Abstract
Hedgehog (HH) signaling is activated in various lymphoid malignancies, but conflicting results exist about its role in chronic lymphocytic leukemia (CLL). Here, we demonstrate that the expression of essential HH pathway components like GLI1, PTCH1, and the HH ligands is highly diverse in CLL. A subset of 36.7% of 60 tested CLL samples responded to all 3 SMOOTHENED (SMO) inhibitors, whereas 40% were completely resistant. Responsiveness correlated with elevated GLI1 and PTCH1 transcript levels and the presence of trisomy 12, whereas no other karyotype correlated with responsiveness. All trisomy 12 CLLs displayed constitutive HH pathway activation driven by autocrine DESERT HH (DHH) ligand secretion, which could be blocked by the HH-blocking Ab 5E1. Cocultures with DHH-expressing BM stromal cells reduced sensitivity of CLLs to SMO-inhibitor treatment by activation of noncanonical ERK phosphorylation directly downstream of the PTCH1 receptor without involvement of SMO and could be overcome by the HH-blocking Ab 5E1 or a combination of SMO and ERK inhibitors. Our results demonstrate that the HH-signaling pathway is an interesting therapeutic target for a subset of patients with CLL, characterized by high GLI1 and PTCH1 transcript levels, and all patients with trisomy 12 and indicate HH-blocking Abs to be favorable over SMO inhibitors in overcoming stroma-mediated protective effects.
Introduction
B-cell chronic lymphocytic leukemia (CLL) is the most prevalent adult leukemia in Western countries and is characterized by a progressive accumulation of functionally incompetent B-lymphocytes in the peripheral blood, BM, and lymphoid organs.1,2 Several prognostic markers such as Rai and Binet staging systems,3,4 immunoglobulin VH gene mutational status,5 ζ-associated protein 70 (ZAP70) expression6,7 and cytogenetic abnormalities8 such as trisomy 12 and del 13q14 can be used to predict the survival outcome and the need for treatment of patients with CLL.9 Despite encouraging new therapeutic strategies10 CLL cannot be cured by conventional therapy at present, and only hematopoietic stem cell transplantation has led to complete and ongoing remissions in some patients.11 The exact pathogenesis of this clinically heterogenous leukemia is still unknown,12 but it is well established that B-CLL cells are habitually interacting with stromal cells, indicating the importance of the microenvironment for survival of CLL cells.13-15
In our previous work, we demonstrated that the hedgehog (HH) ligands sonic (Shh), indian (Ihh), and desert (Dhh) are produced by stromal cells in lymphoid organs and that the BM promote expansion of murine lymphomas in vitro and in vivo.16
HH ligands activate the transmembrane receptor PATCHED (PTCH), which abrogates the suppression of the 7-transmembrane-helix protein SMOOTHENED (SMO), the key player for signal transduction of the canonical HH pathway.17 SMO activation leads to production of activating forms of the glioma-associated oncoproteins 1-3 (GLI1-3), the HH transcription factors. With exception of GLI1, which functions exclusively as a transcriptional activator, the HH transcription factors exist as full-length transcriptional activators or as truncated transcriptional repressors.18 The activation process of GLI leads to expression of HH target genes such as PTCH1, GLI1, BCL2, BMI1, and Cyclin D1 and hence to increased proliferation, survival, and self-renewal.19-21 Sustained canonical HH pathway activation has been associated with cancers of the brain,22,23 the skin,24-26 the gastrointestinal tract,27 the prostate,28,29 the pancreas,30,31 and the lung.32 Noncanonical HH signaling was described in mammary gland cells, inducing direct ERK activation downstream of the PTCH1 receptor without involvement of the SMO receptor.33
Diverse roles of HH signaling are described in hematologic malignancies. Although HH signaling mediated by SMO is required for maintenance of the leukemic stem cell population in chronic myeloid leukemia,34,35 it is dispensable for the development of acute leukemias induced by the MLL-AF936 fusion gene or T-cell acute lymphoblastic leukemia induced by an activated form of NOTCH.37 In our previous work, we could show that HH pathway inhibition by SMO inhibitors could block the expansion of murine lymphomas in vitro and in vivo.16 In addition, human lymphoid malignancies, such as diffuse large B-cell lymphoma,38 NPM-ALK–driven anaplastic large cell lymphoma,39 mantle cell lymphoma,40 and multiple myeloma16,41 show activation of HH signaling and responsiveness to SMO inhibitors in vitro. Conflicting results exist about its role in CLL. Although Hedge et al argue for activation of HH signaling on the level of SMO in all CLL cases,42 Desch et al reported dependence of CLL cells on the GLI1 transcription factor, but not on the SMO receptor.43
Therefore, the aim of the present study was to identify biomarkers and clinical parameters, which can discriminate in between SMO-inhibitor responsive and resistant CLLs and to identify mechanisms of HH pathway activation in CLL.
Methods
CLL patient samples
This study was approved by the Institutional Review Board of the University Medical Center Freiburg. Peripheral blood samples were obtained with informed consent in accordance with the Declaration of Helsinki from patients with B-CLL who were either untreated or off therapy for ≥ 6 months (supplemental Table 2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). CLL cases were characterized for IgVH mutational status, ZAP70 expression, disease stage according to Binet and Rai criteria, and history of treatment. Furthermore, genetic aberrations were analyzed by chromosomal analysis and FISH analysis and copy number changes were verified by single nucleotide polymorphism arrays44 (patient characteristics are shown in supplemental Table 2). PBMCs were separated by Ficoll gradient centrifugation and either used fresh or cryopreserved in FCS with 10% DMSO until use. Cells were maintained in DMEM/10% FCS and 1% penicillin-streptomycin. PBMCs for quantitative PCR (qPCR) contained > 88% CD20+/CD5+ CLL cells.
