the results. *P < .05; **P < .01; ***P < .001.
Discussion
Management of PTCL patients is challenging in terms of prognostication and treatment (a CHOP-based regimen remains the gold standard despite dismal survival rates). Therefore, intensification (allogeneic BM transplantation) or novel agents should be proposed at the frontline for the “not low risk” (PIT = 0) patients to increase the 20%-30% 5-year OS rates currently predicted with CHOP. The results of the present study suggest a potential prognosticator based on snoRNA analysis, which is able to identify patients with a good OS after CHOP (without intensification), even among intermediate-/high-risk patients.
snoRNAs are types of small noncoding RNAs that were discovered decades ago. Apart from their role in ribosomal RNA maturation, only a few studies have investigated their pathologic implications.20,23,25,33 We have demonstrated herein that snoRNA-expression profiles are relevant for PTCL prognostication. As miRNAs, snoRNAs are widely underexpressed in lymphoma cells compared with 2 sets of normal T cells. Indirect evidence suggests that activation through CD3+ selection may affect snoRNA expression profiles. Compared with the CD4+ and CD8+ subpopulations presumably not activated by CD3+ selection, we observed variations in expression. These variations had no impact on the segregation between tumor and control cells, because more than 60% of the snoRNAs were still down-regulated in tumors (63% vs 71%). Except for miRNAs that act as tumor suppressors, we found no real explanation as to the decreased expression of both miRNAs and snoRNAs in cancer cells. Nevertheless, we observed that snoRNA expression analysis may be useful in the context of PTCL in terms of prognostication and, to a lesser extent, diagnosis (leading to a better distinction between ALK− ALCL and PTCL-NOS). Previous attempts to delineate PTCL subtypes by gene-expression profiling have been reported. Despite a good delineation of most entities, borderline/overlapping features or signatures make it difficult to distinguish AITL from some cases of PTCL-NOS enriched in the TFH signature, and ALK+ ALCL from CD30+ PTCL-NOS.2,6,8,36,37 Therefore, supervised analyses are often required to obtain a clear delineation between PTCL variants.2,6,36,37 We show herein that snoRNA signatures are robust enough to differentiate ALCL from other PTCL entities using unsupervised clustering. However, these signatures were unable to separate ALK+ from ALK− ALCL cases: for this, one must resort to supervised analyses.38 Supervised analyses revealed that snoRNA U3 as a single marker is sufficient to distinguish ALK+ from ALK− ALCL because it is significantly up-regulated in ALK+ tumors. Interestingly, this particular snoRNA belongs to a subgroup of rare snoRNA genes controlled by their own promoter. It would be interesting to determine whether ALK has any impact on U3 gene regulation. Like classic snoRNAs, U3 is involved in rRNA processing through interactions with processome proteins such as UTP14, which is itself involved in the p53 pathway.39-41 A recent study showed the prognostic impact of morphologic and phenotypical features of childhood ALK+ ALCL.42 However, our series of patients was too small to confirm these data on the basis of snoRNA expression.
Predicting prognostic outcome with a simple and reliable biologic approach remains challenging in PTCL. Despite the availability of clinical scoring techniques (only the mPIT includes an estimation of proliferating tumor cells through Ki67 staining), isolating low-risk patients is difficult, and even then the OS rate at 5 years is less than 40% (an estimated proportion of 20% of patients may be cured with CHOP; this cannot be predicted with any prognostic tool). Based on snoRNA expression, we have constructed a molecular prognosticator for both AITL and PTCL-NOS. The snoRNA signatures described herein were obtained from patients who presented with high IPI and PIT scores (therefore representing a classic poor outcome T-cell lymphoma cohort). As such, delineation of the 2 subgroups of patients with differential prognosis underlines the robustness of the snoRNA signatures. Among the snoRNA signatures for AITL and PTCL-NOS, the orphan HBII-239 snoRNA was the strongest biomarker. HBII-239 is different from the other snoRNAs because its genomic sequence overlaps with a previously described miRNA-768 precursor.43 After analysis of the degree of conservation between species, miRNA-768 was removed from miRNA base because it was considered that the presence of consensus C and D box sequences in the primary-miRNA argued in favor of a snoRNA sequence. However, using the TaqMan miRNA assay, we observed that miRNA-768 was well processed and produced a mature miRNA corresponding to miRNA-768-3p (the 5p strand was almost undetectable). Therefore, we focused our attention on the phenotype associated with miRNA-768 overexpression. In vitro experiments demonstrated that miRNA-768-3p negatively affected T-cell lymphoma growth through deregulation of the S phase. This effect reinforces the notion that overexpression of HBII-239 and HBII-239–processed miRNA-768 is associated with better prognosis. Currently, there is no known specific target of this miRNA, and we used bioinformatics to sort through more than 1000 potential targets of miRNA-768, some involved in cell-cycle regulation.
Two major conclusions can be drawn from the results of the present study. First, snoRNAs are differentially regulated in healthy compared with malignant T-cell populations. Second, in addition to a global down-regulation of these molecules, specific signatures may have a prognostic significance in PTCL. The different snoRNAs that can be regarded as potential biomarkers in these tumors may play a direct role in different cellular pathways. Deciphering the functions of these biomarkers will be the subject of future investigations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients who participated in this study, their referring physicians, Marion Travert, and Jean-José Maoret (Genotoul platform, Toulouse, France).
This work was supported by the Trans-Pyrenean Cooperation Research in Innovative Therapy for Leukemia, the Association pour la Recherche sur le Cancer, the PAIR lymphoma project, and the TENOMIC project (Program Hospitalier de Recherche Clinique). P.B. is supported by the Institut Universitaire de France and the LABEX TOUCAN (analyse intégrée de la résistance dans les cancers hématologiques).
Authorship
Contribution: W.V., L.B. and C.Q. performed the experiments; W.V., L.Y., P.G., and P.B. conceived and designed the study; V.F., A.M., M.P., L.L., L.d.L., and P.G. collected and assembled the clinical records; W.V., L.Y., and C.Q. analyzed and interpreted the data; W.V., L.Y., and P.B. prepared the first draft of the manuscript; W.V., C.G., P.G., and P.B. finalized the manuscript; and all authors contributed to the writing of the manuscript and gave final approval.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pierre Brousset, MD, PhD, Département de Pathologie, Centre Hospitalier Universitaire Purpan, Place Baylac, 31059 Toulouse Cedex, France; e-mail: brousset.p@chu-toulouse.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal