Abstract
Mutations in the all-trans retinoic acid (ATRA)–targeted ligand binding domain of PML-RARα (PRα/LBD+) have been implicated in the passive selection of ATRA-resistant acute promyelocytic leukemia clones leading to disease relapse. Among 45 relapse patients from the ATRA/chemotherapy arm of intergroup protocol C9710, 18 patients harbored PRα/LBD+ (40%), 7 of whom (39%) relapsed Off-ATRA selection pressure, suggesting a possible active role of PRα/LBD+. Of 41 relapse patients coanalyzed, 15 (37%) had FMS-related tyrosine kinase 3 internal tandem duplication mutations (FLT3-ITD+), which were differentially associated with PRα/LBD+ depending on ATRA treatment status at relapse: positively, On-ATRA; negatively, Off-ATRA. Thirteen of 21 patients (62%) had additional chromosome abnormalities (ACAs); all coanalyzed PRα/LBD mutant patients who relapsed off-ATRA (n = 5) had associated ACA. After relapse Off-ATRA, ACA and FLT3-ITD+ were negatively associated and were oppositely associated with presenting white blood count and PML-RARα type: ACA, low, L-isoform; FLT3-ITD+, high, S-isoform. These exploratory results suggest that differing PRα/LBD+ activities may interact with FLT3-ITD+ or ACA, that FLT3-ITD+ and ACA are associated with different intrinsic disease progression pathways manifest at relapse Off-ATRA, and that these different pathways may be short-circuited by ATRA-selectable defects at relapse On-ATRA. ACA and certain PRα/LBD+ were also associated with reduced postrelapse survival.
Introduction
Acute promyelocytic leukemia (APL) is initiated by formation of the PML-RARα fusion gene, which is cytogenetically identifiable by at t(15;17) in > 90% of cases.1,2 Depending on which of 3 breakpoint cluster regions (bcr's) in the PML gene on chromosome 15 is transected, 1 of 3 possible PML-RARα transcript types is formed by joining to a single bcr in the RARα gene on chromosome 17: the long (L), variable (V), or short (S)–isoform, which is invariant in each APL case.3 At diagnosis, 2 other genetic abnormalities are frequently associated with APL, mutations of the FMS-related tyrosine kinase 3 (FLT3) gene,4 and chromosomal abnormalities in addition to the t(15;17).5 Two types of FLT3 mutations occur that constitutively activate the molecule's receptor tyrosine kinase: internal tandem duplication mutations (FLT3-ITD+) in the juxtamembrane region and missense mutations primarily affecting aspartate-835 (FLT3-D835+) in the carboxy-terminal receptor tyrosine kinase domain.6 The more common FLT3-ITD+ have been associated with 3 APL characteristics: high white blood count (WBC), S-isoform type, and microgranular morphology.4 Many additional chromosome abnormalities (ACAs) have been identified of which a minor fraction are recurrent, most commonly trisomy 8.5 Studies in transgenic mice indicate that secondary mutations, including FLT3 mutations or those linked to certain ACAs, are required for the emergence of fully developed APL after initiation by PML-RARα.1 In addition, evidence in mice and/or humans suggests that FLT3 mutations and ACAs or cytogenetically occult gene copy number variations may be associated with alternative molecular pathways of disease progression to frank leukemia.7-9
Studies associated with clinical trials using all-trans retinoic acid (ATRA)/chemotherapy (CT) have investigated whether the afore-described genetic abnormalities at diagnosis affect the incidence of posttreatment disease progression and relapse. In some trials, an association has been found between increased relapse risk and the S-isoform and/or FLT3-ITD+.4,10 However, after adjustment for the associated WBCs by multivariate analysis (WBC ≥ 10 000/μL being the most consistent indicator of increased relapse risk after ATRA/CT therapy11 ), the independent prognostic significance of the S-isoform and/or FLT3-ITD+ has most often been lost or marginalized.3,8,10,12-14 Similarly, in some ATRA/CT trials, ACAs at diagnosis were associated with increased relapse risk by univariate but not multivariate analysis.8,10,15 In protocol C9710, patients analyzed at diagnosis for FLT3 mutations and ACAs, the only association with reduced disease-free survival (DFS) was a complex karyotype (≥ 2 ACAs; X.P. and W.S., unpublished data, June 2012).
ATRA/CT therapy usually reduces the presenting APL cell population by ≥ 5 orders of magnitude.16 Such reduction can result in the emergence of rare leukemic subclones at relapse that are hierarchically and genetically distinct from the leukemic cell population at presentation.17,18 Hence, examination of APL cells at relapse may provide more information than at diagnosis about molecular aberrations contributory to relapse and also about variables that may affect prognosis after postrelapse salvage therapy. In a previous study associated with Intergroup APL trial INT0129,19 we found that the predominant relapse clone harbored mutations in the ATRA-targeted ligand binding domain (LBD) of PML-RARα (PRα/LBD+) in one-third of ATRA-treated relapse patients.20,21 Two results were notable in the small mutation-positive group (n = 6): (1) in 4 patients, the predominant mutation-harboring clone emerged Off-ATRA treatment, suggesting that PRα/LBD+ has an alternative role to that of passively selecting mutant ATRA-resistant clones; and (2) the relapse mutation was detected in a low-level subclone in 1 of 3 patients studied, indicating that posttreatment selection rather than de novo acquisition of PRα/LBD+ can occur. An initial objective of this study was to determine if the aforementioned relationships of PRα/LBD+ were altered by the changed treatment of intergroup trial C9710, which included 2 induction/consolidation treatment arms: an ATRA/CT-arm in which the 2 agent types were administered differently than in INT0129; and an arsenic trioxide (ATO)–arm, in which ATO consolidation was added to the ATRA/CT regimen.22 When it became apparent that the incidence and distribution of PRα/LBD+ on the ATRA/CT-arm were similar to those on INT0129 (see “PML-RARα LBD mutations: nature and posttreatment clonal emergence”), we additionally queried in the larger C9710 relapse cohort analyzed for PRα/LBD+ (n = 45) how the other common genetic aberrations in APL, FLT3 mutations, and ACAs, relate to PRα/LBD+ and to other APL characteristics. The objective was to determine whether the inter-relationships of these parameters provide further insights into the undefined mechanisms of posttreatment disease progression and relapse.
Methods
Protocol C9710 relapse patients and samples
The 45 patients included in this study were derived from the ATRA/CT-arm of protocol C9710,22 based solely on the availability after consolidation therapy of a BM and/or peripheral blood sample evaluable for a PRα/LBD mutation at relapse. Up to the cut-off date for sample acquisition (May 2007), 81 of 294 (28%) ATRA/CT-arm patients had relapsed, including 62 of 237 (26%) adults and 19 of 57 (33%) children (< 15 years old). Sample recovery was: 34 of 62 (55%) for adults, 11 of 19 (58%) for children, and 45 of 81 (56%) overall. After May 2007, there were 8 additional relapses in 7 adults and 1 child (April 2012), which were not analyzed. Too few patients from the alternative adult C9710 ATO consolidation arm (ATO-arm) relapsed (n = 7) for analysis.22 Relapse signified clinical relapse; however, 2 patients, who were reclassified after central review from clinical relapse to nonrelapse were included based on detection of molecular relapse by quantitative RT-PCR. Subsequently, samples or data were collected at relapse and diagnosis to assess FLT3-ITD+, FLT3-D835+, and ACA; available samples from patients with a PRα/LBD mutation at relapse were also retrospectively tested at diagnosis. All patients signed consent for correlative laboratory studies according to the Declaration of Helsinki, and all participating institution's review boards approved this study.
The sodium metrizamide-fractionated, low-density mononuclear cells from all diagnostic samples had ≥ 70% blasts/promyelocytes (X.P. and W.S., unpublished data, June 2012). Relapse samples were assessed by the PML-RARα transcript level.16 Total cellular RNA and genomic DNA were extracted as described elsewhere (X.P. and W.S., unpublished data, June 2012).
PML-RARα methods
Conventional RT-PCR to identify PML-RARα isoform type and nested PCR and DNA sequence analysis to identify mutations in the RARα-region of PML-RARα were performed as described (1 primer substitution; supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).16,20 Procedures for quantitative RT-PCR measurement of PML-RARα transcript levels by normalization to transcripts for the housekeeping gene glyceraldehyde-3′-phosphate dehydrogenase (GAPDH) have also been reported.16 A modification of the amplification refractory mutational system procedure23 was developed, using mutation-specific primers, to identify PRα/LBD+ at high sensitivity (10−3-10−4) in diagnostic or prerelapse samples (supplemental Methods; supplemental Tables 1 and 2; supplemental Figure 1).
