Abstract
Improvements in neutrophil chemotaxis assays have advanced our understanding of the mechanisms of neutrophil recruitment; however, traditional methods limit biologic inquiry in important areas. We report a microfluidic technology that enables neutrophil purification and chemotaxis on-chip within minutes, using nanoliters of whole blood, and only requires a micropipette to operate. The low sample volume requirements and novel lid-based method for initiating the gradient of chemoattractant enabled the measurement of human neutrophil migration on a cell monolayer to probe the adherent and migratory states of neutrophils under inflammatory conditions; mouse neutrophil chemotaxis without sacrificing the animal; and both 2D and 3D neutrophil chemotaxis. First, the neutrophil chemotaxis on endothelial cells revealed 2 distinct neutrophil phenotypes, showing that endothelial cell-neutrophil interactions influence neutrophil chemotactic behavior. Second, we validated the mouse neutrophil chemotaxis assay by comparing the adhesion and chemotaxis of neutrophils from chronically inflamed and wild-type mice; we observed significantly higher neutrophil adhesion in blood obtained from chronically inflamed mice. Third, we show that 2D and 3D neutrophil chemotaxis can be directly compared using our technique. These methods allow for new avenues of research while reducing the complexity, time, and sample volume requirements to perform neutrophil chemotaxis assays.
Introduction
Chemotaxis, the ability of a cell to directionally migrate toward a chemical stimulus, is central to biologic processes, such as wound healing,1 innate immunity,2 and cancer progression.3,4 The Boyden chamber assay, developed in 1962,5 marked the first advance in engineering platforms that enabled systematic in vitro measurements of a cell's response to chemotactic factors. Since the introduction of the Boyden chamber, other methods have been developed for studying cell chemotaxis, such as the Dunn,6 Zigmond,7 under-agarose,8 and micropipette-based9 assays; however, these techniques limit the types of biologic and clinical questions that can be addressed. The Boyden chamber develops unstable chemical gradient profiles,10 filters cells by size and deformability,11 and delivers ambiguous results as chemotaxis and chemokinesis are difficult to differentiate in this system.12 The under-agarose assay allows for visualization of the cell migration path but has an unstable chemical gradient that forms in all directions.10 Micropipette-based assays offer significant technical advances, such as high resolution real-time imaging, over these older techniques.13 However, the method is best suited for qualitative analysis because its onerous labor requirements limit investigators to low throughput sampling. Furthermore, these assays require lengthy blood draws and cell purification protocols14 that depend on tens of milliliters of blood to conduct an experiment. This limitation makes assaying neutrophil chemotaxis challenging for circumstances in which milliliters of blood are not easily obtainable, such as for infants or small animals.
Within the last decade, microfluidic platforms have demonstrated the ability to control the spatiotemporal properties of chemical gradients in a precise and reproducible way.15-18 In some microfluidic applications, the measurement of neutrophils has been used for clinical diagnostics,19,20 demonstrating the potential for translation of microfluidic methods into a clinical setting. However, few examples exist of microfluidic techniques being used in hematology research. Although the published microfluidic platforms meet the technical requirements for neutrophil chemotaxis assays, they can be difficult to operate, require specialized equipment not typically found in biology laboratories,21 are tailored for specific applications, and can be limiting in experimental throughput. Furthermore, the majority of microfluidic chemotaxis assays do not leverage the ability to use minute volumes of reagents and still rely on time-consuming and extensive cell purification methods.14 The persistence of traditional chemotaxis techniques in the hematology research community is perhaps evidence of some of these limitations for modern microfluidic approaches. Consequently, no dominant microfluidic chemotaxis platform has emerged that facilitates new avenues of research and is accessible to hematology investigators.
Here we present a comprehensive microfluidic solution, dubbed Kit-on-a-lid-assay (KOALA) for chemotaxis, that performs neutrophil purification from nanoliter volumes of blood in minutes, generates repeatable chemotactic gradients, and does not require specialized equipment to operate. The platform makes possible the study of neutrophil chemotaxis in infants or small mammals and is particularly useful for dynamic phenomena that need repeated sampling over multiple time points. These features permit new applications, make the assay accessible to a broad audience of investigators, and significantly shorten the time from the start of sample collection to analysis (Figure 1A). The versatility of the KOALA platform was demonstrated through several applications that are challenging or impossible using traditional techniques. Importantly, all the work presented here was performed using only a micropipette and optical microscope, and no other external equipment was required.
Methods
Additional methods
Additional methods on fabrication of the KOALA microfluidic devices, characterization of the KOALA platform, mixing the chemoattractant and hydrogel, the culture of endothelial cells, staining protocols for immunocytochemistry, description of statistical analysis, mouse blood draw and neutrophil capture, and details regarding cell tracking techniques can be found in supplemental Methods (see the Supplemental Materials link at the top of the article).
Microscopy
For all time-lapse experiments, phase-contrast images were taken using an Olympus IX-81 optical microscope (Olympus) with a 10× objective that had a numerical aperture (NA) of 0.30; the images were captured using a SPOT RT Monochrome CCD camera (Diagnostic Instruments). For characterization of the chemical gradient using fluorescent dye, a 4× objective with NA = 0.13 was used. The time-lapse experiments were conducted in an incubation chamber at 37°C; before the start of an imaging session, the samples were allowed to warm to 37°C for a minimum of 15 minutes. Slidebook Version 5.0.0.14 software (Intelligent Imaging Innovations) was used to capture the time-lapse images. The data were exported into the .tif format before cell tracking. The imaging medium was dry for all images shown in this work. For HUVEC monolayer immunostaining, images were acquired on a Nikon Eclipse Ti inverted fluorescence microscope coupled to a Nikon DS-QiMc CCD camera (Nikon Instruments); see additional details in supplemental Methods. For higher-resolution imaging of neutrophil morphologies, an Olympus IX-70 optical microscope (Olympus) with 20× objective set at 1.5× (30× effective) was used. The NA of the objective was 0.45, and a Hamamatsu camera (model C4742-80-12AG; Hamamatsu Photonics) was used to capture the images using Metamorph Version 7.5.0.0 software (Molecular Devices).
Neutrophil migration on HUVECs
After the neutrophil capture, a lid containing 100nM N-formyl-methionyl-leucyl-phenylalanine (fMLP) was placed onto the base of the device, contacting the hydrogel-chemoattractant mixture (H-CA) to the output port of the migration channel. An image was taken every 30 seconds in each microchannel for 90 minutes.
Mouse neutrophil migration
After the neutrophil capture, a lid containing various doses of H-CA was placed onto the base of the device, contacting the H-CA drop with the output port of the migration channel. Phase-contrast images were taken with a 10× objective using an Olympus IX-81 optical microscope. An image was taken every 30 seconds in each microchannel for 120 minutes. For dose-response experiments, fMLP concentrations of 1mM, 100μM, 10μM, and 0M control (gel only) were used. For TNF-Tg and wild-type (WT) mice migration experiments, 1mM fMLP was used.
Human neutrophil purification
We purified peripheral blood neutrophils from human blood using Polymorphprep according to the manufacturer's recommendations (Nycomed). All donors were self-reportedly healthy, and we obtained informed consent at the time of the blood draw. The human subject protocol was approved by the University of Wisconsin Center for Health Sciences Human Subjects Committee. This study was conducted in accordance with the Declaration of Helsinki.
3D neutrophil migration
After purification, human neutrophils were suspended in a 3D matrix by mixing 3 μL of cells at a density of ∼ 1 × 106 cells/mL into 18 μL of type I rat collagen (354249; BD Biosciences) to a final concentration of 5 mg/mL. The cell-collagen mixture was incubated at 37°C for 15 minutes, alternating the microfluidic device orientation between upside-down and right-side-up, until gel was solidified. Sacrificial deionized water was placed around the device to mitigate evaporation. A lid containing 3 μL H-CA mixture was placed onto the base and time-lapse image acquisition was performed as previously described.
Results
In the KOALA platform, the neutrophil purification and gradient generation are decoupled in a microfluidic base and a multifunction lid, respectively. The neutrophil purification is performed in microchannels that are located in the base by using a capture technique first reported in a flow-based system.22 Briefly, neutrophils are captured on a polystyrene surface functionalized by P-selectin, and subsequent washing steps remove the other components of the whole blood. Fluid handling is based on passive pumping techniques, avoiding the need for external active pumping systems.23 The lid has 2 primary functions: (1) to house the reagents required to generate the gradient of chemoattractant and (2) to prevent evaporation and flow, which is detrimental to the proper formation of a soluble gradient in live-cell microscopy experiments.24 After the neutrophil capture, the chemotaxis assay is initiated by placing the lid onto the base, thereby allowing a chemoattractant in the lid to controllably diffuse into the microchannel. During the experiment, the lid can easily be removed, providing fluidic access to the channels or allowing for the application of different lids.
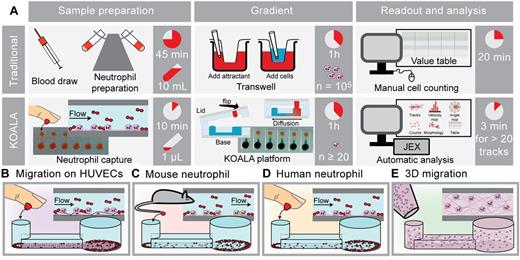
The versatility and enabling potential of the KOALA platform were demonstrated through several applications that are challenging or impossible using traditional techniques. Flexibility of the experimental design was significantly improved with the ability to perform different functions of the assay in different locations, such as capturing mouse neutrophils in the vivarium, culturing cell monolayers in a cell culture hood and incubator, and initiating the gradient at a microscopy facility. The ease of use, variability of applications, and time savings are summarized in Figure 1. First, a monolayer of HUVECs was cultured and subsequently used to capture neutrophils from whole blood (Figure 1B). Second, the KOALA platform enabled repeated neutrophil chemotaxis experiments on small mammals using ∼ 150 nL of whole blood per microchannel (Figure 1C). This sample volume requirement is the lowest reported for leukocyte purification and chemotaxis and at least 4 orders of magnitude lower than traditional purification methods currently used in hematology research.14 Finally, traditional 2D or 3D migration (Figure 1D-E) are both possible with the KOALA platform because the gradient generation from the lid is functionally independent of the cell preparation that is performed in the base microchannels.
Overview of the KOALA platform. (A) Comparison of the purification and chemotaxis protocols for the KOALA platform and Transwell assay. Other traditional assays that use live-cell imaging (ie, micropipette-based assays) take many hours to manually track the same number of cells that can be tracked with automated JEX tracking in 3 minutes. (B-E) Applications of the KOALA platform for neutrophil chemotaxis, including neutrophils migrating on endothelial cells (B); a 2D chemotaxis assay for neutrophils obtained from mice (C); a 2D chemotaxis assay for human primary neutrophils captured and sorted from a drop of whole blood (D); and neutrophil chemotaxis in a 3D matrix (E). Traditional purification methods and sample volumes shown in panel A are required for 3D neutrophil chemotaxis.
Overview of the KOALA platform. (A) Comparison of the purification and chemotaxis protocols for the KOALA platform and Transwell assay. Other traditional assays that use live-cell imaging (ie, micropipette-based assays) take many hours to manually track the same number of cells that can be tracked with automated JEX tracking in 3 minutes. (B-E) Applications of the KOALA platform for neutrophil chemotaxis, including neutrophils migrating on endothelial cells (B); a 2D chemotaxis assay for neutrophils obtained from mice (C); a 2D chemotaxis assay for human primary neutrophils captured and sorted from a drop of whole blood (D); and neutrophil chemotaxis in a 3D matrix (E). Traditional purification methods and sample volumes shown in panel A are required for 3D neutrophil chemotaxis.
Modeling fluid dynamics in microchannels
The method for purifying neutrophils from whole blood relies on pumping media through the KOALA microchannels after the capture of neutrophils on the functionalized plastic or activated endothelium. Therefore, we used a previously developed analytical model25 to characterize the range of shear stress generated by passive pumping in the microchannels. In brief, the flow rate in the microchannels is controlled by the pressure generated by the surface tension of the liquid at the input port and the fluidic resistance of the microchannel. The flow rate was calculated for both microchannel geometries presented in this work using the Laplace equation (supplemental Equation 1) to define the pressure drop at the input port, combined with the Washburn equation (supplemental Equation 2) for determining the fluidic resistance of a microchannel with rectangular cross-section. The flow rate in the microchannels was calculated using supplemental Equations 1 and 2 and numerical simulation (supplemental Figure 1). For the 180-μm-tall microchannels, which were used for neutrophil capture on endothelial cells, the shear stress averaged 12 dyne/cm2. This value is in the physiologic range of the shear stresses measured in aorta and capillaries,26 and below shear stress values causing damage to the cell monolayer.27 We found that for the 80-μm-tall microchannels, which were used for neutrophil capture on P-selectin-coated polystyrene, the shear stress averaged 0.1 dyne/cm2. These results show that the KOALA microchannels are well suited for use with cell monolayers and neutrophil sorting from whole blood.
Developing and characterizing KOALA and automated tracking
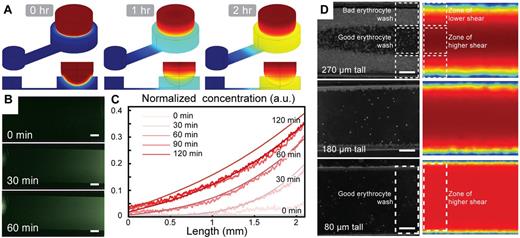
The ability to generate a stable and controllable chemical gradient for the duration of the experiment in a user-friendly way was central to the development of the KOALA platform. Computer simulation suggested that we would achieve reliable diffusion and gradient formation of our chemoattractant using the proposed hydrogel-based delivery method (Figure 2A). Using fluorescent dye to monitor the profile of the gradient, we found that the KOALA platform generates and maintains a chemical gradient for at least 2 hours (Figure 2B). These results correlated closely with the predicted gradient determined using numerical simulation (Figure 2C). We found that delivering the source of chemoattractant using a laden hydrogel was a simple and repeatable method. Furthermore, we observed that the lid and hydrogel delivery method nearly eliminates flow and evaporation during a typical neutrophil chemotaxis experiment (supplemental Video 1), which are common problems for static flow chemotaxis devices.
Characterization of the chemical gradient generation and neutrophil sorting for the KOALA platform. (A) COMSOL simulations of the diffusion of a chemoattractant loaded in a hydrogel sphere in the lid of the assay, dipping into a microfluidic channel in the base. (B) Fluorescent microscopy images of an AlexaFluor dye diffusing into the microchannel in the base of the KOALA platform. (C) Comparison of the concentration profiles between the experiments and the numerical simulations. (D) COMSOL Version 3.3 simulation of the shear stress in microchannels of different heights and comparison with the efficacy of the whole blood washing. (B,D) Scale bars represent 100 μm. Phase-contrast images acquired using Slidebook Version 5.0.0.14 software with an Olympus IX-81 microscope using 4× (NA = 0.13; B) and 10× (NA = 0.30; D) objectives at 37°C. Additional details on imaging are given in “Microscopy.”
Characterization of the chemical gradient generation and neutrophil sorting for the KOALA platform. (A) COMSOL simulations of the diffusion of a chemoattractant loaded in a hydrogel sphere in the lid of the assay, dipping into a microfluidic channel in the base. (B) Fluorescent microscopy images of an AlexaFluor dye diffusing into the microchannel in the base of the KOALA platform. (C) Comparison of the concentration profiles between the experiments and the numerical simulations. (D) COMSOL Version 3.3 simulation of the shear stress in microchannels of different heights and comparison with the efficacy of the whole blood washing. (B,D) Scale bars represent 100 μm. Phase-contrast images acquired using Slidebook Version 5.0.0.14 software with an Olympus IX-81 microscope using 4× (NA = 0.13; B) and 10× (NA = 0.30; D) objectives at 37°C. Additional details on imaging are given in “Microscopy.”
A second requirement of the KOALA device was the ability to purify neutrophils from whole blood on-chip, after the capture of neutrophils on the P-selectin–coated microchannels. This eliminates the need for extensive purification protocols. To minimize undesirable erythrocyte contamination, we characterized the washing efficiency in the microchannels for various aspect ratios (Figure 2D). We determined that the aspect ratio of the microchannel has a strong impact on the washing efficiency, and we found that channel dimensions of 800 μm × 80 μm (aspect ratio of 10:1) was an optimal channel geometry that minimized erythrocyte contamination (Figure 2D). Furthermore, we found that the KOALA microchannels captured a sufficient number of neutrophils in the field of view of a microscope to obtain statistically relevant tracking information (supplemental Figure 2). In addition, we calculated the efficiency of neutrophil capture using KOALA by counting fluorescently tagged neutrophils in whole blood before and after washing (supplemental Figure 2). We found that the KOALA neutrophil sorting technique captures ∼ 80% of human primary neutrophils.
Finally, to manage the large quantities of cell migration data, we developed a tracking algorithm, dubbed Je'Xperiment (JEX), to analyze large sets of phase-contrast time-lapse data (supplemental Figure 3; source code available on request). Within 3 minutes, the software was able to track the neutrophils and output properties of each cell, such as its speed, chemotactic index, and directional velocity (supplemental Equations 3-5, respectively). The automated algorithm produced comparable results to manual tracking using the “cell tracking” plugin of ImageJ in only a fraction of the time (supplemental Figure 4).
Neutrophil chemotaxis for different slopes of chemical gradient
To determine how the slope of the chemical gradient affected neutrophil chemotaxis, we developed a KOALA device with multiple channel lengths to vary the steepness of the chemical gradient (supplemental Figure 5A); furthermore, a dose-response varying the source chemoattractant concentration was performed to measure the chemotaxis response for each channel length (supplemental Figure 5B; supplemental Tables 1-3). As expected, the random migration speed increased as the channel length decreased, and little neutrophil movement was observed for shallow slope chemical gradients in the longer channels (supplemental Figure 5C). A similar trend was observed with the chemotactic index; however, this output quickly reached a roughly constant value as the channel length decreased (supplemental Figure 5D). Interestingly, the chemotaxis velocity reached a maximum value at an intermediate channel length for higher chemoattractant dose conditions but then decreased as the channel length decreased (supplemental Figure 5E). These results demonstrate that a microchannel length of 3 mm, the length chosen for the experiments shown in this work, enables robust neutrophil chemotaxis under a range of chemoattractant doses. Supplemental Videos 2 and 3 show examples of 2D human neutrophil chemotaxis in 3-mm-long microchannels, using source concentrations of 500nM fMLP and 0nM fMLP (control), respectively.
Neutrophil chemotaxis on an endothelial cell substrate
To study the different adherent and migratory states neutrophils exhibit under inflammatory conditions in the blood vessel, we developed the KOALA platform to conduct neutrophil capture and chemotaxis on an endothelial cell substrate (ECS). Because the KOALA approach only requires a micropipette to operate and is easily transportable, we were able to integrate a confluent endothelial cell monolayer into the chemotaxis assay. The confluent endothelial cell monolayer could then be activated by IL1-β to express E-selectin (Figure 3A; supplemental Figure 6). This process mimics the inflammatory cascade in vivo and allows for the capture of circulating neutrophils on the endothelial surface as the whole blood passes through the microchannel. Unstimulated HUVECs did not capture neutrophils using our KOALA platform, as expected (Figure 3A).
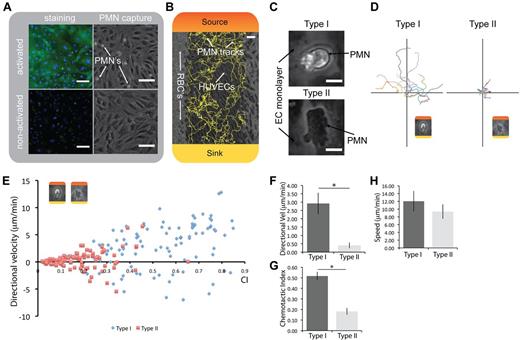
Capture and migration of human primary neutrophils on endothelial cells. (A) Staining for E-selectin on a HUVEC monolayer and corresponding polymorphonuclear leukocyte (PMN) capture from whole blood. (B) PMN migration on HUVECs with red blood cells (RBCs) on the side; automated tracks generated with JEX. Colors represent the formation of the chemical gradient: red represents maximum chemoattractant source concentration; and yellow, the chemoattractant concentration in the sink. (C) Micrographs showing type I and type II neutrophil morphologies (also see supplemental Video 5). (D) Representative tracks of each neutrophil phenotype transposed to the origin. (E) Representative individual neutrophil tracks of chemotactic index and directional velocity toward the chemoattractant, illustrating the differences in chemotactic function. (F-G) Type I phenotype is significantly more directional and has a higher velocity toward the chemoattractant than type II. *P < .05. n = 3. (H) No significant difference in random migration speed between the 2 neutrophil phenotypes (n = 3). Direct comparisons of neutrophil chemotaxis with controls shown in supplemental Figure 8. (A-B) Scale bars represent 100 μm. (C) Scale bar represents 10 μm. Error bars represent SEM. Phase-contrast images acquired using Slidebook Version 5.0.0.14 software with an Olympus IX-81 microscope using a 10× objective (NA = 0.30) at 37°C, except for micrographs shown in panel C. Additional details on imaging are given in “Microscopy.”
Capture and migration of human primary neutrophils on endothelial cells. (A) Staining for E-selectin on a HUVEC monolayer and corresponding polymorphonuclear leukocyte (PMN) capture from whole blood. (B) PMN migration on HUVECs with red blood cells (RBCs) on the side; automated tracks generated with JEX. Colors represent the formation of the chemical gradient: red represents maximum chemoattractant source concentration; and yellow, the chemoattractant concentration in the sink. (C) Micrographs showing type I and type II neutrophil morphologies (also see supplemental Video 5). (D) Representative tracks of each neutrophil phenotype transposed to the origin. (E) Representative individual neutrophil tracks of chemotactic index and directional velocity toward the chemoattractant, illustrating the differences in chemotactic function. (F-G) Type I phenotype is significantly more directional and has a higher velocity toward the chemoattractant than type II. *P < .05. n = 3. (H) No significant difference in random migration speed between the 2 neutrophil phenotypes (n = 3). Direct comparisons of neutrophil chemotaxis with controls shown in supplemental Figure 8. (A-B) Scale bars represent 100 μm. (C) Scale bar represents 10 μm. Error bars represent SEM. Phase-contrast images acquired using Slidebook Version 5.0.0.14 software with an Olympus IX-81 microscope using a 10× objective (NA = 0.30) at 37°C, except for micrographs shown in panel C. Additional details on imaging are given in “Microscopy.”
After the neutrophil capture, a gradient of chemoattractant was formed in the microchannels by placing the lid onto the base, initiating the ECS neutrophil chemotaxis assay (Figure 3B; supplemental Video 1; control, ie, no chemoattractant, in supplemental Video 4). When analyzing the area of randomly selected neutrophils that were captured on the ECS, we observed a bimodal distribution of the neutrophils that could be characterized by distinct morphologic differences (supplemental Figure 7A). One neutrophil morphology appeared darker and more spread on the endothelium when observed under phase-contrast microscopy, whereas the other neutrophil morphology was more compact and lighter in appearance (Figure 3C). We termed these 2 neutrophil morphologies as “type I” and “type II” henceforth for reference. The type I neutrophil morphology had less than half the area of type II neutrophils as they appeared under phase-contrast (supplemental Figure 7B). Analysis of the migration behavior of type I and II neutrophils showed significantly different migratory phenotypes (Figure 3D-H). Type I cells displayed a significantly more directional chemotactic response than type II cells (Figure 3D,G). By contrast, the type II morphology was highly spread, had limited directionality (Figured 3C-D,G), and was predominantly observed at the endothelial cell junctions (supplemental Video 5). The type I phenotype had a significantly higher directional velocity than type II (Figure 3E-F); however, type I neutrophils did not have a significantly higher random migration speed than type II (Figure 3H). In control experiments, neutrophils migrated randomly on the ECS, which is in contrast with the 2D and 3D applications of the KOALA platform where neutrophils were immobile when no chemical gradient was present. As expected, neutrophils migrated significantly less directionally, with lower speed and much lower chemotaxis velocity than cells in a chemical gradient (supplemental Figure 8; supplemental Videos 1 and 4). Finally, we observed that neutrophils exhibit both type I and type II phenotypes in roughly even proportions at the beginning of the time lapse, and type II neutrophils transition to type I progressively during the time lapse over a span of 90 minutes (supplemental Figure 9).
Neutrophil adhesion and migration from arthritic mice
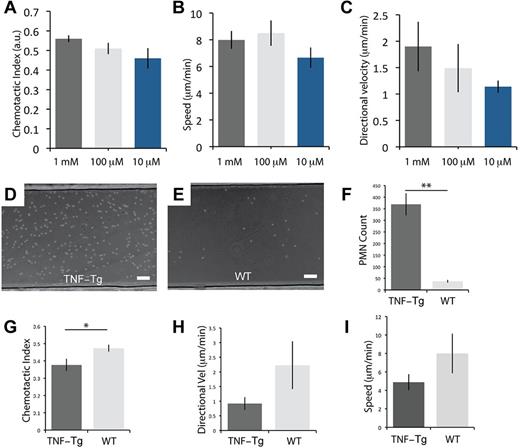
The protocol for neutrophil sorting and migration was modified to probe the adhesion and chemotactic behavior of neutrophils obtained from mice (see supplemental Methods). We show that the KOALA approach allows for repeated evaluation of the neutrophil adhesion and chemotaxis properties of the animal by acquiring whole blood from a small tail vein puncture without requiring animal sacrifice. Consistent with neutrophil chemotaxis assays for mice reported in the literature,28 mouse neutrophils required higher concentrations of chemoattractant than their human analog to activate and migrate (Figure 4A). In addition, we did not observe a significant difference in the migration speed, chemotactic index, or chemotaxis velocity between the doses of chemoattractant (Figure 4A-C).