Apoptosis and viability assays
PBMCs were plated into 96-well plates at a concentration of 1 × 105 cells/well with or without support of the murine stromal cell line M2-10B4 (ATCC) or the human stromal cell line HS-5 (ATCC). In case of stromal coculture, on day −1, 5 × 103 stromal cells/well were plated. After 24 hours, CLL cells were added and treated with 3 different SMO inhibitors cyclopamine45,46 (Calbiochem), NVP-LDE22547 (Novartis), or IPI-92648,49 (Infinity Pharmaceuticals) at indicated concentrations. For apoptosis detection, CLL cells with or without stromal coculture were plated as indicated earlier and treated with SMO inhibitors, 10-40 μg/mL HH-blocking Ab 5E150 (Developmental Studies Hybridoma Bank), 1μM ERK inhibitor UO126 (Sigma-Aldrich) or combinations. After 24, 48, and 72 hours of incubation at 37°C in 5% CO2, cells were stained with a CD19-allophycocyanin Ab (BD Biosciences), followed by annexin V/7-amino-actinomycin D (7-AAD) staining with anti–annexin V–PE and 7-AAD (BD Biosciences) according to the manufacturer's instructions. Cells were analyzed with the CyanADP flow cytometer (Beckman Coulter). Flow cytometric data were analyzed with the FlowJo 7.6 software (TreeStar). To verify the annexin V/7-AAD assay results, we performed a second assay with the first 60 patients after incubation with compounds for 48 and 72 hours and assessed vital cell numbers with the use of Guava ViaCount technology. The Guava ViaCount assay is a fluorescence-based assay, which is performed in a 96-well format and therefore allows the use of the compounds in a broad range of concentrations. Besides distinguishing among necrotic, apoptotic, and viable cells (1 membrane-permanent dye stains all nucleated cells, and a membrane-impermanent dye stains only damaged cells with breaches in the plasma membrane), the results also include total cells per volume, which allows one to calculate total numbers of viable or apoptotic or necrotic cells per well (Millipore; Guava Technologies; Guava ViaCount Assay). Measurements were performed with the Guava Technologies PCA-96 system.
Intracellular phospho-ERK staining
CLL cells or stromally cocultured CLL cells were plated as in “Apoptosis and viability assays” and treated with 5μM cyclopamine, 1μM UO126, or a combination of both. HH-blocking Ab 5E1 was added to stromal cells before adding CLL cells, to block stroma-produced ligands, before they can bind to the PTCH1 receptor of the CLL cells. After 4 hours of incubation, cells were stained with CD19 Ab, fixed with 1.85% formalin, and permeabilized with 90% methanol. Endogenous levels of p44 and p42 MAP kinase were detected with the phospho-p44/42 MAPK mouse mAb (no. 9106; Cell Signaling) and anti–mouse IgG Alexa Fluor-conjugated Ab (no. 4408; Cell Signaling). Cells and data were analyzed as described in “Apoptosis and viability assays.”
Immunohistochemistry staining
Paraffin-embedded BM slides were acquired from the pathology department of the University of Freiburg. Diaminobenzidine immunoperoxidase staining (EnVision+System-HRP/DAB; Dako) on paraffin sections was performed for 10 minutes. Primary Abs to SHH/IHH (IgG rabbit polyclonal; N-19; Santa Cruz Biotechnology) 1:50, DHH (IgG goat polyclonal; H-85; Santa Cruz Biotechnology) 1:250, GLI1 (N-16; Santa Cruz Biotechnology) 1:100, and SMO 1:50 (H-300; Santa Cruz Biotechnology) were incubated overnight, followed by a 30-minute incubation time with secondary Abs (EnVision+System-HRP [DAB]; Dako). Immunodetection was performed with 3,3′ diaminobencidin (EnVision+System-HRP [DAB]; Dako). Photographs were taken with Axioplan 2 (Zeiss). Staining intensity was scored in categories (grade 0 = < 10%; 1 = 10%-20%; 2 = 20%-50%, and 3 = > 50% positive cells; supplemental Table 3).
qPCR
Total cellular RNA was isolated from PBMCs from patients with CLL containing > 88% of CD20+CD5+ CLL cells (single patient information; supplemental Table 4), reverse transcribed, and amplified for 50 cycles at an annealing temperature of 60°C. PCR primers and probes for human GLI1, PTCH1, SMO, and GAPDH and mouse Gli1, Ptch1, Dhh, and Gapdh were obtained from Applied Biosystems. qPCR from these genes was assessed by TaqMan PCR (summary of primers and probes in supplemental Table 1). Results were quantified according to the δ-δ-CT method that was based on the relative expression of a target gene versus a reference gene (GAPDH) and normalized to the median of all samples.
Statistical analysis
Data are represented as the mean ± SEM. Comparisons between parameters were performed with a 2-tailed, paired Student t test. For all analyses, P < .05 was considered statistically significant. Correlations were assessed with the Spearman rank correlation coefficient. The Cochran-Armitage test for trend is a modified χ2 test, which was used for calculation of the association of different genetic subgroups or risk factors with response to SMO-inhibitor treatment.