Determination of FLT3 mutation status
FLT3-ITD+ and FLT3-D835+ were detected in genomic DNA using the FLT3 Mutation Assay (InVivoScribe) or in total cellular RNA using published procedures.24 Quantitative measurement of FLT3-ITD+ and FLT3-D835+ allele levels relative to total FLT3 alleles used genomic DNA (X.P. and W.S., unpublished data, June 2012). FLT3 mutation status at relapse, in which the percentage of APL cells was unknown, is only reported for samples in which the GAPDH-normalized PML-RARα copy number was ≥ 10−5 to exclude possible false-negative results.
Cytogenetic methods
Cytogenetic analysis used standard G- or Q-banding techniques. Karyotypes were designated according to the International System for Cytogenetic Nomenclature. ACAs were defined as chromosome changes in addition to the APL-specific t(15;17)(q22;q21).
Statistical methods
Genetic aberrations were treated as dichotomous variables and were examined for their relationships to one another and other dichotomous variables using Fisher exact test. Continuous variables were assessed using the Satterthwaite test.25 The multiple P values for the many parameters evaluated by univariate analysis in this study are considered exploratory and descriptive in nature to indicate the relative strength of the relationship between variables and not as observed significance levels for hypothesis testing. Consequently, corrections for multiple testing were not applied, and we have used the terminology “association not indicated” for P values > .10 unless there is some other consideration for citation. Multivariate analysis of the genetic aberrations relative to all parameters was performed by logistic regression, but stable results were only achieved at diagnosis related to small sample size. The following parameters were assessed: age, sex, presenting WBC count (pWBC), M3 versus M3 variant cytology, time to relapse (TTR; days from complete remission), relapse On- or Off-ATRA, postrelapse death, PML-RARα isoform type, and all 3 alternative genetic aberrations. The pWBC was assessed for P values ≤ .05 as high (pWBChigh) for pWBCs ≥ 10 000/μL or as low (pWBClow) for pWBCs < 5000/μL in overall analyses or as > or < the median pWBC (6800/μL), respectively, for smaller ATRA-stratified subsets.
Results
Relapse study group parameters
Table 1 summarizes the clinicopathologic parameters assessed for a relationship to the 4 tested genetic aberrations (individual data in supplemental Table 3). Notably, the study group included 11 pediatric cases (< 15 years old) not included in reports of adult C9710 cases (X.P. and W.S., unpublished data, June 2012).22 Compared with all 294 ATRA/CT-arm patients, the 45-patient relapse cohort had an increased proportion of patients with a high relapse-risk pWBC (≥ 10 000/μL), 41% versus 24%. This is consistent with lower DFS in adults with high pWBCs on the ATRA/CT-arm (X.P. and W.S., unpublished data, June 2012). Similarly, the proportion of S-isoform cases was increased in the 45-patient set, 64% versus 39% overall. This is also consistent with trends on the ATRA/CT-arm toward an increased relapse incidence and reduced DFS in S-isoform compared with L-isoform patients (X.P. and W.S., unpublished data, June 2012). In contrast, there was no increase in the incidence of cases with the hypogranular M3 variant cytology in relapse (13%) compared with overall ATRA/CT patients (17%; X.P. and W.S., unpublished data, June 2012).
Relapse study group parameters (n = 45)
| Parameter . | No. (%) or median (range) . |
|---|---|
| Sex | Male, 25 (56%); female, 20 (44%) |
| Age, y | 32 (1-68)* |
| WBC, × 103/μL | 6.8 (0.6-143)† |
| APL cytologic type‡ | M3, 37 (82%); M3 variant, 6 (13%) |
| PML-RARα type | S-isoform, 29 (64%); L-isoform, 16 (36%) |
| Days to relapse | 528 (134-1431) |
| ATRA status at relapse§ | On-ATRA, 20 (44%); Off-ATRA, 25 (56%) |
| Postrelapse death‖ | Alive, 30 (67%); dead, 15 (33%) |
| Parameter . | No. (%) or median (range) . |
|---|---|
| Sex | Male, 25 (56%); female, 20 (44%) |
| Age, y | 32 (1-68)* |
| WBC, × 103/μL | 6.8 (0.6-143)† |
| APL cytologic type‡ | M3, 37 (82%); M3 variant, 6 (13%) |
| PML-RARα type | S-isoform, 29 (64%); L-isoform, 16 (36%) |
| Days to relapse | 528 (134-1431) |
| ATRA status at relapse§ | On-ATRA, 20 (44%); Off-ATRA, 25 (56%) |
| Postrelapse death‖ | Alive, 30 (67%); dead, 15 (33%) |
For complete data on all 45 relapse patients, see supplemental Table 3.
Distribution: < 15 years, 11; 15-60 years, 28; and > 60 years, 6.
Distribution: < 5 × 103/μL, 43%; 5-10 ×103/μL, 16%; > 10 ×103/μL, 41%.
French-American-British subclassification ×103; 2 patients could not be subclassified.
On-ATRA, relapse within 30 days of last ATRA dose; Off-ATRA, relapse > 30 days after last ATRA dose (actual range, 86-966 days).
Median follow-up of surviving patients = 71 months.
PML-RARα LBD mutations: nature and posttreatment clonal emergence
At relapse, 19 amino acid-altering nucleotide base changes in the RARα region of PML-RARα (all in the LBD) were identified in 18 of 45 (40%) patients (Figure 1). Seventeen PRα/LBD+ were single base substitutions (missense) and 2 were small deletions that maintained the open reading frame. One patient had 2 missense mutations (L224P and R276Q), which were shown to be in separate clones by recombinant isolation and sequence analysis (not shown). The mutations were localized to 3 subregions (zones) of the LBD, which are primarily involved in ATRA-dependent transcriptional regulatory activity.26,27 The intervening LBD areas involved in heterodimer formation with RXR were free of mutations. The predominance of mutations in zone I, which forms the ATRA binding pocket and central binding cleft for transcriptional cofactors, is consistent with previous findings.28 However, the greater number of mutations in zone III (n = 6) than zone II (n = 1) is contrary to previous reports of PRα/LBD+ in both relapse patients and ATRA-resistance–selected APL cell lines (additional mutation information in supplemental Table 4).28,29
Mutation sites in the ligand binding domain of PML-RARαafter relapse on protocol C9710 (ATRA/CT-arm). ▴ represents site of 17 missense mutations; and ▵, site of 2 deletion (Δ) mutations. Δ207-208, ΔK207Y208/T207 indicates deletion of lysine and tyrosine corresponding to amino acids 207 and 208 of normal RARα with maintenance of the normal downstream reading frame beginning with RARα threonine 209, now 207; similarly, Δ412-414, ΔE412M413L414/E415→E412.
Mutation sites in the ligand binding domain of PML-RARαafter relapse on protocol C9710 (ATRA/CT-arm). ▴ represents site of 17 missense mutations; and ▵, site of 2 deletion (Δ) mutations. Δ207-208, ΔK207Y208/T207 indicates deletion of lysine and tyrosine corresponding to amino acids 207 and 208 of normal RARα with maintenance of the normal downstream reading frame beginning with RARα threonine 209, now 207; similarly, Δ412-414, ΔE412M413L414/E415→E412.
The PRα/LBD+ affected ≥ 50% of PML-RARα transcripts in all cases and were the only forms detectable by allele-specific sequence analysis in 16 of 18 cases (individual case data in supplemental Table 5). These observations applied whether relapse occurred On-ATRA– or Off-ATRA–containing therapy. Conversely, no mutant allele was detectable by this method (sensitivity ≤ 10−1) at diagnosis in 16 cases tested. Using a higher sensitivity mutation-specific primer assay (sensitivity 10−3-10−4), a subclone harboring the relapse mutation was detected at diagnosis in 2 of 7 cases tested (supplemental Figures 2 and 3; supplemental Table 5). In the 2 mutation-positive cases, it was further determined that the mutation-harboring subclone emerged as a major clone only as relapse neared (Table 2; supplemental Figure 3). In case 27 (relapse On-ATRA), synchronous emergence of the mutant subclone occurred in the BM and peripheral blood. In case 31 (relapse Off-ATRA), a slowly expanding wild-type PML-RARα clone predominated at molecular relapse in the BM, whereas a later-emerging, more rapidly expanding clone harboring the PRα/LBD mutation detected at diagnosis was highly penetrant (> 90%) in peripheral blood. These observations confirm a previous single case report21 that the predominant PRα/LBD mutant clone at relapse is derived by selection of a preexisting mutant subclone rather than by posttreatment acquisition in some cases (cannot exclude additional cases with mutant subclones at < 10−3-10−4 frequency among up to 1012 APL cells at disease presentation) and suggest that a variable relationship of PRα/LBD mutation-harboring subclones to ATRA selection pressure is operative, leading to relapse On- or Off-ATRA treatment (see “Discussion”).
Serial monitoring of 2 patients with detectable PRα/LBD mutations at diagnosis by high-sensitivity mutation-specific primer assay
| . | PRα/LBD mutation level . | PML-RARα transcript level* . | ||
|---|---|---|---|---|
| Marrow . | Blood . | Marrow . | Blood . | |
| Patient 27 (R276Q)† | ||||
| Diagnosis | < 1/1000 | ∼ 1/800‡ | 400 | 40 |
| Postinduction | < 1/1000 | ∼ 1/300‡ | 1 | NA |
| Postconsolidation | < 1/1000 | < 1/1000 | 0.003 | 0.007 |
| Maintenance 4 months | < 1/70 | < 1/70 | < 0.001 | < 0.001 |
| Maintenance 9 months | < 1/10 | NA | 0.01 | NA |
| Relapse/postmaintenance 1 month | > 90/100‡ | > 90/100‡ | 1000 | 10 |
| Patient 31 (K238E)† | ||||
| Diagnosis | ∼ 1/900‡ | ∼ 1/300‡ | 900 | 600 |
| Postinduction | ∼ 1/900‡ | < 1/1000 | 3 | 1 |
| Postconsolidation | < 1/1000 | < 1/800 | 0.007 | NA |
| Postmaintenance | < 1/1000 | < 1/1000 | < 0.001 | < 0.001 |
| Postmaintenance 8 months | < 1/1000 | < 1/1000 | 0.008 | < 0.001 |
| Postmaintenance 20 months | < 1/1000 | ∼ 1/1000‡ | 0.9 | < 0.001 |
| Relapse/postmaintenance 28 months | ∼ 1/700‡ | > 90/100‡ | 200 | 4 |
| . | PRα/LBD mutation level . | PML-RARα transcript level* . | ||
|---|---|---|---|---|
| Marrow . | Blood . | Marrow . | Blood . | |
| Patient 27 (R276Q)† | ||||
| Diagnosis | < 1/1000 | ∼ 1/800‡ | 400 | 40 |
| Postinduction | < 1/1000 | ∼ 1/300‡ | 1 | NA |
| Postconsolidation | < 1/1000 | < 1/1000 | 0.003 | 0.007 |
| Maintenance 4 months | < 1/70 | < 1/70 | < 0.001 | < 0.001 |
| Maintenance 9 months | < 1/10 | NA | 0.01 | NA |
| Relapse/postmaintenance 1 month | > 90/100‡ | > 90/100‡ | 1000 | 10 |
| Patient 31 (K238E)† | ||||
| Diagnosis | ∼ 1/900‡ | ∼ 1/300‡ | 900 | 600 |
| Postinduction | ∼ 1/900‡ | < 1/1000 | 3 | 1 |
| Postconsolidation | < 1/1000 | < 1/800 | 0.007 | NA |
| Postmaintenance | < 1/1000 | < 1/1000 | < 0.001 | < 0.001 |
| Postmaintenance 8 months | < 1/1000 | < 1/1000 | 0.008 | < 0.001 |
| Postmaintenance 20 months | < 1/1000 | ∼ 1/1000‡ | 0.9 | < 0.001 |
| Relapse/postmaintenance 28 months | ∼ 1/700‡ | > 90/100‡ | 200 | 4 |
See supplemental Methods and supplemental Figure 3 for serially plotted data. The “<” symbol indicates mutation not detected at indicated assay sensitivity limit.
NA indicates not available.
Numbers indicate the number of PML-RARα transcript copies per 105 copies of GAPDH.
Maintenance therapy for 1 year was single-agent ATRA in patient 27 and ATRA plus 6-mercaptopurine and methotrexate in patient 31.
Detectable at the indicated level (eg, 1 mutant copy detected per 800 total PML-RARα copies).
FLT3 mutations: minor changes from diagnosis to relapse
FLT3-ITD+ were present in 46% and 37% of cases and FLT3-D835+ in 22% and 12% of cases, respectively, at diagnosis and relapse (n = 41; 4 patients different at the 2 time points). Among 37 patients with paired pretreatment and relapse analyses, the corresponding incidences were 43% and 41% (FLT3-ITD+) and 22% and 14% (FLT3-D835+; Table 3). In the paired subset, changes in mutation status occurred from diagnosis to relapse in 5 of 18 patients with FLT3-ITD+ and in 5 of 9 patients with FLT3-D835+, yielding a net loss of 1 and 3 mutant cases, respectively. Loss of FLT3 mutations often occurred in cases with a low level of the mutation-harboring clone at diagnosis. There were no differences in FLT3-ITD mutation status or change related to relapse On-ATRA– or Off-ATRA–containing therapy; 4 of 5 FLT3-D835 cases relapsed On-ATRA (Figure 2). Quantitative results available in a subset of patients indicated that the level of the mutant allele relative to total FLT3 allele was always < 50% and that differences were modest in 5 cases with paired FLT3-ITD+ determinations at diagnosis and relapse.
Changes in FLT3 mutation status from diagnosis (D) to relapse (R) in 37 patients with paired sample analysis
| FLT3 mutation . | Stable negative . | Loss D → R . | Gain D → R . | Stable positive . | Net change . | Percent positive . | |
|---|---|---|---|---|---|---|---|
| Diagnosis . | Relapse . | ||||||
| FLT3-ITD+ | 19 | 3 | 2 | 13 | −1 | 43 | 41 |
| FLT3-D835+ | 28 | 4 | 1 | 4 | −3 | 22 | 14 |
| FLT3 mutation . | Stable negative . | Loss D → R . | Gain D → R . | Stable positive . | Net change . | Percent positive . | |
|---|---|---|---|---|---|---|---|
| Diagnosis . | Relapse . | ||||||
| FLT3-ITD+ | 19 | 3 | 2 | 13 | −1 | 43 | 41 |
| FLT3-D835+ | 28 | 4 | 1 | 4 | −3 | 22 | 14 |
Qualitative and quantitative variations in FLT3 mutations from diagnosis to relapse: majority stable; loss of minor subclones; unbiased relapse distribution of FLT3-ITD+ On-ATRA/Off-ATRA; possible On-ATRA relapse bias of FLT3-D835+; and coincidence of FLT3-ITD+ and PRα/LBD+ after relapse On-ATRA. (A) FLT3-ITD mutation-positive cases at diagnosis and/or at relapse. (B) FLT3-D835 mutation-positive cases at diagnosis and/or at relapse. White represents mutation not detected; gray, mutation detected; black, no result available; and darker gray, coincident PRα/LBD mutation. Numbers indicate the percentage of mutant FLT3 allele relative to total FLT3 allele; nonbold indicates minor subclone (< 15%). Four cases each (3 in the same case; 1 each in different cases; supplemental Table 6) were not assayed at diagnosis for FLT3-ITD and/or FLT3-D835 mutations and, additionally, did not have detectable mutations at relapse; thus, these cases are not represented in the figure panels.
Qualitative and quantitative variations in FLT3 mutations from diagnosis to relapse: majority stable; loss of minor subclones; unbiased relapse distribution of FLT3-ITD+ On-ATRA/Off-ATRA; possible On-ATRA relapse bias of FLT3-D835+; and coincidence of FLT3-ITD+ and PRα/LBD+ after relapse On-ATRA. (A) FLT3-ITD mutation-positive cases at diagnosis and/or at relapse. (B) FLT3-D835 mutation-positive cases at diagnosis and/or at relapse. White represents mutation not detected; gray, mutation detected; black, no result available; and darker gray, coincident PRα/LBD mutation. Numbers indicate the percentage of mutant FLT3 allele relative to total FLT3 allele; nonbold indicates minor subclone (< 15%). Four cases each (3 in the same case; 1 each in different cases; supplemental Table 6) were not assayed at diagnosis for FLT3-ITD and/or FLT3-D835 mutations and, additionally, did not have detectable mutations at relapse; thus, these cases are not represented in the figure panels.
Karyotype abnormalities: ACA nonselectively increased at relapse
ACAs were present in 10 of 39 patients at diagnosis (26%) and in 13 of 21 patients at relapse (62%). All 21 relapse cases were also analyzed at diagnosis, and 6 (29%) had ACAs (Table 4). All 6 ACAs at diagnosis were retained at relapse, and the only case with < 100% ACA in t(15;17)–positive metaphase cells at diagnosis acquired 100% status at relapse. Two ACA-positive cases and 7 ACA-negative cases at diagnosis had newly present ACA at relapse. Single ACAs were newly present in the predominant relapse clone in 7 patients, whereas in 2 patients with ≥ 2 ACA the clonal penetration was mixed or uncertain.
Chromosomal abnormalities in addition to t(15;17) (ACAs) identified at diagnosis and relapse
| Patient no. . | ACA at diagnosis* . | ACA at relapse† . |
|---|---|---|
| 5 | None | t(3;11)(q28;q23), 10/19‡ |
| 6 | None | t(3;12)(p12;q24.3), 20/20 |
| 15 | None | t(4;5)(q21;q31), 4/4 |
| 9 | None | add(6)(q21), 2/2 |
| 12 | None | +8, 19/19 |
| 4 | None | i(8)(q10), 20/20 |
| 10 | None | Clone 1: add(22)(p11.2), 8/16; clone 2: idem,add(7)(q33), 2/16 |
| 1 | t(4;5)(q23;q31), 11/11ठ| t(4;5)(q23;q31); del(20)(q11.2q13.3), 3/3 |
| 7 | +8, 19/19 | +8, 9/9 |
| 8 | +8, 3/3 | +8; ?del(4)(q25),der(15)t(15;17)(q22;q21), −13, der(?16)t(13;16)(q12;q11), ider(17)(q10)t(15;17)(q22;q21), 1/1 |
| 20 | +8, 11/15 | +8, 20/20 |
| 3 | t(10;11)(q11.2;p11.2), 19/19§ | t(10;11)(q11.2;p11.2), 19/19 |
| 19 | der(15),ider(17)(q10) t(15;17)(q22;q21), 16/16 | der(15),ider(17)(q10) t(15;17)(q22;q21), 3/3 |
| 35 | add(8)(p23), 20/20 | Not available |
| 45 | +8, 20/20 | Not available |
| 43 | add(12)(p13), 19/19 | Not available |
| 40 | Clone 1: t(14;22)(p11.2;q11.2), 11/13; Clone 2: idem,ider(17)(q10) t(15;17)(q22;q21), 2/13 | Not available |
| Patient no. . | ACA at diagnosis* . | ACA at relapse† . |
|---|---|---|
| 5 | None | t(3;11)(q28;q23), 10/19‡ |
| 6 | None | t(3;12)(p12;q24.3), 20/20 |
| 15 | None | t(4;5)(q21;q31), 4/4 |
| 9 | None | add(6)(q21), 2/2 |
| 12 | None | +8, 19/19 |
| 4 | None | i(8)(q10), 20/20 |
| 10 | None | Clone 1: add(22)(p11.2), 8/16; clone 2: idem,add(7)(q33), 2/16 |
| 1 | t(4;5)(q23;q31), 11/11ठ| t(4;5)(q23;q31); del(20)(q11.2q13.3), 3/3 |
| 7 | +8, 19/19 | +8, 9/9 |
| 8 | +8, 3/3 | +8; ?del(4)(q25),der(15)t(15;17)(q22;q21), −13, der(?16)t(13;16)(q12;q11), ider(17)(q10)t(15;17)(q22;q21), 1/1 |
| 20 | +8, 11/15 | +8, 20/20 |
| 3 | t(10;11)(q11.2;p11.2), 19/19§ | t(10;11)(q11.2;p11.2), 19/19 |
| 19 | der(15),ider(17)(q10) t(15;17)(q22;q21), 16/16 | der(15),ider(17)(q10) t(15;17)(q22;q21), 3/3 |
| 35 | add(8)(p23), 20/20 | Not available |
| 45 | +8, 20/20 | Not available |
| 43 | add(12)(p13), 19/19 | Not available |
| 40 | Clone 1: t(14;22)(p11.2;q11.2), 11/13; Clone 2: idem,ider(17)(q10) t(15;17)(q22;q21), 2/13 | Not available |
Results from 39 cases analyzed at diagnosis; see supplemental Table 6 for complete karyotype information.
Results from 21 cases analyzed at relapse; see supplemental Table 5 for complete karyotype results.
Number of metaphase cells with ACA (numerator); total number of t(15;17)–positive metaphase cells (denominator); applies to all fractions in columns.
These translocations cannot represent constitutional rearrangements because normal karyotypes were also present.
Table 4 summarizes all ACA observed in the study (complete karyotypic data in supplemental Tables 5 and 6). Chromosome 8 abnormalities were most frequent. Trisomy 8 was present at diagnosis in 4 of 39 (10%) and at relapse in 4 of 21 (19%) cases analyzed. All 3 patients with trisomy 8 at diagnosis, who were analyzed at relapse, retained the +8, 2 as the sole ACA and 1 in a complex karyotype. In 1 case each, trisomy 8 or i(8)(q10) was newly present at relapse, bringing the total relapse cases with 8q duplication to 5 of 21 (24%). The second most frequent ACA was ider(17q), which was present at diagnosis in 2 of 39 (5%) and at relapse in 2 of 21 (10%) cases analyzed; the latter included 1 retained from diagnosis and 1 newly present at relapse in a complex karyotype with trisomy 8. Balanced translocations increased from diagnosis to relapse both in incidence, 8% (3/39) to 29% (6/21), and as a fraction of all ACA, 30% (3 of 10) to 46% (6 of 13). A t(4;5) occurred in 2 cases with a common breakpoint in 5q31; the chromosome 4 breakpoints affected neighboring bands, 4q21 and 4q23. All other structural chromosome changes were nonrecurrent. A complex karyotype was present in 1 of 39 (3%) cases at diagnosis and 3 of 21 (14%) patients at relapse.
Association analysis of genetic aberrations at diagnosis
Table 5 summarizes the test parameters for which an indication of association was identified by univariate or multivariate analysis (supplemental Tables 7 and 8 for all data). By univariate analysis, FLT3-ITD+ were strongly associated with a pWBChigh and with an increased incidence of the S-isoform; ACAs were associated with a longer TTR. By multivariate analysis, all 3 aberrations had a different set of relationships, except for weaker associations of FLT3-ITD+ with the S-isoform and pWBChigh. FLT3-D835+ were also associated with pWBChigh, an association also found in overall C9710 patients (X.P. and W.S., manuscript submitted, June 2012) and also showed a weak trend toward association with the S-isoform. FLT3-ITD+ and FLT3-D835+ were reciprocally negatively associated, and ACAs were associated with older age.
Associations at diagnosis of patients with FLT3 mutations or ACAs by univariate and multivariate analyses
| Test parameter . | FLT3-ITD mutation . | FLT3-D835 mutation . | ACA . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Descriptor . | Univariate* . | Multivariate† . | Descriptor . | Univariate* . | Multivariate† . | Descriptor . | Univariate‡ . | Multivariate§ . | |
| Age | ANI | ANI | ANI | ANI | Older | ANI | .05 | ||
| Presenting WBC | High | .002 | .07 | High | ANI | .02 | — | ANI | ANI |
| Time to relapse | ANI | ANI | ANI | ANI | Longer | .03 | ANI | ||
| PML-RARα type | ↑ S-isoform incidence | < .001 | .04 | ↑ S-isoform incidence | ANI | .10 | — | ANI | ANI |
| FLT3-ITD+ | — | — | — | Negative associaton | ANI | .01 | — | ANI | ANI |
| FLT3-D835+ | Negative association | ANI | .06 | — | — | — | — | ANI | NE |
| Test parameter . | FLT3-ITD mutation . | FLT3-D835 mutation . | ACA . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Descriptor . | Univariate* . | Multivariate† . | Descriptor . | Univariate* . | Multivariate† . | Descriptor . | Univariate‡ . | Multivariate§ . | |
| Age | ANI | ANI | ANI | ANI | Older | ANI | .05 | ||
| Presenting WBC | High | .002 | .07 | High | ANI | .02 | — | ANI | ANI |
| Time to relapse | ANI | ANI | ANI | ANI | Longer | .03 | ANI | ||
| PML-RARα type | ↑ S-isoform incidence | < .001 | .04 | ↑ S-isoform incidence | ANI | .10 | — | ANI | ANI |
| FLT3-ITD+ | — | — | — | Negative associaton | ANI | .01 | — | ANI | ANI |
| FLT3-D835+ | Negative association | ANI | .06 | — | — | — | — | ANI | NE |
Data for all parameters are listed in “Statistical methods” by univariate and multivariate analysis in supplemental Tables 7 and 8. Descriptor indicates the nature of the association of the test parameter with the aberration in the heading.