Capture and migration of mouse neutrophils from whole blood for TNF-Tg and WT mice. (A-C) Chemoattractant (fMLP) dose-response for neutrophils obtained from WT mice from a tail vein puncture, showing the chemotactic index, speed, and directional velocity (n = 3). (D-E) Representative micrographs of neutrophil capture out of whole blood for arthritic (TNF-Tg) and WT mice. (F-I) Difference in the number of neutrophils adhering to the P-selectin substrate, chemotactic index, directional velocity, and speed for neutrophils from TNF-Tg and WT mice. **P < .01. *P < .05. (F) n = 5. (G-I) n = 3. No observable migration for control (no fMLP). (D-E) Scale bars represent 100 μm. Error bars represent SEM. Phase-contrast images acquired using Slidebook Version 5.0.0.14 software with an Olympus IX-81 microscope using a 10× (NA = 0.30) objective at 37°C. Additional details on imaging are given in “Microscopy.”
Capture and migration of mouse neutrophils from whole blood for TNF-Tg and WT mice. (A-C) Chemoattractant (fMLP) dose-response for neutrophils obtained from WT mice from a tail vein puncture, showing the chemotactic index, speed, and directional velocity (n = 3). (D-E) Representative micrographs of neutrophil capture out of whole blood for arthritic (TNF-Tg) and WT mice. (F-I) Difference in the number of neutrophils adhering to the P-selectin substrate, chemotactic index, directional velocity, and speed for neutrophils from TNF-Tg and WT mice. **P < .01. *P < .05. (F) n = 5. (G-I) n = 3. No observable migration for control (no fMLP). (D-E) Scale bars represent 100 μm. Error bars represent SEM. Phase-contrast images acquired using Slidebook Version 5.0.0.14 software with an Olympus IX-81 microscope using a 10× (NA = 0.30) objective at 37°C. Additional details on imaging are given in “Microscopy.”
Once we optimized the KOALA platform for mouse neutrophils, we demonstrated its utility and sensitivity for the characterization of neutrophil adhesion and chemotaxis properties in a mouse disease model. We obtained neutrophils from mice that spontaneously develop arthritis because of overexpression of human TNF-α29,30 (supplemental Videos 6 and 7). We found that neutrophils from these mice displayed a strong increase in adhesion (Figure 4D-F) and a significant decrease in directionality (Figure 4G) compared with controls. In addition, we observed a trend toward decreased speed and directional velocity for neutrophils from arthritic mice compared with WT (Figure 4H-I). We observed that roughly 1 order of magnitude more neutrophils from arthritic mice adhered to P-selectin–coated microchannels, as would be expected (Figure 4D-F). Furthermore, we saw a significant decrease in directionality (Figure 4G) of neutrophils from arthritic mice compared with controls, as well as a trend toward decreased speed and directional velocity for neutrophils from arthritic mice compared with WT mice (Figure 4H-I).
3D neutrophil chemotaxis
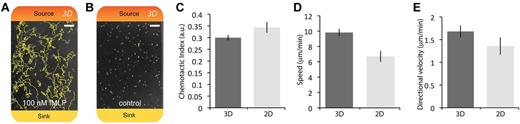
In addition to 2D embodiments of neutrophil chemotaxis, the KOALA platform can be used to study migration in 3D matrices with minimal modification of loading protocols. This was accomplished by flowing and polymerizing a hydrogel containing purified neutrophils into the microchannels. We show that neutrophil migration and automated tracking are readily achievable by analyzing the 2D projected migration path of the cells (Figure 5A; supplemental Video 8). We observed that neutrophils in the presence of a chemoattractant could migrate directionally in the 3D matrix, whereas cells in the control channels (no chemoattractant) were completely immobile (Figure 5A-B). In addition, we compared the 2D and 3D migration of human neutrophils in the presence of 100nM fMLP gradient (Figure 5C-E). Neutrophils migrating in 3D showed a trend of migrating with a slightly higher speed than the 2D case while being slightly less directional in their migration path. These results demonstrate that KOALA can be used to directly compare 2D and 3D neutrophil chemotaxis, which may be relevant for different physiologic conditions.
Migration of purified human neutrophils in a 3D collagen matrix. (A-B) Neutrophils tracked using JEX analysis, migrating in a 100nM gradient of fMLP compared with no observable migration in the control (gel with no chemoattractant). Colors represent the formation of the chemical gradient: red represents maximum chemoattractant source concentration; and yellow, the chemoattractant concentration at the sink. (C-E) Comparison of neutrophil migration in a 100nM gradient of fMLP for the 2D and 3D embodiments of the KOALA platform, showing the chemotactic index, speed, and directional velocity. Standard purification protocols were used for 3D migration experiments (see “3D neutrophil migration”). (A-B) Scale bars represent 100 μm. Error bars represent SEM. Phase-contrast images acquired using Slidebook Version 5.0.0.14 software with an Olympus IX-81 microscope using a 10× objective (NA = 0.30) at 37°C. Additional details on imaging are given in “Microscopy.”