Results
Level of HH pathway activation is highly diverse in CLL
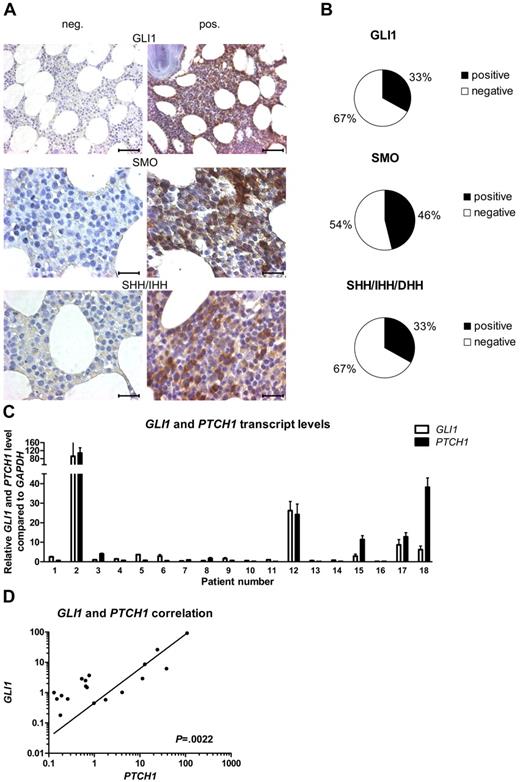
To evaluate the presence of the HH-signaling pathway in CLL, we performed IHC stainings for the downstream target GLI1, the receptor SMO and the HH ligands SHH, IHH, and DHH, in BM samples of 24 patients with CLL. Expression levels were scored from grade 0 to grade 3 (grade 0 = 0%-10%; 1 = 10%-20%; 2 = 20%-50%, and 3 = > 50% positive cells). IHC stainings for the HH-downstream target gene GLI1 showed expression (grades 1-3) of this protein in 33% of the analyzed patient samples (8 of 24), whereas in the rest of the samples the GLI1 transcription factor was completely absent (Figure 1A-B; supplemental Table 3). The receptor SMO, which plays a central role in signal transduction of the ligand-mediated canonical HH signal, was present in 46% (11 of 24) of the tested CLL samples (Figure 1A-B; supplemental Table 3). At least 1 of the 3 HH ligands SHH, IHH, and DHH was present in 33% of the patient samples (SHH/IHH 4 of 24; DHH 6 of 24), which might indicate an autoregulatory activation loop in this CLL subset (Figure 1A-B; supplemental Table 3). There is a direct positive correlation between GLI1 expression and SMO expression (n = 24; P = .0204, r = 0.4801, Spearman correlation coefficient) and between GLI1 expression and ligand expression (GLI1 vs SHH/IHH, P = .0111, r = 0.5194; GLI1 vs DHH, P = .0020, r = .6103). To validate the activation status of the HH-signaling pathway, we performed qPCR for the direct HH target genes GLI1 and PTCH1 on isolated white blood cells (WBCs) from patients with CLL (n = 18; supplemental Table 4). In confirming the IHC results, we found a great heterogeneity for the expression levels of the HH-target genes GLI1 and PTCH1 within different CLL patient samples but a direct positive correlation between the transcript levels of the 2 target genes (n = 18; P = .0022, r = 0.67321; Figure 1C-D). In individual patient samples the GLI1 and PTCH1 transcript levels were elevated > 100-fold compared with the median of all samples. This indicates a highly diverse activation level of HH signaling in CLL, with a subset of ∼ 30% of CLL samples showing high transcript levels (> 3-fold than the median) for GLI1 and PTCH1.
The activation status of HH signaling is highly diverse in CLL. (A) IHC stainings from CLL BM samples for GLI1, SMO, and the HH ligands SHH/IHH. Negative (left) and positive (right) examples for expression of GLI1, SMO, and the HH ligands in CLL. Original magnification ×40 for GLI1 (scale bar, 50 μm) and ×100 for others (scale bar, 25 μm). Microscope: Axioplan 2 (Zeiss); GLI1 40×/0.95 NA; SMO + SHH/IHH 100×/1.25 NA oil objective, room temperature; microscope software: AxioVision LE. (B) Percentage of samples positive or negative for IHC staining for GLI1, SMO, and the HH ligands SHH/IHH and DHH in 24 CLL cases (single patient information, including scoring; supplemental Table 3). (C) qPCR for GLI1 and PTCH1 in peripheral blood samples from patients with CLL. Transcript levels are shown relative to GAPDH and compared with the median of all samples (patient single data; supplemental Table 4). (D) Correlation in between GLI1 and PTCH1 transcript levels in same patients with CLL as in panel C shows a positive correlation between both genes (n = 18; P = .0022, Spearman correlation coefficient, r = 0.67321).
The activation status of HH signaling is highly diverse in CLL. (A) IHC stainings from CLL BM samples for GLI1, SMO, and the HH ligands SHH/IHH. Negative (left) and positive (right) examples for expression of GLI1, SMO, and the HH ligands in CLL. Original magnification ×40 for GLI1 (scale bar, 50 μm) and ×100 for others (scale bar, 25 μm). Microscope: Axioplan 2 (Zeiss); GLI1 40×/0.95 NA; SMO + SHH/IHH 100×/1.25 NA oil objective, room temperature; microscope software: AxioVision LE. (B) Percentage of samples positive or negative for IHC staining for GLI1, SMO, and the HH ligands SHH/IHH and DHH in 24 CLL cases (single patient information, including scoring; supplemental Table 3). (C) qPCR for GLI1 and PTCH1 in peripheral blood samples from patients with CLL. Transcript levels are shown relative to GAPDH and compared with the median of all samples (patient single data; supplemental Table 4). (D) Correlation in between GLI1 and PTCH1 transcript levels in same patients with CLL as in panel C shows a positive correlation between both genes (n = 18; P = .0022, Spearman correlation coefficient, r = 0.67321).
SMO-inhibitor treatment of primary human CLL samples results in 3 different response groups
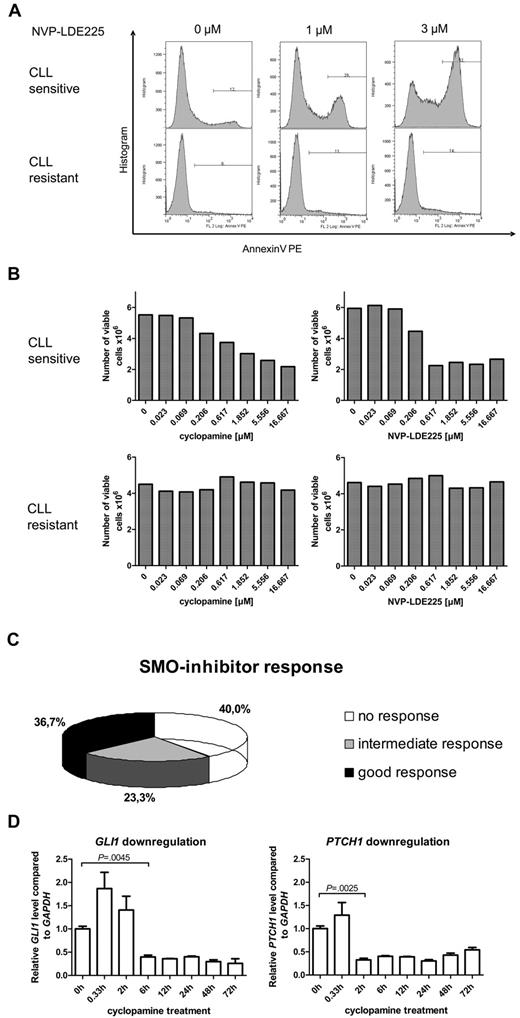
To further investigate the role of the HH-signaling pathway in CLL, we examined the effect of 3 different SMO inhibitors (cyclopamine, NVP-LDE225, IPI-926) on apoptosis induction (annexin V/7-AAD staining) and viable cell counts in 60 different CLL patient samples. Responses to SMO-inhibitor treatment were strongly diverse, ranging from dose-dependent apoptosis induction by SMO inhibitors at already low inhibitor concentrations to complete apoptosis resistance to SMO inhibition (Figure 2A, 1 example for responsive [P1] and 1 for nonresponsive [P9] patient). Those results could be reconfirmed by analysis of viable cell counts after 24 and 48 hours of treatment with the use of the Guava ViaCount Assay (Figure 2B, 1 responsive [P1] and 1 nonresponsive [P9] patient). IC50 values for NVP-LDE225 and IPI-926 were generally lower than for cyclopamine. Responsiveness to SMO-inhibitor treatment was defined as “good” if the CLL sample responded to all 3 SMO inhibitors and with IC50s for cyclopamine < 5μM and for NVP-LDE225 and IPI-926 < 2.5μM. The intermediate response group was represented by patients who responded to only 1 or 2 of the 3 used SMO inhibitors, and nonresponders showed no response to any SMO inhibitor or only responded at very high inhibitor concentrations. Of the CLL patient samples 36.7% were classified in the good response group, 23.3% in the intermediate response group, and 40% of the samples showed no response to SMO-inhibitor treatment (Figure 2C). SMO inhibition by cyclopamine and NVP-LDE225 in responsive samples resulted in a time-dependent and significant down-regulation of the HH target genes GLI1 and PTCH1, indicating that the SMO inhibitors target the HH-signaling pathway in the CLL cells (Figure 2D; supplemental Figure 1). Although treatment with cyclopamine significantly suppressed GLI1 and PTCH1 transcript levels for > 72 hours, NVP-LDE225 suppressed GLI1 transcript levels for only 24 hours and PTCH1 transcript levels for only 12 hours, indicating that this compound might be less stable under in vitro culture conditions than cyclopamine.
SMO inhibition in primary human CLL samples (n = 60) resulted in 3 different response groups. (A) Apoptosis assay showing annexin V–positive cells after treatment of primary CLL cells with the SMO inhibitor NVP-LDE225 (0μM, 1μM, and 3μM) for 24 hours. Top panel shows one representative example for a SMO inhibitor–sensitive CLL (patient 2), bottom panel an example for a SMO inhibitor–resistant CLL (patient 9). (B) Viable cell counts from the same 2 patients with the use of Guava ViaCount after 24 hours of treatment with the SMO inhibitors cyclopamine and NVP-LDE225 in increasing concentrations. Left side shows CLL cells treated with cyclopamine; right side CLL cells treated with NVP-LDE225. Top panel shows a SMO inhibitor–sensitive example (patient 2), bottom panel shows a SMO inhibitor–resistant CLL (patient 9). (C) Diagram displays the distribution of 60 CLL patient samples (supplemental Table 2; patients 1-60) among the 3 different response groups to SMO inhibitor treatment. Response was determined as good if the CLL sample responded to all 3 SMO inhibitors and with IC50s for cyclopamine < 5μM and for NVP-LDE225 and IPI-926 < 2.5μM. The intermediate response group was represented by patients who responded to only 1 or 2 of the 3 used SMO inhibitors, and nonresponders showed no response to any SMO inhibitor or only responded at very high concentrations. (D) Treatment of CLL cells with 5μM cyclopamine and extraction of RNA after defined treatment periods. qPCR for GLI1 and PTCH1 in comparison to GAPDH and relative to the time point before treatment were performed in triplicates and show a significant reduction of GLI1 transcript levels after 6 hours (n = 3; P = .0045, Student t test) and of PTCH1 transcript levels after 2 hours (n = 3; P = .0041, Student t test). Target gene reduction stays significant until 72 hours.
SMO inhibition in primary human CLL samples (n = 60) resulted in 3 different response groups. (A) Apoptosis assay showing annexin V–positive cells after treatment of primary CLL cells with the SMO inhibitor NVP-LDE225 (0μM, 1μM, and 3μM) for 24 hours. Top panel shows one representative example for a SMO inhibitor–sensitive CLL (patient 2), bottom panel an example for a SMO inhibitor–resistant CLL (patient 9). (B) Viable cell counts from the same 2 patients with the use of Guava ViaCount after 24 hours of treatment with the SMO inhibitors cyclopamine and NVP-LDE225 in increasing concentrations. Left side shows CLL cells treated with cyclopamine; right side CLL cells treated with NVP-LDE225. Top panel shows a SMO inhibitor–sensitive example (patient 2), bottom panel shows a SMO inhibitor–resistant CLL (patient 9). (C) Diagram displays the distribution of 60 CLL patient samples (supplemental Table 2; patients 1-60) among the 3 different response groups to SMO inhibitor treatment. Response was determined as good if the CLL sample responded to all 3 SMO inhibitors and with IC50s for cyclopamine < 5μM and for NVP-LDE225 and IPI-926 < 2.5μM. The intermediate response group was represented by patients who responded to only 1 or 2 of the 3 used SMO inhibitors, and nonresponders showed no response to any SMO inhibitor or only responded at very high concentrations. (D) Treatment of CLL cells with 5μM cyclopamine and extraction of RNA after defined treatment periods. qPCR for GLI1 and PTCH1 in comparison to GAPDH and relative to the time point before treatment were performed in triplicates and show a significant reduction of GLI1 transcript levels after 6 hours (n = 3; P = .0045, Student t test) and of PTCH1 transcript levels after 2 hours (n = 3; P = .0041, Student t test). Target gene reduction stays significant until 72 hours.
Elevated GLI1 and PTCH1 transcript levels, trisomy 12, and DHH expression are biomarkers for SMO-inhibitor responsiveness in CLL
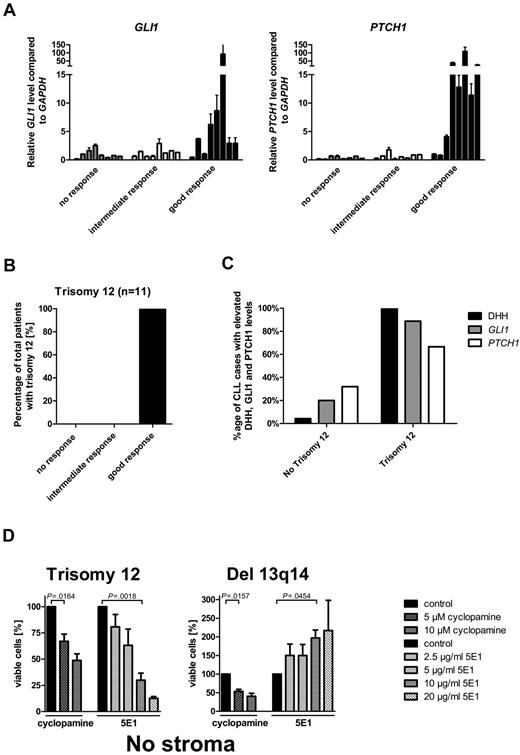
To identify biomarkers or clinical parameters, which can discriminate between SMO inhibitor-responsive and -resistant CLLs, we correlated transcript levels from HH pathway members and clinical characteristics with the 3 different response groups. Elevated transcript levels for the 2 HH target genes GLI1 and PTCH1 strongly correlated with the good response group (Figure 3A; supplemental Table 4). Increases > 3-fold over the median of all samples in either PTCH1 or GLI1 transcription was present in 87% of the CLL samples in the good response group, but only in 13% in the intermediate response group and in 0% in the nonresponder group, which indicates this cutoff as a good parameter to discriminate between good and intermediate/no response. Transcript levels for other pathway members and target genes such as SMO, STK36, BMI1, PTCH2, SHH, and IHH showed no association with any response group (data not shown). In addition, clinical parameters and risk factors, such as ZAP70 expression, mutational status, WBC counts, stage of disease, or previous therapy did not correlate with any response group (supplemental Figure 2; supplemental Table 5). The comparison of chromosomal aberrations in the first 60 patients with response groups identified no correlation between response and presence of del 13q14 (n = 29), no chromosomal aberrations (n = 16), del 11q (n = 7), or del 17p (n = 6; Table 1). But all patients with CLL with trisomy 12 (n = 4 within the first 60 patients), which is present in 12%-16% of all CLL cases, were represented in the good response group (Table 1). We therefore increased the number of patients with trisomy 12 to 11 and found the presence of trisomy 12 to be a highly predictive marker for good response to SMO inhibitors in CLL (Cochran Armitage trend test P = .0006; Figure 3B, Table 1). Interestingly, chromosome 12 contains the gene locus for GLI1 and DHH, which are both positive regulators for the HH-signaling pathway. According to the DHH IHC staining from 24 randomly taken CLL cases, only 2 contained > 20% DHH-positive CLL cells (grading 2 and 3), with the only trisomy 12–positive case to have > 50% DHH-positive cells. Therefore, we added 5 more cases for trisomy 12. All 5 additional cases with trisomy 12 showed DHH staining of grades 2 and 3, including 4 of 5 with > 50% positive cells (grade 3). Taken together, high DHH expression is nearly exclusively present in cases positive for trisomy 12 (Figure 3C; supplemental Figure 3). Furthermore, in most trisomy 12 patient samples the GLI1 transcript levels were elevated > 3-fold compared with the median of all samples (8 of 9; Figure 3C; supplemental Table 4), indicating activation of the HH-signaling pathway in those samples. PTCH1 transcript levels were elevated > 3-fold in 66.6% of the measured samples (6 of 9; Figure 3C; supplemental Table 3). A previous study from Desch et al had already shown an up-regulation of SMO transcripts in trisomy 12 cases compared with other genotypes.43 To further verify that the autocrine DHH expression and secretion might be the reason for HH-pathway activation in patients with trisomy 12 and might be important for the survival of those cells, we used the HH ligand–blocking Ab 5E1 to block the autocrine loop, that means the interaction between the DHH ligand and the PTCH1 receptor. All 4 patients with trisomy 12 treated with increasing amounts of the HH ligand–blocking Ab showed a dose-dependent apoptosis induction, which was even more effective than treatment with the SMO inhibitor (Figure 3D). In contrast, treatment of patients with del 13q14 (n = 3), which were highly responsive to SMO-inhibitor treatment, showed no response to the Ab treatment, but an adverse reaction with a dose-dependent increase in viable cells (Figure 3D). Those results strongly support the theory that patients with trisomy 12 in contrast to other genetic subgroups have an autocrine active HH-signaling pathway, which is essential for their survival.
Elevated GLI1 and PTCH1 transcript levels and trisomy 12 and DHH expression are biomarkers for SMO inhibitor responsiveness in CLL. (A) qPCR for GLI1 and PTCH1 in CLL samples of the different response groups. High GLI1 (n = 24; P = .0260, Cochran-Armitage trend test) or PTCH1 (n = 24; P = .0006, Cochran-Armitage trend test) transcript levels (> 3-fold than the median) compared with GAPDH and relative to the median of all samples are correlated with good response to SMO inhibitor treatment in CLL (single data information in supplemental Table 4, patients 2-11, 13-17, 23-25, 36, 37, 42, 46, 47, and 60). (B) Distribution of trisomy 12 between the different response groups. Presence of trisomy 12 (n = 11; P = .0006, Cochran-Armitage trend test) correlates with good response to SMO inhibitor treatment. (C) Comparison of DHH expression (> 20% positive CLL cells by IHC) or elevated relative PTCH1 and GLI1 transcript levels compared with GAPDH (> 3-fold than the median) between patients positive for trisomy 12 and other patients. Single data for IHC are included in supplemental Table 3, and single data for TaqMan results are included in supplemental Table 4. (D) Percentage of viable cells (annexin V/7-AAD staining) after treatment of primary CLL cells with the SMO inhibitor cyclopamine or the HH-blocking Ab 5E1 for 48 hours compared with control. Patients positive for trisomy 12 (n = 4) show a dose-dependent decrease in viable cells after treatment with SMO inhibitors or the 5E1 Ab, whereas samples positive for del 13q14 (n = 3) show only a decrease in viable cells after SMO inhibitor treatment, but an increase in viable cells after treatment with the 5E1 Ab (analyzed patients 1, 7, 18, 60, 66, 67, and 68).
Elevated GLI1 and PTCH1 transcript levels and trisomy 12 and DHH expression are biomarkers for SMO inhibitor responsiveness in CLL. (A) qPCR for GLI1 and PTCH1 in CLL samples of the different response groups. High GLI1 (n = 24; P = .0260, Cochran-Armitage trend test) or PTCH1 (n = 24; P = .0006, Cochran-Armitage trend test) transcript levels (> 3-fold than the median) compared with GAPDH and relative to the median of all samples are correlated with good response to SMO inhibitor treatment in CLL (single data information in supplemental Table 4, patients 2-11, 13-17, 23-25, 36, 37, 42, 46, 47, and 60). (B) Distribution of trisomy 12 between the different response groups. Presence of trisomy 12 (n = 11; P = .0006, Cochran-Armitage trend test) correlates with good response to SMO inhibitor treatment. (C) Comparison of DHH expression (> 20% positive CLL cells by IHC) or elevated relative PTCH1 and GLI1 transcript levels compared with GAPDH (> 3-fold than the median) between patients positive for trisomy 12 and other patients. Single data for IHC are included in supplemental Table 3, and single data for TaqMan results are included in supplemental Table 4. (D) Percentage of viable cells (annexin V/7-AAD staining) after treatment of primary CLL cells with the SMO inhibitor cyclopamine or the HH-blocking Ab 5E1 for 48 hours compared with control. Patients positive for trisomy 12 (n = 4) show a dose-dependent decrease in viable cells after treatment with SMO inhibitors or the 5E1 Ab, whereas samples positive for del 13q14 (n = 3) show only a decrease in viable cells after SMO inhibitor treatment, but an increase in viable cells after treatment with the 5E1 Ab (analyzed patients 1, 7, 18, 60, 66, 67, and 68).
Responsiveness to SMO inhibitor treatment in different CLL subgroups
| Responsiveness to SMO inhibitor treatment . | Good, % (n/N) . | Intermediate, % (n/N) . | Resistant, % (n/N) . |
|---|---|---|---|
| Del 13q14 | 37.9 (11/29) | 37.9 (11/29) | 24.1 (7/29) |
| Trisomy 12 | 100 (11/11) | 0 (0/11) | 0 (0/11) |
| Del 17p | 33 (2/6) | 16.6 (1/6) | 50 (3/6) |
| Del 11q | 28.5 (2/7) | 28.5 (2/7) | 42.8 (3/7) |
| Normal karyotype | 31.2 (5/16) | 12.5 (2/16) | 56.2 (9/16) |
| Responsiveness to SMO inhibitor treatment . | Good, % (n/N) . | Intermediate, % (n/N) . | Resistant, % (n/N) . |
|---|---|---|---|
| Del 13q14 | 37.9 (11/29) | 37.9 (11/29) | 24.1 (7/29) |
| Trisomy 12 | 100 (11/11) | 0 (0/11) | 0 (0/11) |
| Del 17p | 33 (2/6) | 16.6 (1/6) | 50 (3/6) |
| Del 11q | 28.5 (2/7) | 28.5 (2/7) | 42.8 (3/7) |
| Normal karyotype | 31.2 (5/16) | 12.5 (2/16) | 56.2 (9/16) |
Response rates to SMO inhibitor treatment in the different genetic subgroups in CLL are shown. Some patients have several chromosomal aberrations and are therefore represented in several subgroups (single patient information, including risk factors and response group, are shown in supplemental Table 2). Cochran-Armitage trend test for trisomy 12 was highly statistically significant (P = .0006; n = 11). No other genetic subgroup showed a statistically significant correlation with any response group.
Altogether, elevated GLI1 and PTCH1 transcript levels, presence of trisomy 12, and elevated expression of DHH are valuable parameters to discriminate between SMO inhibitor-responsive and -resistant samples and might be useful biomarkers for future clinical trials.
Stromal cells reduce SMO-inhibitor sensitivity of CLL cells
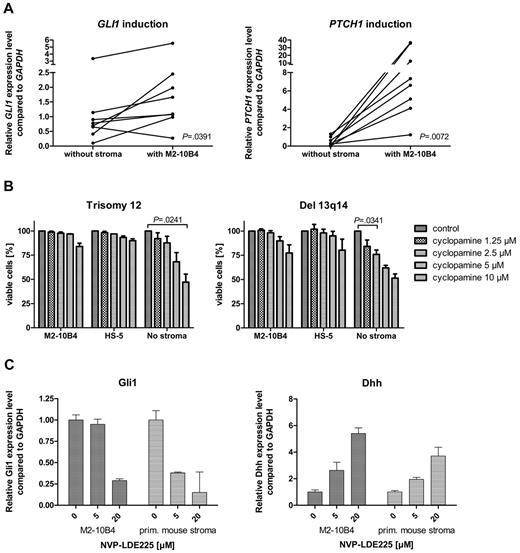
To investigate the effect of the microenvironment on the activation status of the HH-signaling pathway in CLL cells, we performed stroma-CLL coculture assays with the use of the murine stroma cell line M2-10B4. We examined the transcription of the HH target genes GLI1 and PTCH1 in CLL cells (n = 8) either cultivated alone or in coculture with the stromal cell line M2-10B4, which produces high amounts of DHH. PTCH1 was up-regulated in all (8 of 8) and GLI1 in 7 of 8 CLL cells in the presence of the stromal cells (Figure 4A). Mean GLI1 transcript levels were increased 1.9-fold compared with control, and mean PTCH1 transcript levels were increased > 30.7-fold, indicating induction of HH signaling in CLL cells by stromal cells. This activation of HH target genes was seen in trisomy 12 patient samples (n = 4) and in 13q14 patient samples (n = 4; Figure 4A), which indicates that the paracrine ligand stimulation is dominant over the autocrine ligand stimulation present in patients with trisomy 12. Comparison of transcript levels of Dhh in the murine stromal cell line M2-10B4 versus DHH in primary human trisomy 12 CLLs showed a > 20-fold increase of Dhh transcript levels in the stroma. In contrast to our expectations, the presence of murine (M2-10B4) or human (HS-5) stromal cells reduced the sensitivity of CLL cells to all 3 SMO inhibitors and in some cases even resulted in complete resistance to SMO inhibition (Figure 4B). This stroma-induced resistance was seen in both patients with trisomy 12 and patients with del 13q14. To identify the mechanism of apoptosis resistance induced by stromal cells, we examined the HH-signaling pathway in the stromal cell line M2-10B4 and in primary mouse stroma from Cdkn2a−/− mice. SMO inhibition with NVP-LDE225 for 24 hours resulted in a reduction of Gli1 in both M2-10B4 and primary mouse stromal cells and interestingly in a simultaneous up-regulation of the HH ligand Dhh in the stromal cells (Figure 4C). Those results indicate that SMO inhibition induces a negative autoregulatory loop with enhanced secretion of Dhh by stromal cells on SMO inhibition, which results in an at least continuous or even increased stimulation of the PTCH1 receptor in the CLL cells.
