Abstract
The polycomb repressive complex 2 (PRC2) is a highly conserved histone H3 lysine 27 methyltransferase that regulates the expression of developmental genes. Inactivating mutations of the catalytic component of PRC2, EZH2, are seen in myeloid disorders. We reasoned that the other 2 core PRC2 components, SUZ12 and EED, may also be mutational targets in these diseases, as well as associated factors such as JARID2. SUZ12 mutations were identified in 1 of 2 patients with myelodysplastic syndrome/myeloproliferative neoplasms with 17q acquired uniparental disomy and in 2 of 2 myelofibrosis cases with focal 17q11 deletions. All 3 were missense mutations affecting the highly conserved VEFS domain. Analysis of a further 146 myelodysplastic syndrome/myeloproliferative neoplasm patients revealed an additional VEFS domain mutant, yielding a total mutation frequency of 1.4% (2 of 148). We did not find mutations of JARID2 or EED in association with acquired uniparental disomy for chromosome 6p or 11q, respectively; however, screening unselected cases identified missense mutations in EED (1 of 148; 1%) and JARID2 (3 of 148; 2%). All 3 SUZ12 mutations tested and the EED mutation reduced PRC2 histone methyltransferase activity in vitro, demonstrating that PRC2 function may be compromised in myeloid disorders by mutation of distinct genes.
Introduction
Single nucleotide polymorphism (SNP) arrays have revealed regions of acquired uniparental disomy (aUPD) as recurrent events in hematologic malignancies.1 Some of these regions are associated with the acquisition of somatic mutations in specific genes, but the presumptive targets of many stretches of aUPD remain to be identified.2-5 We and others recently identified EZH2 as a target for the 7q aUPD in myeloid malignancies, specifically myelodysplastic syndrome (MDS), myeloproliferative neoplasm (MPN), and overlapping MDS/MPN.5,6 EZH2 mutations in this heterogeneous group of diseases are associated with a poor prognosis and appear to be early events in the disease process, at least in some cases.5,7-9
EZH2 is a key component of the polycomb repressive complex 2 (PRC2), which regulates the expression pattern of developmental genes in both hemopoietic and nonhemopoietic systems. PRC2 is a histone methyltransferase (HMT) that trimethylates histone H3 lysine 27, resulting in a mark (H3K27me3) that specifies transcriptional repression. PRC2 consists of 3 core subunits, of which SUZ12 and EED are required for complete function and stability of the complex, whereas EZH2 is the catalytic component (reviewed in Bracken and Helin10 ). Other proteins, such RBBP4/7 and Jarid2, are generally considered to be cofactors that help to recruit and modulate the activity of PRC2.11-14
PRC2 is known to play important, and sometimes apparently contradictory, roles in both stem-cell renewal and cancer. EZH2 is overexpressed in several epithelial and hematologic malignancies, and its overexpression is associated with an adverse prognosis in prostate and breast cancer (reviewed in Sparmann and van Lohuizen15 ). Furthermore, overexpression of EZH2 has been linked to genomic loss of miRNA-101 in prostate cancer.16 Missense mutations at EZH2 tyrosine 641 have been found in B-cell lymphomas of germinal center origin that synergize with wild-type (WT) EZH2 to result in increased levels of H3K27me3.17-19 In contrast, the EZH2 mutations seen in myeloid disorders are inactivating, a finding that was unexpected but is consistent with models suggesting that a critical balance of polycomb activity is essential for normal stem cell activity, with either loss or gain of polycomb function being potentially tumorigenic.11
Given the finding of inactivating EZH2 mutations in myeloid neoplasms, we hypothesized that other PRC2 components might also be mutational targets in these disorders. We therefore focused on an analysis of the core PRC2 components SUZ12 and EED plus the cofactors RBBP4 and JARID2.
Methods
Subjects
Peripheral blood or BM samples were obtained from patients diagnosed with a hematologic malignancy according to standard morphological, hematologic, and laboratory criteria. Our primary study group consisted of a previously described cohort of MDS/MPN cases (n = 148),5 of which 67 had chronic myelomonocytic leukemia (CMML), 57 had atypical chronic myeloid leukemia, and 24 had MDS/MPN-unclassified. We also analyzed 2 patients (UPN23 and UPN30) from a previously described cohort of MPN cases (n = 151).20 The study was approved by the internal review boards of all participating institutions and informed consent was provided according to the Declaration of Helsinki.
Genomic analysis
Analysis of 148 MDS/MPN with Affymetrix SNP 6.0 arrays and 151 MPN cases with Affymetrix 250K Nsp or Sty arrays has been reported previously, along with the identification and definitions of regions of aUPD.5,20 Patient UPN23, with JAK2 V671F− postessential thrombocythemia myelofibrosis, and patient UPN30, with JAK2 V671F+ postpolycythemia vera myelofibrosis, were described as having deletions at 17q11.2 that included NF1.20 Deletions of NF1 in the 2 MDS/MPD cases with 17q aUPD were investigated by multiplex ligation probe amplification using the P081 and P082 kits according to the manufacturer's instructions (MRC Holland).
Mutational screening
Mutation analysis of MDS/MPN patients was performed by direct sequencing or high-resolution melting (HRM) analysis on genomic DNA extracted from total peripheral blood or BM leukocytes after RBC lysis. Analysis was performed initially on DNA that had been amplified using an Illustra GenomiPhi Version 2 DNA Amplification kit (GE Healthcare); all variants were confirmed by amplification and resequencing of the relevant region from unamplified genomic DNA. Standard Sanger sequencing was performed using an ABI 3130 genetic analyzer (Applied Biosystems) and Mutation Surveyor DNA Variant Analysis Version 3.30 software (SoftGenetics). HRM was performed as described previously21 using a Rotor-Gene 6000 (QIAGEN) and primers as detailed in supplemental Tables 1 through 6 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). SUZ12 exons 1-9 were screened using direct sequencing because these exons have high homology to the pseudogene, SUZ12P; the remaining exons were screened using HRM. HRM PCR conditions were as follows: 95°C for 30 seconds, 45 cycles of 95°C for 30 seconds, annealing temperature for 30 seconds, 72°C for 30 seconds, and 1 cycle at 72°C for 10 minutes, followed by HRM. Products showing abnormal melt patterns were sequenced directly. Details about which exons were analyzed and whether direct sequencing or HRM was performed are given in supplemental Tables 1 through 6. The numbering system for SUZ12, EED, and JARID2 mutations refer to Ensembl transcripts ENST00000322652, ENST00000263360, and ENST00000341776, respectively. All mutations were checked to determine whether they were rare SNPs in the Ensembl 1000 genome Release 8 database (http://www.1000genomes.org/), and all mutations were run through SIFT Version 2 (http://sift.jcvi.org/) and PolyPhen Version 2.1.0 (http://genetics.bwh.harvard.edu/pph2/) protein-prediction software algorithms to determine whether variant residues were likely to affect protein function.
Baculovirus expression and HMT assay
Hemagglutinin-tagged murine Eed (isoform II) and murine Suz12 cloned into pFastBac were generously provided by S. Orkin (Howard Hughes Medical Institute, Boston, MA). Flag-tagged murine Ezh2 cloned into pFastBac was generously provided by D. Reinberg (Howard Hughes Medical Institute, New York, NNY).22,23 The QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) was used to create murine equivalents of the human SUZ12 F603L, D605V, E610G, and EED G255D mutants. Baculoviruses were generated using the Bac-to-Bac Expression system (Invitrogen). Sf9 cells were cultured in Sf-900 II serum-free medium (Invitrogen). Sf9 insect cells were coinfected with Ezh2, Eed, and Suz12 baculovirus. To compare the activity of PRC2 with WT and mutant Suz12 or Eed, and to control for different titers, separate cultures were set up with different amounts of baculovirus for WT and mutant cells. For each WT/mutant combination, the most appropriate condition for comparison was chosen based on the levels of protein in either the Suz12 or Eed (for the Suz12 mutant and Eed mutant, respectively) Western blot. Baculovirus-infected cells were harvested 70 hours after infection, and proteins were purified using anti-hemagglutinin agarose for the Suz12 mutants or anti–FLAG-agarose for the EED mutants (Sigma-Aldrich). The presence of Ezh2, Eed, and Suz12 was confirmed by Coomassie blue staining and Western blotting. Histone methylation activity was assessed as described previously.5 Methylated histones were detected by gel autoradiography using an enlightening autoradiography enhancer (PerkinElmer). All experiments were performed twice using 2 independent clones.