Migration of purified human neutrophils in a 3D collagen matrix. (A-B) Neutrophils tracked using JEX analysis, migrating in a 100nM gradient of fMLP compared with no observable migration in the control (gel with no chemoattractant). Colors represent the formation of the chemical gradient: red represents maximum chemoattractant source concentration; and yellow, the chemoattractant concentration at the sink. (C-E) Comparison of neutrophil migration in a 100nM gradient of fMLP for the 2D and 3D embodiments of the KOALA platform, showing the chemotactic index, speed, and directional velocity. Standard purification protocols were used for 3D migration experiments (see “3D neutrophil migration”). (A-B) Scale bars represent 100 μm. Error bars represent SEM. Phase-contrast images acquired using Slidebook Version 5.0.0.14 software with an Olympus IX-81 microscope using a 10× objective (NA = 0.30) at 37°C. Additional details on imaging are given in “Microscopy.”
Discussion
In this study, we report a microfluidic neutrophil chemotaxis platform that reduces sample volume requirements by orders of magnitude compared with traditional techniques, such as Boyden and Dunn assays. The KOALA platform provides increased functionality and allows for new applications by decoupling the gradient generation and migration aspects of the microfluidic platform. Using this approach, the chemoattractant conditions and the neutrophil migration environment can be controlled without the protocol for one negatively influencing the other. The KOALA chemotaxis platform uses a hydrogel to controllably diffuse the chemoattractant into the microchannels without inducing undesired flows, thereby increasing reliability of the gradient generation. The use of hydrogels has been shown to provide a semipermeable structural material for microfluidic devices that can mediate the transfer of soluble factors.31 In the KOALA platform, the hydrogel prevents unintended flow of the chemoattractant into the microchannel during the initial contact of the lid and base. Instead, the chemoattractant controllably diffuses into the microchannels because of the concentration gradient that exists between the source and sink.
The techniques used by the KOALA platform to form a chemical gradient and purify neutrophils differ significantly from traditional neutrophil chemotaxis methods, making direct quantitative comparisons challenging. For example, the gradient profile and cell microenvironment in KOALA devices vary greatly from those of traditional methods, such as the Boyden or micropipette-based assays. In addition, the purification method used by KOALA removes potentially harmful steps that are common in traditional purification protocols, such as high-shear centrifugation, which has been shown to affect neutrophil function,32 and the cell lysis step for eliminating erythrocytes,33 which can induce harmful osmolarity shock. We show that our microfluidic approach reliably captures neutrophils efficiently (supplemental Figure 2) and produces a linear chemical gradient (Figure 2A-C), enabling neutrophil chemotaxis in a range of chemical microenvironments (supplemental Figure 5). This novel lid-based approach of delivering the chemotactic gradient makes the KOALA platform amendable to studying neutrophil chemotaxis in multiple contexts, including 2D and 3D systems, over an endothelium, and in both human and mouse models.
We report the first in vitro microfluidic assay that enables the direct monitoring of neutrophil chemotaxis on an ECS. This embodiment of the KOALA platform permits investigators to probe endothelial-leukocyte interactions that result in different phenotypes adopted by neutrophils. Importantly, the decoupling of the cell manipulation and gradient generation (base and lid, respectively) enables the investigator to culture endothelial cell monolayers and access the neutrophils and endothelial cells for further immunocytochemistry at any point during the assay by simply removing the lid. The ECS neutrophil chemotaxis assay revealed that neutrophils adopt 2 primary phenotypes in contact with endothelial cells, which had the appearance of being more compact or highly spread under phase-contrast microscopy (type I and type II, respectively). We observed that most of the type II cells transitioned to type I cells during the course of a time-lapse session, whereby they adopted the migratory behavior and morphology observed in the initially type I cells (supplemental Video 5; supplemental Figure 9). The type I phenotype was significantly more directional than type II and displayed higher migration velocity in the direction of the chemoattractant (Figure 3D-G). Type II phenotypes were commonly found at intercellular junctions, whereas the type I phenotype was observed in all locations on the cell membrane. This spatial organization may be the result of increased cell stiffness at the intercellular junctions, which is known to elicit a more spread morphology.34 Another possibility is that the presence of transmigratory molecules (eg, ICAM1 and VCAM1), which are preferentially expressed at these locations,35 may cause the neutrophils to adopt a more adherent phenotype, resulting in the observed spread morphology. These experiments demonstrate that neutrophils exhibit multiple morphologies and variable chemotactic behavior when they are captured on cytokine-stimulated endothelial cells, and further studies are needed to determine the mechanisms of these neutrophil phenotypes. The KOALA platform equips investigators with a simple tool to explore these interactions, which were difficult or impossible to study in the past.