ANI indicates association not indicated; —, not applicable; and NE, not evaluable.
Univariate analysis of FLT3 mutations had a sample size of n = 41, which differed by 1 case each for the FLT3-ITD and FLT3-D835 sets (see supplemental Table 6).
Multivariate analysis of FLT3 mutations had a sample size of n = 39.
Univariate analysis of ACA had a sample size of n = 39.
Multivariate analysis of ACA had a sample size of n = 35.
Association analysis of genetic aberrations at relapse
Univariate association analysis at relapse was made in a 41-patient subset evaluable for both PRα/LBD+ and FLT3 mutations (positive results summarized in Table 6; all results in supplemental Table 9). PRα/LBD+ were associated with pWBClow and tended to occur more frequently if relapse occurred On-ATRA treatment. Conversely, FLT3-ITD+ were associated with a pWBChigh and the S-isoform. FLT3-ITD+ were also inversely (negatively) associated with ACAs. The small number of FLT3-D835+ (n = 5) impeded analysis, although the TTR in patients with FLT3-D835+ was reduced; correspondingly, 4 of 5 patients with FLT3-D835+ relapsed On-ATRA. ACAs in a 21-patient subset were associated with the absence of FLT3-ITD+ and increased post-relapse death and with a trend toward association with the L-isoform. Multivariate analysis did not yield stable results.
Associations at relapse of patients with PRα/LBD and FLT3 mutations (n = 41*) or ACAs (n = 21) by univariate analysis
| Test parameter . | PRα/LBD mutations (18 vs 23*) . | FLT3-ITD mutations (15 vs 26*) . | FLT3-D835 mutations (5 vs 36*) . | ACA (13 vs 8*) . | ||||
|---|---|---|---|---|---|---|---|---|
| Descriptor . | P . | Descriptor . | P . | Descriptor . | P . | Descriptor . | P . | |
| Presenting WBCs | Low | .03 | High | .006 | — | ANI | — | ANI |
| Time to relapse | — | ANI | ANI | ANI | Shorter | < .001 | — | ANI |
| Relapse On-ATRA† | Increased incidence | .06 | — | ANI | — | .15‡ | — | ANI |
| Postrelapse death | — | ANI | — | ANI | — | ANI | Increased Incidence | .05 |
| PML-RARα type | — | ANI | ↑ S-isoform incidence | < .001 | — | ANI | ↑ L-isoform incidence | .07 |
| FLT3-ITD+ | — | ANI | — | — | — | ANI | Negative association | .003 |
| ACA | — | ANI | Negative association | .003 | — | ANI | — | ANI |
| Test parameter . | PRα/LBD mutations (18 vs 23*) . | FLT3-ITD mutations (15 vs 26*) . | FLT3-D835 mutations (5 vs 36*) . | ACA (13 vs 8*) . | ||||
|---|---|---|---|---|---|---|---|---|
| Descriptor . | P . | Descriptor . | P . | Descriptor . | P . | Descriptor . | P . | |
| Presenting WBCs | Low | .03 | High | .006 | — | ANI | — | ANI |
| Time to relapse | — | ANI | ANI | ANI | Shorter | < .001 | — | ANI |
| Relapse On-ATRA† | Increased incidence | .06 | — | ANI | — | .15‡ | — | ANI |
| Postrelapse death | — | ANI | — | ANI | — | ANI | Increased Incidence | .05 |
| PML-RARα type | — | ANI | ↑ S-isoform incidence | < .001 | — | ANI | ↑ L-isoform incidence | .07 |
| FLT3-ITD+ | — | ANI | — | — | — | ANI | Negative association | .003 |
| ACA | — | ANI | Negative association | .003 | — | ANI | — | ANI |
Detailed data for all parameters are listed in “Statistical methods” and in supplemental Table 9. Descriptor indicates the nature of the association of the test parameter with the aberration in the heading.
— indicates not applicable; and ANI, association not indicated.
Number positive vs number negative.
Relapse On-ATRA, relapse within 30 days of last dose of ATRA.
Four of 5 patients (80%) with FLT3-D835+ versus 14 of 36 patients (39%) without FLT3-D835+ relapsed On-ATRA (P = 0.15).
ATRA treatment status at relapse impacts genetic aberration associations
We assessed the potential effect of ATRA treatment status at relapse because defects in ATRA-targeted molecular pathways might be interactive with differences in intrinsic disease characteristics and/or the genetic aberrations studied. Remarkably, virtually all of the associations noted in the overall analysis at relapse (Table 6) were restricted to relapse Off-ATRA: PRα/LBD with pWBClow; FLT3-ITD+ with pWBChigh, S-isoform and ACA (negatively; Table 7; details in supplemental Table 10). Exceptionally, FLT3-ITD+ remained weakly associated with the S-isoform after relapse On-ATRA, related to the 100% co-occurrence of these 2 parameters. Although case numbers were very small (n = 12), ACAs were associated with pWBClow, the L-isoform and FLT3-ITD+ (reciprocally). FLT3-D835+ were too few for analysis (n = 5). This analysis also revealed a reciprocal negative association of FLT3-ITD+ and PRα/LBD+ after relapse Off-ATRA.
Associations of PRα/LBD and FLT3-ITD mutations and ACAs after relapse On-ATRA or Off-ATRA therapy
| Test parameter . | PRα/LBD mutations . | FLT3-ITD mutations . | ACA . | |||||
|---|---|---|---|---|---|---|---|---|
| Descriptor . | On-ATRA (11 vs 7) . | Off-ATRA (7 vs 16) . | Descriptor . | On-ATRA (6 vs 12) . | Off-ATRA (9 vs 14) . | Descriptor . | Off-ATRA (9 vs 3) . | |
| Presenting WBCs* | Low | ANI | .001 | High | ANI | < .001 | Low | .02 |
| PML-RARα type | — | ANI | ANI | ↑ S-isoform incidence | .05 | .002 | ↑ L-isoform incidence | .05 |
| PRα/LBD+ | — | — | — | Negative association | ANI | .02 | — | ANI |
| FLT3-ITD+ | Negative association | ANI | .02 | — | — | — | Negative association | .005 |
| ACA | — | ANI | ANI | Negative association | ANI | .005 | — | — |
| Test parameter . | PRα/LBD mutations . | FLT3-ITD mutations . | ACA . | |||||
|---|---|---|---|---|---|---|---|---|
| Descriptor . | On-ATRA (11 vs 7) . | Off-ATRA (7 vs 16) . | Descriptor . | On-ATRA (6 vs 12) . | Off-ATRA (9 vs 14) . | Descriptor . | Off-ATRA (9 vs 3) . | |
| Presenting WBCs* | Low | ANI | .001 | High | ANI | < .001 | Low | .02 |
| PML-RARα type | — | ANI | ANI | ↑ S-isoform incidence | .05 | .002 | ↑ L-isoform incidence | .05 |
| PRα/LBD+ | — | — | — | Negative association | ANI | .02 | — | ANI |
| FLT3-ITD+ | Negative association | ANI | .02 | — | — | — | Negative association | .005 |
| ACA | — | ANI | ANI | Negative association | ANI | .005 | — | — |
FLT3-D835 mutations and ACAs after relapse. On-ATRA too few for analysis. Detailed data for all parameters are listed in “Statistical methods” and supplemental Table 10.
— indicates not applicable; and ANI, association not indicated.
WBC high or low P values were calculated from above or below the median WBC (6800/μL).
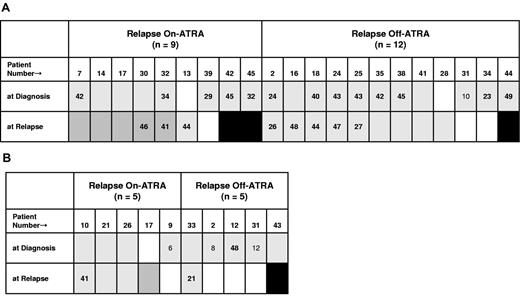
We performed an alternative, secondary analysis in the patient subsets positive for FLT3-ITD+ and PRα/LBD+ using relapse On- or Off-ATRA as the variable; and, notably, this revealed a positive association of these 2 mutations in the On-ATRA relapse set: among patients with FLT3-ITD+, 5 of 6 who relapsed On-ATRA versus 0 of 9 who relapsed Off-ATRA had coincident PRα/LBD+ (P = .002; Figure 2A; supplemental Table 11). Similarly, we noted that all 5 cytogenetically analyzed patients with PRα/LBD+, who relapsed Off-ATRA had coincident ACAs (Figure 3). Although PRα/LBD+ and ACAs were not associated in the overall or On-ATRA/Off-ATRA relapse analyses (Tables 6 and 7), among patients with PRα/LBD+, 4 of 6 Off-ATRA relapse versus 1 of 11 On-ATRA relapse patients had ACAs at diagnosis (P = .03; Figure 3). These observations, as well as the common association of PRα/LBD+ and ACAs with pWBClow and FLT3-ITD+ (negatively), suggest a nonreciprocal association of PRα/LBD+ with ACA at relapse Off-ATRA.
In patients with PRα/LBD+, coincident ACA occurred more frequently after relapse Off-ATRA in patients with ACA at diagnosis. White represents ACA absent; gray, ACA present; darker gray, newly detected ACA at relapse; and black, no cytogenetic analysis.
In patients with PRα/LBD+, coincident ACA occurred more frequently after relapse Off-ATRA in patients with ACA at diagnosis. White represents ACA absent; gray, ACA present; darker gray, newly detected ACA at relapse; and black, no cytogenetic analysis.
A summary of the inter-parameter associations described above at diagnosis and relapse is presented in Table 8, and data for all study parameters related to each genetic aberration are depicted on an individual case basis in supplemental Figure 4. We acknowledge that the assigned associations are based on small case numbers and that they must be considered provisional.
Summary of genetic aberration associations
| Overall associations |
| At diagnosis |
| FLT3-ITD+ with S-isoform, pWBChigh |
| FLT3-D835+ with pWBChigh |
| FLT3-ITD+ and FLT3-D835+ negatively |
| At relapse |
| FLT3-ITD+ with S-isoform, pWBChigh, ACA negatively |
| ACA with FLT3-ITD+ negatively, L-isoform, postrelapse death |
| PRα/LBD+ with pWBClow |
| ATRA treatment stratified associations at relapse |
| Relapse On-ATRA |
| FLT3-ITD+ with S-isoform, PRα/LBD+ |
| Relapse Off-ATRA |
| FLT3-ITD+ with S-isoform, pWBChigh, ACA and PRα/LBD+ negatively |
| ACA with pWBClow, L-isoform, FLT3-ITD+ negatively |
| PRα/LBD+ with pWBClow, ACA, FLT3-ITD+ negatively |
| Overall associations |
| At diagnosis |
| FLT3-ITD+ with S-isoform, pWBChigh |
| FLT3-D835+ with pWBChigh |
| FLT3-ITD+ and FLT3-D835+ negatively |
| At relapse |
| FLT3-ITD+ with S-isoform, pWBChigh, ACA negatively |
| ACA with FLT3-ITD+ negatively, L-isoform, postrelapse death |
| PRα/LBD+ with pWBClow |
| ATRA treatment stratified associations at relapse |
| Relapse On-ATRA |
| FLT3-ITD+ with S-isoform, PRα/LBD+ |
| Relapse Off-ATRA |
| FLT3-ITD+ with S-isoform, pWBChigh, ACA and PRα/LBD+ negatively |
| ACA with pWBClow, L-isoform, FLT3-ITD+ negatively |
| PRα/LBD+ with pWBClow, ACA, FLT3-ITD+ negatively |
TTR assessments
TTR can be an indicator of the intrinsic malignant properties of individual leukemias. Thus, we examined relations between TTR and specific PRα/LBD+ and/or ACAs. In the patient subset, including all PRα/LBD+ and/or ACA (n = 24), the TTRs stratified into 2 groups: 134-546 days (n = 13) or 644-1431 days (n = 11), corresponding to relapse On-ATRA or Off-ATRA (Table 9). The TTRs were representative of the amount of ATRA administered up to the completion of maintenance with the exception of patient 1, who relapsed nearly 2 years after completing ATRA-containing consolidation. Several points seem noteworthy. First, the 3 shortest TTR (134-218 days) occurred in patients with a PRα/LBD+ in zone III (Y208N, R217Hx2; Figure 1). Second, the 5 patients with the shortest TTRs (134-265 days) were pediatric cases (ages 1-11 years). Of note, the Y208N mutation in a 1-year-old (patient 14) was not detectable at diagnosis with > 10−4 sensitivity (supplemental Figure 2), excluding the preexistence of this mutation as a cause of early neonatal relapse. Third, although almost all zone I mutations occurred during or within 6 months of finishing maintenance, zone III mutations occurred either soon after finishing consolidation or after prolonged follow-up. Fourth, a higher proportion of ACAs occurred in the long/Off-ATRA TTR group (9/9 vs 4/7; P = .06), including 4/5 + 8/i (8q), 2/2 ider (17q), 4/5 balanced translocations, and 2/3 complex karyotypes. There was, however, no apparent difference in the proportion of patients with newly present ACA who experienced short/On-ATRA (3/4) or long/Off-ATRA (6/9) times to relapse. Finally, the co-occurrence of FLT3 mutations with either PRα/LBD+ and/or ACA was confined to relatively short-term relapses On-ATRA.
Relationship of TTR to specific PRα/LBD+ and ACA, age, and FLT3 mutations
| Patient no. . | TTR, days . | Days of maintenance . | PRα/LBD+ . | ACA . | Age . | FLT3 mutation . |
|---|---|---|---|---|---|---|
| 14 | 134 | 0 | Y208N† | None | 1§ | ITD+ |
| 32 | 175 | 0 | R217H† | NA | 4§ | ITD+ |
| 39 | 218 | 107 | R217H† | NA | 11§ | — |
| 5 | 265 | 118 | N | t(3;11)‡ | 11§ | — |
| 10 | 265 | 130 | N | add(22p)‡ add(7q)‡ | 3§ | D835+ |
| 22 | 309 | 179 | Δ412–414 | NA | 44 | — |
| 30 | 339 | 196 | I273F | NA | 43 | ITD+ |
| 9 | 359 | 231 | G289E | add(6)(q21)‡ | 35 | — |
| 17 | 363 | 218 | R276Q and L224P† | None | 65 | ITD+ and D835+‡ |
| 7 | 401 | 279 | G289E | +8 | 42 | ITD+ |
| 11 | 520 | 365 | T285A | None | 28 | — |
| 27 | 528 | 365 | R276Q | NA | 32 | — |
| 37 | 546 | 365 | G289R | NA | 15 | — |
| 15 | 644* | 365 | R272Q | t(4;5)‡ | 13§ | — |
| 4 | 655* | 365 | N | i(8)(q10)‡ | 31 | — |
| 20 | 657* | 365 | R276W | +8 | 64 | — |
| 12 | 678* | 365 | N | +8‡ | 31 | — |
| 8 | 682* | 365 | G289R | +8 and 3ACA‡ | 49 | — |
| 40 | 686* | 365 | S287W | NA | 49 | — |
| 3 | 718* | 365 | N | t(10;11) | 17 | — |
| 1 | 728* | 0 | S287L | t(4;5),del(20)‡ | 64 | — |
| 6 | 874* | 365 | Δ207–208† | t(3;12)‡ | 18 | — |
| 19 | 1057* | 365 | N | ider(17q) | 36 | — |
| 31 | 1431* | 365 | K238E† | NA | 32 | — |
| Patient no. . | TTR, days . | Days of maintenance . | PRα/LBD+ . | ACA . | Age . | FLT3 mutation . |
|---|---|---|---|---|---|---|
| 14 | 134 | 0 | Y208N† | None | 1§ | ITD+ |
| 32 | 175 | 0 | R217H† | NA | 4§ | ITD+ |
| 39 | 218 | 107 | R217H† | NA | 11§ | — |
| 5 | 265 | 118 | N | t(3;11)‡ | 11§ | — |
| 10 | 265 | 130 | N | add(22p)‡ add(7q)‡ | 3§ | D835+ |
| 22 | 309 | 179 | Δ412–414 | NA | 44 | — |
| 30 | 339 | 196 | I273F | NA | 43 | ITD+ |
| 9 | 359 | 231 | G289E | add(6)(q21)‡ | 35 | — |
| 17 | 363 | 218 | R276Q and L224P† | None | 65 | ITD+ and D835+‡ |
| 7 | 401 | 279 | G289E | +8 | 42 | ITD+ |
| 11 | 520 | 365 | T285A | None | 28 | — |
| 27 | 528 | 365 | R276Q | NA | 32 | — |
| 37 | 546 | 365 | G289R | NA | 15 | — |
| 15 | 644* | 365 | R272Q | t(4;5)‡ | 13§ | — |
| 4 | 655* | 365 | N | i(8)(q10)‡ | 31 | — |
| 20 | 657* | 365 | R276W | +8 | 64 | — |
| 12 | 678* | 365 | N | +8‡ | 31 | — |
| 8 | 682* | 365 | G289R | +8 and 3ACA‡ | 49 | — |
| 40 | 686* | 365 | S287W | NA | 49 | — |
| 3 | 718* | 365 | N | t(10;11) | 17 | — |
| 1 | 728* | 0 | S287L | t(4;5),del(20)‡ | 64 | — |
| 6 | 874* | 365 | Δ207–208† | t(3;12)‡ | 18 | — |
| 19 | 1057* | 365 | N | ider(17q) | 36 | — |
| 31 | 1431* | 365 | K238E† | NA | 32 | — |
— indicates not applicable.
Relapse Off-ATRA.
Zone III LBD mutations.
Newly detected at relapse.
Pediatric cases.
Postrelapse survival assessments
At a median follow-up of 71 months among surviving patients (April 2012), 15 of 45 patients (33%) had died and 2 were lost to follow-up. Of 28 surviving patients with postrelapse follow-up data, all had longer follow-up time (range, 371-3564 days) than the median time to death in 14 nonsurvivors (368 days; Table 10). In this dataset, ACA at relapse was the only genetic aberration associated with reduced postrelapse survival (P = .05), consistent with the overall case analysis based on postrelapse death (Table 6). However, the data were insufficient to assess whether any particular ACA or type of ACA affected postrelapse survival. PRα/LBD+ data were also limited, but, notably, 3 of 4 mutations affecting G289 and both deletion mutations occurred in and constituted 5 of 6 PRα/LBD+ in nonsurvivors. FLT3-ITD+ occurred 2 times more often in survivors than nonsurvivors (46% vs 21%), and, although not significant, this observation makes it improbable that FLT3-ITD+ would be associated with reduced postrelapse survival in a larger sample.
Relationship of genetic aberrations and recurrent subtype aberrations to postrelapse survival
| Parameter . | Nonsurvivors (n = 15) . | Survivors (n = 28) . | P . |
|---|---|---|---|
| Median postrelapse survival, d (range) | 368* (6-1104) | 1497* (371-3564) | < .001* |
| ACA, % positive | 100* (6) | 50* (12) | .05* |
| +8/i(8q), cases | 3 | 2 | — |
| Balanced translocation, cases | 2 | 3 | — |
| Complex karyotype, cases | 1 | 1 | — |
| PRα/LBD mutations, % positive | 40 | 39 | NS |
| Missense mutations, cases | 4 | 11 | NS |
| G289E/R, cases | 3* | 1* | — |
| R276Q/W, cases | 1 | 2 | — |
| Deletion mutations, cases | 2* | 0* | — |
| FLT3-ITD mutations, % positive | 21* | 46* (26) | .18 |
| FLT3-D835 mutations, % positive | 7 | 13 (24) | NS |
| Parameter . | Nonsurvivors (n = 15) . | Survivors (n = 28) . | P . |
|---|---|---|---|
| Median postrelapse survival, d (range) | 368* (6-1104) | 1497* (371-3564) | < .001* |
| ACA, % positive | 100* (6) | 50* (12) | .05* |
| +8/i(8q), cases | 3 | 2 | — |
| Balanced translocation, cases | 2 | 3 | — |
| Complex karyotype, cases | 1 | 1 | — |
| PRα/LBD mutations, % positive | 40 | 39 | NS |
| Missense mutations, cases | 4 | 11 | NS |
| G289E/R, cases | 3* | 1* | — |
| R276Q/W, cases | 1 | 2 | — |
| Deletion mutations, cases | 2* | 0* | — |
| FLT3-ITD mutations, % positive | 21* | 46* (26) | .18 |
| FLT3-D835 mutations, % positive | 7 | 13 (24) | NS |
Individual case data are detailed in supplemental Table 12. Numbers in brackets indicate the number of cases tested if less than total cases indicated by “n” in the NS corresponding column heading.
— indicates too few for analysis; and NS, no significant difference.
Distribution differences of possible interest between nonsurvivors and survivors.
Discussion
Our prestudy hypothesis was that the change in therapy on trial C9710 vis-à-vis the previous intergroup trial INT0129 would reduce the incidence of relapse with clones harboring PRα/LBD+. This was based, in part, on a report that concurrent (C9710) rather than sequential (INT0129) administration of ATRA and CT reduces the incidence of relapse22,30 and on indications of clinical synergy of ATO with ATRA.31 After release of C9710 treatment-arm outcome data, it became apparent that the addition of ATO to ATRA/CT must have been highly effective in eliminating PRα/LBD+-harboring clones because only 7 relapses occurred on the ATO-arm.22 Conversely, we found no substantive differences between the ATRA/CT-arm of C9710 and INT0129 regarding the incidence of PRα/LBD+ at relapse (40% and 33%, respectively) or distribution of PRα/LBD+ in relapses On-ATRA/Off-ATRA selection pressure (11/7 and 2/4, respectively; P = .36). We cannot exclude the possibility, however, that alternative ATRA-CT treatment, which remains standard of care in much of the world,32 might be more effective in PRα/LBD mutant clone reduction.
A rationale for the involvement of PRα/LBD+ in relapse clone selection is that PRα/LBD+-harboring APL cells are resistant to ATRA-induced differentiation, resulting in their passive selection because of the elimination of ATRA-sensitive APL cells containing wild-type PML-RARα.28 Consistent with this notion, the majority of C9710 cases harboring PRα/LBD+ (61%) relapsed On-ATRA. Nevertheless, the fraction of PRα/LBD+–positive patients who relapsed long after the cessation of ATRA therapy (39%) is notable. Further, the relapse PRα/LBD mutation detected in a low-level subclone at diagnosis in 2 cases in this study and 1 case in a previous study21 only emerged late in proximity to relapse. These observations suggest that PRα/LBD+ may have an active, ATRA-independent role, as well as an ATRA-dependent passive role, in relapse clone selection. A potential mechanism by which this might occur is augmentation of the fundamental self-renewal–promoting activity of PML-RARα involved in APL initiation and propagation.33,34 Consistent with this possibility, APL cells expressing mutant PML-RARα/LBD produced more aggressive leukemias than those expressing wild-type PML-RARα after transplantation to secondary recipients in a transgenic mouse model.35 Further investigations are needed to assess this speculative gain-of-function activity. Such investigations might further consider whether this activity is related to the site of mutation in the LBD because our study suggests that relapse clones harboring mutations in the amino-proximal portion of the LBD are less tightly linked to ATRA selection pressure than mutations in the central binding cleft.
Unlike PRα/LBD+, FLT3-ITD+ were already prevalent at diagnosis, did not increase at relapse, and were evenly distributed at relapse On- or Off-ATRA. Further, in patients with paired quantitative analyses of FLT3-ITD+ at diagnosis and relapse, there was no increase in the level of the mutant allele relative to total FLT3 alleles (ratio < 50%). Thus, there was no evidence that FLT3 mutations, considered as single lesions, are contributory to post-therapy disease progression in contrast to their established role in disease pathogenesis.1 However, when analyzed in the context of other study parameters, at least one potential contributory mechanism was revealed related to ATRA treatment status at relapse. All FLT3-ITD+ patients who relapsed Off-ATRA lacked PRα/LBD+, whereas most FLT3-ITD+ patients (83%) who relapsed On-ATRA had a coincident PRα/LBD+. The latter positive association suggests an ATRA-dependent complementary interaction between these mutations. A scenario by which this might occur is that ATRA-induced reduction of competition with the bulk APL cell population permits growth of the double-mutant subclone because of constitutive FLT3-ITD+ growth stimulatory activity, which would be further enhanced if PRα/LBD+ have gain-of-function activity. This mechanism does not necessarily require an increase in FLT3-ITD kinase activity, possibly a critical consideration, because FLT3-ITD mutant allele levels did not increase at relapse in contrast to non-APL AML.36 A clinical consideration is that patients who relapse with both mutations might selectively benefit from treatment with FLT3 kinase inhibitors.
Genetic change associated with chromosomal abnormalities has been implicated as an essential complementary factor in driving progression of PML-RARα–initiated disease to frank leukemia in mouse APL models.1,5,37,38 In humans, a similar involvement of ACA in disease development has been postulated but remains unsubstantiated,1,39 while clinical trial studies have found no independent association between ACA at diagnosis and treatment outcome (X.P. and W.S., manuscript submitted, June 2012).10,14 Our study also suggests that ACA present at diagnosis do not predispose to posttreatment progression, since there was no increase in the incidence of ACA at diagnosis in the relapse subset (26%) compared with overall adult C9710 patients (28%; X.P. and W.S., unpublished data, June 2012). The most frequent ACA in APL, trisomy 8/i (8q), which has the most compelling supportive evidence for a possible linkage to pretreatment disease progression,9,38 had the same incidence in both the overall and relapse cohorts (10%). A complex karyotype, which occurred in 9% of overall C9710 adults at diagnosis and was associated with reduced DFS (X.P. and W.S., unpublished data, June 2012), was only present at diagnosis in 1 relapse case (3%), probably because of chance exclusion from the study cohort. At relapse, exclusive ACA gains occurred, resulting in a 2-fold increased incidence of ACA (29% to 62%). Proportionate increases occurred in trisomy 8/i (8q) (10%-24%) and ider17q (5%-10%), the second most frequent ACA in APL.40 These increases seem consistent with hypotheses that gene dose effects, including increased oncogene activity,9,38 might contribute to posttreatment progression in these patients, but the proportionality argues against a posttreatment-specific effect. Balanced translocations also increased in incidence (8%-29%), which included a recurrent t(4;5) with a common 5q31 and neighboring 4q23 and 4q21 breaksites. Although 5q31 has rarely been reported in translocations in APL, rearrangements affecting 4q21, including complex insertion/rearrangements with PML-RARα, have been recurrently reported.5,41,42 More generally, several structural chromosome changes seem of potential functional interest: they were predominantly newly present at relapse; most differ from ACAs commonly reported at diagnosis43 and/or are contiguous to candidate leukemia/cancer genes (supplemental Table 13). Further study is required to determine whether any of the heterogeneous ACAs have clinically relevant functional significance or whether they represent adventitious abnormalities. If functionally relevant, the molecular pathways involved seem likely to be disparate, although linkage to a shared central network process(es) cannot be discounted.9,38
Despite their heterogeneity, ACAs were associated with an APL phenotype characterized by pWBClow and L-isoform, if relapse occurred Off-ATRA. This phenotype was shared by patients who relapsed Off-ATRA with a PRα/LBD+, all whom had coincident ACA at relapse. Further, there was a propensity for PRα/LBD+ to emerge in patients who already had ACAs at diagnosis. This suggests that the association may reflect inherently increased genetic instability in this APL phenotype. As such, the ACA and/or PRα/LBD+ might be coincidental to the relapse process. However, such genetic instability might also generate aberrations that actively participate in the process, which seems more consistent with the otherwise unexplained frequency and clonal penetration of PRα/LBD+ in relapse clones that emerge off of ATRA selection pressure. In this scenario, the augmented self-renewal activity of mutant PML-RARα, as discussed above, might, for example, complement increased oncogene-stimulated clonal growth in a patient with trisomy 8. Another common feature of the shared ACA-PRα/LBD+ phenotype is the negative association with FLT3-ITD+, which is strongly associated with the opposing APL phenotype characterized by pWBChigh and S-isoform. This could indicate that FLT3-ITD+ are associated with an alternative mechanism of posttreatment progression after relapse Off-ATRA, although this study provides no evidence of a possible mechanism. Notably, the above dichotomized associations are not apparent after relapse On-ATRA (Table 7), suggesting that ATRA-specific resistance mechanisms are involved and short-circuit distinct phenotype-linked pathways of disease progression that are manifest after relapse Off-ATRA. We acknowledge that the proposed interactions are speculative and might have alternative explanations, including involvement of an occult mutation(s) unrelated to phenotypic characteristics, as recently reported in a murine APL model,39 or involvement of other unrecognized abnormalities that could affect the timing of relapse independent of ATRA treatment.
Finally, we additionally assessed the relationship of the genetic aberrations to clinical outcome irrespective of potential functional involvement. This was assessed by the incidence of postrelapse death in all 45 relapse patients and by postrelapse survival in 35 patients with sufficient follow-up to provide an indication of response to salvage therapy. No case-specific information is available about salvage therapy in C9710 patients after first relapse, but in all likelihood the great majority received ATO because this is standard of care.44 Of the 4 genetic aberrations, only the presence of ACA at relapse was associated with reduced postrelapse outcome. However, particular ACA either showed no apparent association or because of small numbers and heterogeneity could not be evaluated for an association with postrelapse survival. A subset of patients with PRα/LBD+ had reduced postrelapse survival: among 6 nonsurvivors with PRα/LBD+, 3 had missense mutations of G289 and 2 had deletion mutations. In a previous study, both patients with a deletion in the PRα/LBD were also nonsurvivors.45 In addition, 4 pediatric/juvenile cases with mutations affecting Y208 or R217 in amino-proximal LBD may have had a more aggressive or less treatment-sensitive form of APL because 3 had the shortest TTR and the fourth was a nonsurvivor. There was no indication that FLT3-ITD+ or FLT3/D835+ at relapse had an adverse impact on postrelapse outcome, and 4 of 5 patients evaluable for outcome who relapsed On-ATRA with dual FLT3-ITD and PRα/LBD mutations were long-term survivors. From these findings, we suggest that the collection of further genetic aberration data from first-relapse APL patients would probably identify minor patient subsets that may benefit from nonstandard postrelapse therapy and that such data may be particularly informative in patients who are not responding well to salvage therapy.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Public Health Service Grants CA56771, CA31946, CA33601, CA21115, CA23318, CA32102, and CA38926.
National Institutes of Health
Authorship
Contribution: R.E.G. conceived and designed the research, analyzed and interpreted data, and wrote the manuscript; B.K.M. collected and analyzed data and performed statistical analysis; J.R. and X.P. performed research and analyzed and interpreted data; C.D.B. analyzed and interpreted data and critically reviewed the paper; A.J.C., R.P.K., and D.R. performed research and critically reviewed the paper; E.S.-T. and D.-c.Z. performed research and collected data; I-M.L.C. collected vital materials and data; R.H., G.K., and D.A.S. performed research and collected vital materials and data; J.H.F., M.S.T., and R.A.L. supervised patient data collection and critically reviewed the paper; B.L.P. supervised patient data collection, as principal investigator of protocol C9710, and critically reviewed the paper; F.R.A. participated in conception of the research, supervised patient data collection, and critically reviewed the paper; E.P. supervised research performance, analyzed and interpreted data, and critically reviewed the paper; C.L.W. supervised collection of vital research materials and critically reviewed the paper; and W.S. conceived and designed the research, supervised research performance, analyzed and interpreted data, and critically reviewed the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert E. Gallagher, Department of Oncology, Montefiore Medical Center, Albert Einstein Cancer Center, 111 East 210th St, Bronx, NY 10467; e-mail: robert.gallagher@einstein.yu.edu.