Abs
Abs against Suz12 (sc-46264) and Eed (sc-28701) were purchased from Santa Cruz Biotechnology. Abs against Ezh2 (3147) were purchased from Cell Signaling Technology. Protein blots were performed using standard procedures.
Results
SNP array analysis
We focused our initial analysis on the core PRC2 components EED and SUZ12, and specifically on cases with genomic abnormalities involving chromosome bands 11q14.2 or 17q11.2, where these 2 genes are located. In our previously reported SNP array analysis, we found that 2 of 148 MDS/MPN patients had 17q aUPD (Figure 1A), 3 of 148 MDS/MPN patients had 11q aUPD but were WT for CBL exon 8 or 9, and 2 of 151 MPN patients had 17q11.2 deletions encompassing NF1.5,20
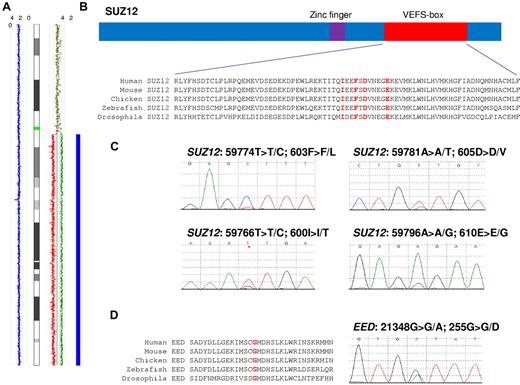
SUZ12 and EED mutations are highly conserved. (A) Chromosome 17 copy number analyzer for GeneChip (CNAG) output for case E7168 indicating homozygous SNP calls (blue rectangle) with a copy number of 2 (blue line) and allele-specific copy number calls (red and green lines). (B) Conservation of residues in the VEFS-box domain. Residues in red correspond to the mutations found. (C) Electropherograms showing the 4 SUZ12 mutations. (D) EED conservation and mutation in patient E1880.
SUZ12 and EED mutations are highly conserved. (A) Chromosome 17 copy number analyzer for GeneChip (CNAG) output for case E7168 indicating homozygous SNP calls (blue rectangle) with a copy number of 2 (blue line) and allele-specific copy number calls (red and green lines). (B) Conservation of residues in the VEFS-box domain. Residues in red correspond to the mutations found. (C) Electropherograms showing the 4 SUZ12 mutations. (D) EED conservation and mutation in patient E1880.
17q abnormalities are associated with SUZ12 mutations
Direct sequencing was used to screen all SUZ12 exons in the 4 patients with 17q abnormalities. Three missense mutations were identified in 3 patients, all in exon 15, which encodes part of the highly conserved VEFS-box domain (Figure 1B). The clinical and genomic details of mutated patients are summarized in Table 1. Mutations were found in 2 patients, one with aUPD and one with a 17q deletion: patient E7168 had a T > C change at nucleotide 59774 that predicts a F603L substitution and patient UPN30 had an A > T change at nucleotide 59781 that predicts a D605V substitution (Figure 1C). The mutations in these 2 patients were predominant on the sequencing traces, with the residual WT alleles only weakly visible. This is consistent with the array results, which suggested that the majority of cells were homozygous (E7168) or hemizygous (UPN30) in this region.
Subjects analyzed, mutations, and clinical details
| Patient . | Diagnosis . | Mutation . | Genomic abnormality . | Sex . | Age, y . | WBCs, ×109/L . | Platelets, ×109/L . | Karyotype . | Disease progression . | Death, mo . |
|---|---|---|---|---|---|---|---|---|---|---|
| E7168 | CMML | SUZ12 F603L | aUPD 17q | F | 80 | 49 | 87 | 46,XX | 55 | |
| UPN 30 | Post-PV MF | SUZ12 D605V | del(17q11.2) | M | 64 | 44 | 164 | Not known | PV at age 57; underwent SCT shortly after progression to MF but relapsed with AML | 6 |
| UPN 23 | Post-ET MF | SUZ12 I600T NF1 528ΔT | del(17q11.2) | M | 54 | 6 | 450 | 46,XY | ET at age 50; SCT at age 60 after evidence of increasing blasts (from 10% to 19%) | |
| E7169 | CMML | SUZ12 E610G | None | F | 84 | 50 | 23 | 46,XX | AML (43 mo) | 43 |
| E1880 | aCML | EED G255D | None | M | 69 | 28 | 46 | 46,XY | 9 |
| Patient . | Diagnosis . | Mutation . | Genomic abnormality . | Sex . | Age, y . | WBCs, ×109/L . | Platelets, ×109/L . | Karyotype . | Disease progression . | Death, mo . |
|---|---|---|---|---|---|---|---|---|---|---|
| E7168 | CMML | SUZ12 F603L | aUPD 17q | F | 80 | 49 | 87 | 46,XX | 55 | |
| UPN 30 | Post-PV MF | SUZ12 D605V | del(17q11.2) | M | 64 | 44 | 164 | Not known | PV at age 57; underwent SCT shortly after progression to MF but relapsed with AML | 6 |
| UPN 23 | Post-ET MF | SUZ12 I600T NF1 528ΔT | del(17q11.2) | M | 54 | 6 | 450 | 46,XY | ET at age 50; SCT at age 60 after evidence of increasing blasts (from 10% to 19%) | |
| E7169 | CMML | SUZ12 E610G | None | F | 84 | 50 | 23 | 46,XX | AML (43 mo) | 43 |
| E1880 | aCML | EED G255D | None | M | 69 | 28 | 46 | 46,XY | 9 |
Post-PV MF indicates post-polycythemia myelofibrosis; post-ET MF, post-essential thrombocythemia myelofibrosis; aCML, atypical chronic myeloid leukemia; and SCT, stem cell transplantation.
A relatively low-level mutation (< 50%) was found in patient UPN23: a T > C change at nucleotide position 59766, predicting an I600T substitution (Figure 1C). This patient had a 17q11.2 deletion that at first sight was inconsistent with the finding of a low-level mutation. However, deletions encompassing NF1 are known to fall into 2 principal classes: type 1 deletions result from recombination between low copy repeats at 17q11 and result in the complete loss of SUZ12, NF1, and several other genes. Type 2 NF1 deletions are smaller and result from recombination between a SUZ12 pseudogene (SUZ12P) and the true SUZ12 gene (supplemental Figure 1).24 Inspection of the SNP array data revealed that patient UPN23 had a type 2 NF1 deletion with break points in SUZ12P and intron 4 of SUZ12, thus leaving the 3′ end of SUZ12 (including exon 15) intact. Constitutional DNA was available for patient UPN23 only, and sequencing confirmed that the I600T mutation was somatically acquired (supplemental Figure 3).
NF1 mutations
Patient UPN30 was previously shown to harbor a truncating NF1 mutation in the nondeleted allele, whereas for patient UPN23, the nondeleted allele appeared to be intact.20 We analyzed NF1 in the 2 patients with 17q aUPD by direct sequencing of all coding exons and multiplex ligation probe amplification to look for deletions, but no abnormalities were found.
Prevalence of SUZ12 mutations
To determine the prevalence of SUZ12 mutations, we used direct sequencing and HRM to cover all SUZ12 exons in the remaining SNP 6.0 cohort of MDS/MPN patients (n = 146). One additional patient, E7169, was found to have an A > G change at nucleotide 59796, predicting an E610G substitution (Figure 1C). Therefore, in total, 2 of 148 (1.4%) MDS/MPN patients had SUZ12 mutations. In addition, the VEFS-box domain (exons 14-16) were screened in 42 patients with acute myeloid leukemia [normal karyotype, n = 21; inv(16), n = 9; t(8;21), n = 8; and t(15;17), n = 4], but no mutations were found.
An EED mutation not associated with 11q aUPD
None of the 3 MDS/MPN patients with 11q aUPD (CBL WT) had EED mutations. We then went on to screen a random subset of the remaining MDS/MPN patients (n = 148, including 5 cases with 11q aUPD and CBL mutations) and identified a single mutated patient, E1880, who had a heterozygous G > A change at nucleotide 21348, predicting a G255D substitution at a highly conserved residue (Figure 1D).
JARID2 and other mutations
Other studies have reported aUPD at chromosome 6p in MDS4,25 and acute myeloid leukemia.2 Although we did not identify 6p aUPD in our MDS/MPN patients using our previous definition of runs of homozygosity > 20 Mb,5 2 patients had runs of homozygosity > 10 Mb, which included JARID2. Screening of these cases for JARID2 mutations did not reveal any abnormality; however, analysis of the remaining MDS/MPN patients (n = 146) revealed 3 heterozygous variants (R717Q, D744N, and I1052V) in 3 patients. We also screened EZH1 in a subset of the MDS/MPN patients (n = 94, including 15 with EZH2 mutations), focusing on exons 16-21 because these encode the catalytic SET domain, but no sequence changes were found. Similarly, no mutations were found in RBBP4 (located at 1p35.1) in the 148 MDS/MPN patients, including 6 with chromosome 1 aUPD. All SUZ12 and EED variants were predicted to be functionally important; however, the mutated JARID2 residues were not in conserved residues and/or were not clearly predicted to affect function (supplemental Figure 2 and supplemental Table 7).
Reduced HMT activity
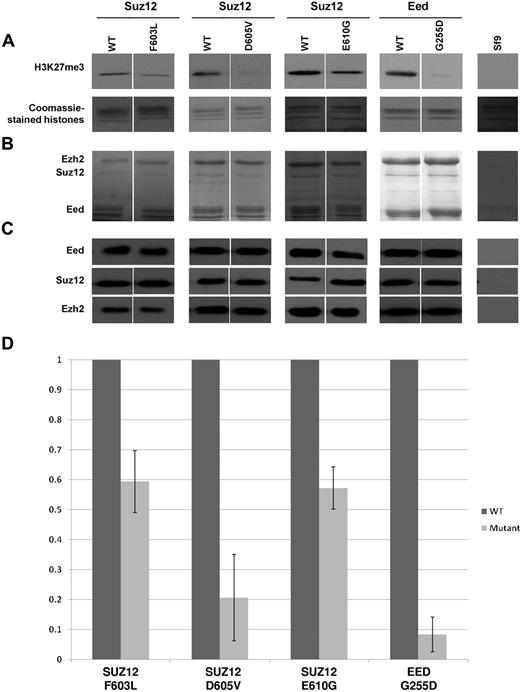
We also investigated whether the SUZ12 and EED mutants affected the HMT activity of the PRC2 complex by infecting Sf9 insect cells with baculovirus expressing the PRC2 components Ezh2, Suz12, and Eed with and without the mutations. After immunoprecipitation, the complexes were assayed for H3K27 methyltransferase activity. All mutants tested (SUZ12 F603L, D605V, E610G, and Eed G255D) showed reduced HMT activity compared with the WT (Figure 2), with the reduction being particularly marked for Suz12 D605V and Eed G255D. These data demonstrate that all 4 mutants we tested compromise PRC2 function.
SUZ12 and EED mutations abrogateHMTactivity. (A) In vitro HMT assay of baculovirus-expressed components demonstrating loss of activity with SUZ12 and EED mutant complexes (top panels) compared with WT controls. The SUZ12 mutants F603L, D605V, and E610G and the EED mutant G255D were identified in this study. Sf9 indicates uninfected insect cells. The bottom panel (Coomassie-stained histones) confirms the presence of the histone substrates in each reaction. (B) Coomassie-stained SDS-PAGE of immunoprecipitated PRC2 components Ezh2, Suz12, and Eed confirming the presence of each of these components in the assay. (C) Protein blots of immunoprecipitated PRC complexes with Abs indicated further confirming the presence of the 3 PRC2 components. All results were obtained in 2 independent clones, however, only clone 1 is shown here. (D) Normalized HMT assay results. The intensity of the H3K27me3 bands was normalized against the protein blot bands (Suz12 bands for Suz12 mutants and Eed bands for the EED mutant) and are shown as a percentage of WT activity. The error bars (SDs from mean values) are derived from 2 separate experiments with 2 independently derived clones.
SUZ12 and EED mutations abrogateHMTactivity. (A) In vitro HMT assay of baculovirus-expressed components demonstrating loss of activity with SUZ12 and EED mutant complexes (top panels) compared with WT controls. The SUZ12 mutants F603L, D605V, and E610G and the EED mutant G255D were identified in this study. Sf9 indicates uninfected insect cells. The bottom panel (Coomassie-stained histones) confirms the presence of the histone substrates in each reaction. (B) Coomassie-stained SDS-PAGE of immunoprecipitated PRC2 components Ezh2, Suz12, and Eed confirming the presence of each of these components in the assay. (C) Protein blots of immunoprecipitated PRC complexes with Abs indicated further confirming the presence of the 3 PRC2 components. All results were obtained in 2 independent clones, however, only clone 1 is shown here. (D) Normalized HMT assay results. The intensity of the H3K27me3 bands was normalized against the protein blot bands (Suz12 bands for Suz12 mutants and Eed bands for the EED mutant) and are shown as a percentage of WT activity. The error bars (SDs from mean values) are derived from 2 separate experiments with 2 independently derived clones.
Discussion
After our recent finding of inactivating EZH2 mutations in approximately 12% of MDS/MPN patients,5 we hypothesized that other polycomb components might also be mutational targets in these disorders. We screened SUZ12 and EED, the other 2 core components of the PRC2 complex, and found that approximately 2%-3% of MDS/MPN patients had inactivating mutations in one of these genes. The finding of EZH2 mutations in myeloid disorders was the first indication that a loss of polycomb function may be an important factor in human leukemia. However, in addition to its role as the catalytic component of PRC2, EZH2 also performs relatively ill-defined functions in the cytoplasm.26 The finding of inactivating mutations in other PRC2 components is important because it provides further weight to the argument that the loss of H2K27 methyltransferase activity is the critical consequence of these changes.
SUZ12 is located at 17q11.2, close to NF1, a gene that encodes a negative regulator of RAS signaling. Several lines of evidence have strongly implicated NF1 in the pathogenesis of myeloid neoplasms. First, deregulated RAS signaling is a recurrent theme in these disorders as a consequence of mutations that directly activate NRAS or KRAS or activate upstream tyrosine kinases (eg, point mutations or gene fusions involving KIT, FLT3, JAK2, ABL1, and others) or inactivate negative regulators of signaling (eg, CBL).27 Second, inherited inactivating mutations of NF1 are associated with neurofibromatosis, but also predispose to the development of juvenile myelomonocytic leukemia.28 Third, although not common, deletions of 17q11.2 are recurrent in myeloid neoplasms and in some cases have been associated with inactivating NF1 mutations in the nondeleted allele.29,30 Finally, somatic inactivation of Nf1 in mice results in a progressive myeloproliferative disorder.31 Despite these considerations, a systematic analysis of NF1 in myeloid neoplasms has not yet been reported, principally because of the very large size of the gene (58 exons).