The KOALA platform is the first demonstration of an assay capable of sorting, capturing, and purifying neutrophils obtained from mouse blood without killing the animal. An immediate consequence of our technique is that animal waste for investigators may be reduced because the sacrifice of several animals is not required to perform each neutrophil chemotaxis experiment. This is achievable because nanoliter volumes of blood are sufficient to purify neutrophils for each microchannel in the KOALA platform, resulting in a total blood draw of 3 μL filling 15-30 microchannels. To the authors' knowledge, this sample volume requirement 1 order of magnitude lower than the lowest achieved for leukocyte sorting and chemotaxis (< 1 μL vs 80 μL of diluted blood22 ). Importantly, the repeated measurement of neutrophil adhesion and chemotaxis for the same animal allows for the monitoring of neutrophil activity on mouse disease models in which the disease phenotypes may take long periods of time to develop or where multiple sampling is advantageous.
As a demonstration of the neutrophil capture technique using a tail vein blood draw, we assessed the function of neutrophils obtained from chronically inflamed mice (TNF-Tg) and compared them with WT. In mouse arthritis models, chronic inflammation is observed, and neutrophils specifically are central to the pathogenesis of this disease.36 Using the KOALA platform, we observed a ∼ 10-fold increase in the number of neutrophils that adhered to the P-selectin–coated surface for TNF-Tg mice compared with WT; furthermore, neutrophils from chronically inflamed mice trended toward slower migration speeds and were significantly less directional compared with neutrophils from WT mice. Investigators have shown that, for mice not experiencing chronic inflammation, ∼ 20-70 neutrophils should be present within 1 field of view of our microchannels.37,38 Our average neutrophil capture falls within that range for WT mice but is exceeded by roughly 1 order of magnitude for TNF-Tg mice (Figure 4D-F). Thus, the neutrophil count data suggests that TNF-Tg mice display strong neutrophilia, consistent with previous reports.36 These experiments demonstrate the pertinence of the KOALA technique and validate the use of the platform for studying neutrophils in the context of mouse inflammatory disease models. For the first time, this technique allows researchers to observe the development of disease phenotypes in mice over time, providing greater insight into the dynamics of symptom development. Furthermore, therapies, such as antileukoproteinase-based treatments that target neutrophil adhesion to the endothelium,39 can be systematically studied while monitoring neutrophil function over time. More generally, the KOALA platform can be used to study any disease where neutrophil adhesion and migration are central to the pathogenesis of the disease.
A growing body of work shows that cellular migration in a 3D matrix displays significantly altered phenotypes compared with 2D migration.40-42 Therefore, an important requirement of the KOALA platform is to have the ability to characterize neutrophil migration in 2D and 3D. Indeed, we demonstrate that the assay can be used to conduct direct comparisons between protein-coated 2D substrates and 3D extracellular matrix environments. We observed that neutrophils migrate faster toward the chemoattractant in 3D compared with 2D, confirming a trend that has been suggested by Hakkinen et al observed for migrating fibroblasts43 ; however, in contrast to the latter study, we do not observe a higher migration directionality for 3D compared with 2D. Using the KOALA platform, investigators have the opportunity to study neutrophil chemotaxis in 2D and 3D while subjecting the cells to comparable chemical gradient profiles and geometries. These features allow hematology researchers to investigate neutrophil chemotaxis in multiple microenvironments and uncover differences in phenotypes that may have been masked by biases inherent to a 2D in vitro system.
In conclusion, we have developed and characterized a microfluidic chemotaxis platform that incorporates on-chip neutrophil purification, a novel lid-based gradient generation, and demonstrates gains in function, operability, and versatility. Importantly, the assay significantly reduces the difficulty of performing challenging experiments involving long-term cell cultures, animal/patient samples, and time-lapse microscopy. To demonstrate this, we analyzed the morphology and migration of neutrophils on a HUVEC monolayer, neutrophil chemotaxis from a tail vein puncture of chronically inflamed mice, and 3D neutrophil migration. The 50- to 10 000-fold reduction in blood requirements we achieved enables small mammal studies, where collection of more than microliters of blood usually requires sacrificing the animal. The KOALA chemotaxis platform achieved increased functionality and simplicity of operation compared with standard techniques, making the assay well suited for widespread adoption in hematology research laboratories.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Kollias and the Alexander Fleming Biomedical Sciences Research Institute for providing the arthritic mouse line and Jay Warrick for his contributions in developing the basis of the JEX software.
The work was supported by an Innovation & Economic Development Research Program grant (E.K.S.), a Morgridge Institute of Research fellowship (E.B.), and the National Institutes of Health (R01 EB010039, D.J.B. and E.K.S.). M.A.S. was supported by the American College of Rheumatology Research and Education Fund (Rheumatology Scientist Development Award).
National Institutes of Health
Authorship
Contribution: E.K.S., E.B., A.H., and D.J.B. designed the study and prepared the manuscript; E.K.S. and E.B. conducted the experiments and analysis in the study; E.B. created the automated tracking software; E.W.K.Y. participated in endothelial cell migration experiments and performed the endothelial monolayer characterization; E.K.S. participated in endothelial cell migration experiments; M.A.S. provided arthritic mice; and M.A.S., S.A.W., and E.K.S. participated in mouse neutrophil chemotaxis experiments.
Conflict-of-interest disclosure: D.J.B. has an ownership interest in BellBrook Labs, LLC, which has licensed technology presented in this manuscript. The remaining authors declare no competing financial interests.
Correspondence: David J. Beebe, Wisconsin Institute Medical Research, 1111 Highland Ave, Madison, WI 53705; e-mail: djbeebe@wisc.edu.
References
Author notes
E.K.S. and E.B. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal