An unexpected revelation of cancer genome studies has been frequent abnormality in genes for factors that modify chromatin, underscored in this issue of Blood by reports from Score et al and Kroeze et al of inactivating mutations and chromosome loss in SUZ12, EED and JARID2 in myelodysplastic syndrome (MDS) and myeloproliferative disease (MPD).1,2
After discovering inactivating EZH2 mutations in myeloid malignancies,3-5 these investigators have taken the logical next step of searching for abnormalities in other components of polycomb repressor complex 2 (PRC2; EZH2, SUZ12, EED, JARID2). While infrequent, mutations in SUZ12, EED, and JARID2 demonstrate the pathogenic importance of PRC2. Inactivating mutations in ASXL1, another polycomb-related gene, are a more common characteristic of the myeloid malignancies. These are recent observations, and the pathways by which decreased PRC2 or ASXL1 function increase clonal fitness remain poorly understood. Bedside-to-bench translation from MDS/MPD may help solve this mystery; important clinical clues include the association of these mutations with transformation of MDS/MPD into fibrotic phases and into acute myeloid leukemia (AML), and allocation of mutations to particular morphologic subtypes of disease: ASXL1 mutations are strongly associated with chronic myelomonocytic leukemia (CMML); EZH2 mutations are more evenly distributed, but are often associated with increased platelet counts.6
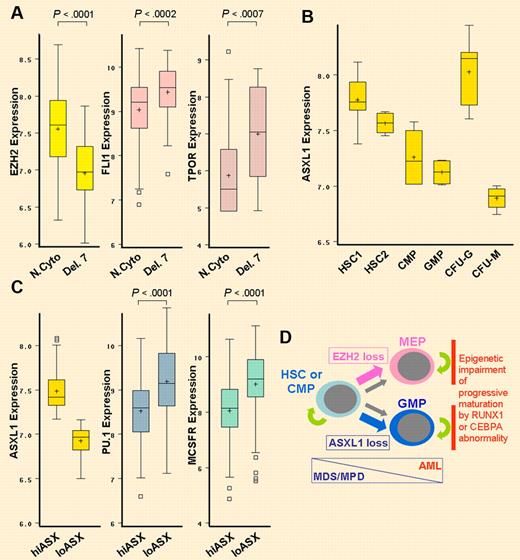
PcG proteins represses key lineage-commitment genes. Accordingly, loss of EZH2 associated with deletion of chromosome 7 is accompanied by significant up-regulation of the key megakaryocyte differentiation driver FLI1, and decreased expression of ASXL1 is accompanied by significant up-regulation of the monocyte differentiation-driver PU.1. (A) Decreased EZH2 expression produced by chromosome 7 loss (Del. 7) is associated with significant up-regulation of FLI1 and thrombopoietin receptor (TPOR). N.Cyto, n = 106; Del. 7, n = 18. Gene expression data extracted from GSE6891. Wilcoxon test. Box plot boundaries = interquartile range; horizontal line = median; + = mean; whiskers = range of values; small boxes = out-lying values. (B) In a normal hematopoietic hierarchy, ASXL1 expression is lowest in colony forming unit monocyte (CFU-M) cells compared with CFU-granulocyte (CFU-G), granulocyte monocyte progenitors (GMP), common myeloid progenitors (CMP), CD38-/CD34+ hematopoietic stem cells (HSC2) and CD133+/CD34dim HSC (HSC1). Gene expression data extracted from GSE24759. (C) In AML with normal cytogenetics, lower ASXL1 expression (ASXlo, n = 53) is significantly associated with higher expression of PU.1 and macrophage colony stimulating factor receptor (MCSFR; higher ASXL1 expression = ASXhi, n = 53). Wilcoxon test. (D) Evolution of MDS/MPD into AML is associated with lineage-restriction of self-renewing leukemia initiating cells,8-10 a differentiation-transition that is favored by EZH2 or ASXL1 inactivation. Mutations in key hematopoietic transcription factor genes such as RUNX1 or CEBPA impair progressive maturation of lineage-committed cells, a critical element in transformation into AML. MEP indicates megakaryocyte-erythroid progenitors.
PcG proteins represses key lineage-commitment genes. Accordingly, loss of EZH2 associated with deletion of chromosome 7 is accompanied by significant up-regulation of the key megakaryocyte differentiation driver FLI1, and decreased expression of ASXL1 is accompanied by significant up-regulation of the monocyte differentiation-driver PU.1. (A) Decreased EZH2 expression produced by chromosome 7 loss (Del. 7) is associated with significant up-regulation of FLI1 and thrombopoietin receptor (TPOR). N.Cyto, n = 106; Del. 7, n = 18. Gene expression data extracted from GSE6891. Wilcoxon test. Box plot boundaries = interquartile range; horizontal line = median; + = mean; whiskers = range of values; small boxes = out-lying values. (B) In a normal hematopoietic hierarchy, ASXL1 expression is lowest in colony forming unit monocyte (CFU-M) cells compared with CFU-granulocyte (CFU-G), granulocyte monocyte progenitors (GMP), common myeloid progenitors (CMP), CD38-/CD34+ hematopoietic stem cells (HSC2) and CD133+/CD34dim HSC (HSC1). Gene expression data extracted from GSE24759. (C) In AML with normal cytogenetics, lower ASXL1 expression (ASXlo, n = 53) is significantly associated with higher expression of PU.1 and macrophage colony stimulating factor receptor (MCSFR; higher ASXL1 expression = ASXhi, n = 53). Wilcoxon test. (D) Evolution of MDS/MPD into AML is associated with lineage-restriction of self-renewing leukemia initiating cells,8-10 a differentiation-transition that is favored by EZH2 or ASXL1 inactivation. Mutations in key hematopoietic transcription factor genes such as RUNX1 or CEBPA impair progressive maturation of lineage-committed cells, a critical element in transformation into AML. MEP indicates megakaryocyte-erythroid progenitors.
Polycomb group (PcG) proteins were originally identified in fruit flies by their critical role in regulating segmentation, by modifying chromatin to repress transcription of hox genes. PcG proteins operate in multiprotein complexes, one of which is PRC2; the PRC2 defining histone methylation mark is tri-methylation of lysine 27 on histone 3 (H3K27me3), a repression mark. PRC2 and H3K27me3 are associated with facultative heterochromatin—chromatin that is plastic in development and differentiation. Master regulators of lineage specification are repressed by PRC2 and H3K27me3 in embryonic stem cells, and lineage commitment and differentiation are associated with removal of these marks.
It would seem, then, that loss of PRC2 function in MDS/MPD should favor differentiation; however, PRC2/ASXL1 mutations are strongly associated with transformation of MDS/MPD into AML,7 a process defined by differentiation block. Perhaps there is no contradiction: MPD is a disease driven by abnormal self-renewal in stem cells or multipotent progenitors,8 whereas AML is driven by abnormal self-renewal in lineage-restricted progenitors.8-10 Thus, evolution of MDS/MPD into blast crisis requires some differentiation, just not too much.8-10 Consistent with PRC2 mutations allowing some differentiation, decreased expression of EZH2 caused by deletion of chromosome 7 is accompanied by significant up-regulation of the transcription factor FLI1, an essential driver of megakaryocyte differentiation, and decreased expression of ASXL1 in AML is associated with significant up-regulation of the monocyte differentiation-driver PU.1 (see figure). Hence, up-regulation of specific cell fate–determining transcription factors by EZH2 or ASXL1 loss explains the associations with high platelet counts and CMML, respectively. Notably, there is a striking increase in ASXL1 inactivation with transformation of polycythemia vera/essential thrombocytosis into myelofibrosis, suggesting a potential role for activation of a monocytic program in this process.
What are the therapeutic implications? For transformation into AML, PRC2/ASXL1 loss that encourages lineage restriction must collude with events that impair progressive maturation. Accordingly, in AML, PRC2/ASXL1 mutations are significantly associated with concomitant mutation in RUNX1 or CEBPA6 ; RUNX1/CEBPA mutations permit lineage commitment and the MYC up-regulation that accompanies this, but impair progressive maturation by epigenetic repression of key late-differentiation genes (eg, CEBPE).9,11,12 Hence, PRC2 dysfunction, because of EZH2 mutations or del(7/7q) and, as shown by the contributions by Score et al and Kroeze et al, also SUZ12, EED, or JARID2 mutations or loss, may encourage cells into the maturation blind alley created by events such as RUNX1 or CEBPA mutation (see figure). This strategy for neoplastic evolution could create a therapeutic vulnerability, because the combination of high expression of lineage-specifying transcription factor, more advanced maturation stage, and the weakening of the epigenetic barrier to activation of late-differentiation genes by removal of H3K27me3 marks, may facilitate the action of drugs such as 5-azacytidine or decitabine, which seek to reactivate genes by depleting DNA-methyltransferase 1 (DNMT1), a central member of the network of chromatin-modifying enzymes that mediate transcription repression. Consistent with this premise, ASXL1 mutation predicts responses to 5-azacytidine or decitabine.13 In short, chromatin-relaxing therapy can potentially exploit the malignant cell context created by PRC2/ASXL1 loss to induce cell cycle exit by p53-independent differentiation pathways, to offer an alternative to frequently ineffective apoptosis-based therapy9,14 (see figure).
Clinical associations of the now extended spectrum of PcG mutations provide valuable bedside-to-bench clues regarding pathogenic pathways. In bench-to-bedside reciprocation, the pathway insights can provide mechanistic guidance and patient selection criteria for 5-azacytidine/decitabine therapy, and in-centive for developing new chromatin-relaxing drugs to treat the myeloid malignancies.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■