In the present study, we identified a total of 4 patients with SUZ12 mutations, 1 CMML with 17q aUPD, 2 MF with focal 17q11.2 deletions, and another CMML with no 17q genomic abnormality. Another MDS/MPN patient had 17q aUPD but no SUZ12 mutation. Neither of the patients with 17q aUPD had NF1 mutations, but one of the patients with del(17q11.2) had mutations of both NF1 and SUZ12. Both of these mutations were clearly inactivating: a frameshift in exon 5 of NF1 and a D605V change in SUZ12 that greatly reduced PRC2 HMT activity (Figure 2). These findings raise the possibility that SUZ12 and not NF1 is the principal target of 17q11.2 abnormalities, although in some cases both genes may be involved. Neurofibromatosis patients with deletions have a higher chance of developing a malignancy and have a worse prognosis than those with point mutations alone.29 This could be a direct effect of SUZ12 haploinsufficiency or it could be the consequence of the deletions effectively predisposing to loss of SUZ12 function along the lines of the classic tumor-suppressor model. Evidence for a suppressive effect of SUZ12 has been provided by previous experimental work. Knockdown of SUZ12 in the G1ME cell line resulted in reduced protein levels of both SUZ12 and EZH2, with the cell line showing concurrent reduction in H2K27 methylation similar to that seen in EZH2 knockdown experiments.30 In addition, in a murine model, loss of Suz12 function by both mutation and shRNA knockdown enhanced hemopoietic stem and progenitor-cell activity.32 However, SUZ12 is overexpressed in a wide variety of tumor types15 and is directly implicated in endometrial stromal cancers by fusion to JAZF1 in more than 50% of patients.33 Like EZH2, SUZ12 therefore appears to have credentials as an oncogene or as a tumor suppressor, depending on the cellular context.
SUZ12 is highly conserved from plants to humans, but in the present study, strikingly, all 4 mutations were seen in exon 15, which encodes part of the VEFS box (Figure 1). This domain is necessary for interaction between SUZ12 and EZH2,34 and is thought to help to stabilize the PRC2 complex. Although we did not find any EED mutations associated with 11q aUPD, we did find 1 patient (1 of 148; 1%) with a heterozygous mutation that almost completely abrogated HMT function in the insect cell assay. The mutation, G255D, is within the fourth WD40 repeat and is adjacent to a dominant-negative M256L change characterized in Drosophila as a residue important for the interaction between EED and EZH2.35 EED is also highly conserved in evolution and was shown to be mutated in 2 of 30 cases of skin cancer.36 Loss of EED in mice caused marked myelo- and lymphoproliferative defects, and indicated that EED is involved in the negative regulation of these pools.37 Taken together, these studies support the hypothesis that the mutations we found in SUZ12 and EED are indeed causative and suggest that they may be acting by interfering with PRC2 complex formation. Although EZH2 mutations are emerging as a poor prognostic factor,5,8,9 the number of SUZ12/EED mutated cases in this study was too small to draw any conclusions with regards to prognosis or clinical phenotype.
The role of JARID2 is less clear. JARID2 has been shown to promote the HMT activity of the PRC2 and may also have a role in facilitating access of the complex to target genes.12-14 Although we found 3 sequence variants, 2 of which mapped to known functional domains (supplemental Figure 2), none of them were clearly predicted to be inactivating and their significance is uncertain.
In summary, we have shown that inactivation of the PRC2 can occur through mutation of any of the 3 core components, and that inactivating mutations of EZH2, SUZ12 or EED are seen in 15% of cases of MDS/MPN. These findings emphasize the importance of PRC2 function in hemopoiesis and suggest that deregulation of the epigenetic control of gene expression plays an important role in the development of myeloid disorders.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by Leukaemia Research (United Kingdom) Specialist Programme Grant 0280. N.W. was supported by grant no. 109590 from the Deutsche Krebshilfe e.V.
Authorship
Contribution: J.S., A.C., and N.C.P.C. designed the study; J.S., C.H.-C., A.V.J., N.W., A.S., D.W., T.E., A.C., and N.C.P.C. performed the laboratory and data analyses; K.Z., F.S., and K.D. provided clinical samples and data; J.S. and N.C.P.C. wrote the draft of the manuscript; and all authors contributed to the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Professor N. C. P. Cross, Wessex Regional Genetics Laboratory, Salisbury District Hospital, Salisbury SP2 8BJ, United Kingdom; e-mail: ncpc@soton.ac.uk.