Stromal cells activate HH signaling in CLL cells and induce SMO inhibitor resistance. (A) GLI1 and PTCH1 transcript levels compared with GAPDH with (right) and without (left) stromal (M2-10B4) support in 8 CLL samples (trisomy 12, n = 4; 13q14, n = 4). Figure shows up-regulation of the HH target genes GLI (n = 8; P = .0391, Student t test) in 7 of 8 samples and PTCH1 (n = 8; P = .0072, Student t test) in 8 of 8 samples (analyzed patients 1, 7, 18, 19, 61, 62, 63, and 64). (B) Percentage of viable cells measured by annexin V/7-AAD staining after treatment with different concentrations of cyclopamine compared with the DMSO control. CLL cells were either cultivated on murine (M2-10B4) or human (HS-5) stroma or without stroma. Presence of stromal cells reduces sensitivity of CLL cells toward SMO inhibitors (trisomy 12, n = 3; responsive del 13q14, n = 3). (C) Inhibition of SMO in M2-10B4 cells and primary mouse stromal cells reduces transcript levels for Gli1 measured by qPCR and relative to Gapdh and results in a simultaneous up-regulation of Dhh.
Stromal cells activate HH signaling in CLL cells and induce SMO inhibitor resistance. (A) GLI1 and PTCH1 transcript levels compared with GAPDH with (right) and without (left) stromal (M2-10B4) support in 8 CLL samples (trisomy 12, n = 4; 13q14, n = 4). Figure shows up-regulation of the HH target genes GLI (n = 8; P = .0391, Student t test) in 7 of 8 samples and PTCH1 (n = 8; P = .0072, Student t test) in 8 of 8 samples (analyzed patients 1, 7, 18, 19, 61, 62, 63, and 64). (B) Percentage of viable cells measured by annexin V/7-AAD staining after treatment with different concentrations of cyclopamine compared with the DMSO control. CLL cells were either cultivated on murine (M2-10B4) or human (HS-5) stroma or without stroma. Presence of stromal cells reduces sensitivity of CLL cells toward SMO inhibitors (trisomy 12, n = 3; responsive del 13q14, n = 3). (C) Inhibition of SMO in M2-10B4 cells and primary mouse stromal cells reduces transcript levels for Gli1 measured by qPCR and relative to Gapdh and results in a simultaneous up-regulation of Dhh.
Noncanonical HH signaling in CLL cells is partially mediated by ERK phosphorylation
In previous reports, Chang et al described a non-canonical HH-signaling pathway in mammary gland cells with direct ERK activation by the PTCH1 receptor without involvement of SMO.33 To investigate whether ERK activation might be involved in noncanonical HH signaling in CLL cells, we investigated the effect of the presence of stromal cells on ERK phosphorylation in CLL cells with the use of intracellular p-ERK staining. Interestingly, baseline levels for ERK phosphorylation measured by intracellular p-ERK staining were significantly higher in patients with trisomy 12, which have an autocrine HH-signaling pathway, compared with the biggest other CLL cohort with del 13q14 (Figure 5A). Coculture of CLL cells with stromal cells or treatment with stroma cell supernatant fluid further induced ERK phosphorylation in both CLL subtypes (Figure 5B; supplemental Figure 4). Although SMO inhibitor treatment with cyclopamine could not significantly reduce stroma-induced ERK phosphorylation in CLL cells (Figure 5B), blocking HH ligand binding with the HH-blocking Ab 5E1 could completely abolish stroma-induced ERK phosphorylation (Figure 5C) and could reduce the phosphorylation status in patients with trisomy 12 even below the baseline levels. Stroma-induced ERK phosphorylation could also be reduced by the addition of the ERK inhibitor U0126 (Figure 5B), which verifies the specificity of our staining method for p-ERK. Figure 5D shows a diagram displaying the interaction of stromal cells with CLL cells mediated by HH ligands in the presence or absence of SMO inhibitors.
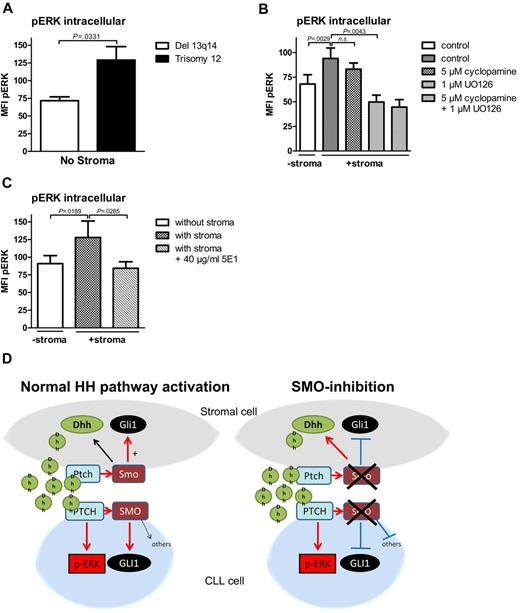
Noncanonical HH signaling in CLL cells is partially mediated by ERK phosphorylation. (A) Intracellular p-ERK staining and measurement by flow cytometry in 4 patients with trisomy 12 and 4 patients with del 13q14 in the absence of stromal cells. Comparison of pERK baseline levels show a statistically significant increase in trisomy 12 patient samples compared with del 13q14 (n = 8; P = .0331, Student t test; analyzed patients 1, 7, 44, 50, 60, 66, 68, 69). (B) Intracellular pERK staining shows induction of ERK phosphorylation in CLL cells on coculture with stromal cells measured by the mean fluorescence intensity (n = 7; P = .0029, Student t test). Although 4 hours of treatment with 5μM cyclopamine could not significantly reduce ERK phosphorylation (n = 7; P = .2173, Student t test), the ERK inhibitor U0126 could significantly reduce stroma-induced ERK phosphorylation (n = 7; P = .0043, Student t test; patients 3, 7, 44, 50, 60, 64, 65). (C) Treatment of stromally cocultured CLL cells with the HH-blocking Ab 5E1 could block stroma-induced ERK phosphorylation in CLL cells (n = 8; P = .0285, Student t test; patients 1, 7, 18, 50, 60, 66, 68, 69). (D) Diagram shows the interaction in between stromal cells and CLL cells mediated by HH ligands with and without SMO inhibitor treatment.
Noncanonical HH signaling in CLL cells is partially mediated by ERK phosphorylation. (A) Intracellular p-ERK staining and measurement by flow cytometry in 4 patients with trisomy 12 and 4 patients with del 13q14 in the absence of stromal cells. Comparison of pERK baseline levels show a statistically significant increase in trisomy 12 patient samples compared with del 13q14 (n = 8; P = .0331, Student t test; analyzed patients 1, 7, 44, 50, 60, 66, 68, 69). (B) Intracellular pERK staining shows induction of ERK phosphorylation in CLL cells on coculture with stromal cells measured by the mean fluorescence intensity (n = 7; P = .0029, Student t test). Although 4 hours of treatment with 5μM cyclopamine could not significantly reduce ERK phosphorylation (n = 7; P = .2173, Student t test), the ERK inhibitor U0126 could significantly reduce stroma-induced ERK phosphorylation (n = 7; P = .0043, Student t test; patients 3, 7, 44, 50, 60, 64, 65). (C) Treatment of stromally cocultured CLL cells with the HH-blocking Ab 5E1 could block stroma-induced ERK phosphorylation in CLL cells (n = 8; P = .0285, Student t test; patients 1, 7, 18, 50, 60, 66, 68, 69). (D) Diagram shows the interaction in between stromal cells and CLL cells mediated by HH ligands with and without SMO inhibitor treatment.
Stroma-induced SMO inhibitor resistance mediate by noncanonical HH signaling can be overcome by the HH-blocking Ab 5E1 or ERK inhibitors
To investigate whether noncanonical HH signaling mediated by high levels of stroma-secreted Dhh might be one of the reasons for stroma-induced SMO inhibitor resistance of otherwise SMO inhibitor–sensitive CLLs, we treated CLL cells with or without stromal support with the HH ligand–blocking Ab 5E1. The 5E1 Ab blocks the interaction of all HH ligands (Shh, Ihh, and Dhh) with the PTCH1 receptor. In contrast to SMO inhibitor treatment, the 5E1 Ab could completely overcome stroma-mediated resistance and induced apoptosis rates > 50% within 48 hours even in the presence of stromal cells (Figure 6A, 1 patient with trisomy 12). In addition, the protective effect induced by the human stromal cell line HS-5 could be completely overcome by the 5E1 Ab (Figure 6A, 1 patient with trisomy 12). Trisomy 12 patient samples were more sensitive to 5E1 treatment than were patients with del 13q14 in the presence of stromal cells (Figure 6B). Comparing the effect of the 5E1 Ab in patients with trisomy 12 with and without stroma support shows that we need ∼ 6-fold concentrations to reach the same effect (50% reduction of viable cells) in the presence of stromal cells, indicating that higher amounts of ligands have to be blocked in the presence of the stromal cells (Figure 3D; Figure 6B). In addition, treatment of CLL cells with stromal supernatant fluid significantly enhanced their survival, and this effect could again be blocked by the HH-blocking Ab 5E1, indicating soluble HH ligands to be responsible for the prosurvival effect of stromal supernatant fluid (supplemental Figure 5). In addition, a combination of SMO and ERK inhibitors could overcome stroma-mediated resistance, and both compounds showed additive effects, indicating that it is necessary to block both canonical and noncanonical HH signaling for apoptosis induction in the presence of stromal cells (Figure 6C). Figure 6D shows the effect of the HH ligand–binding Ab 5E1 and a combination of SMO and ERK inhibitors on noncanonical and canonical HH signaling in CLL cells and stromal cells.
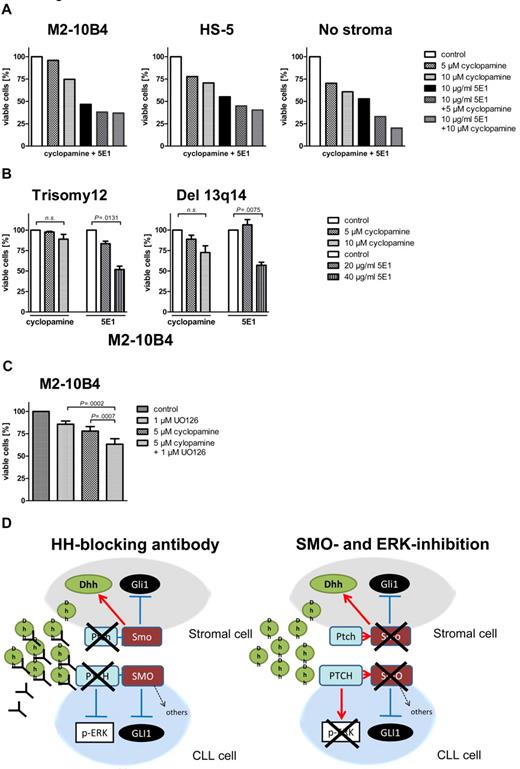
Stroma-induced SMO inhibitor resistance mediated by noncanonical HH signaling can be overcome by the HH-blocking Ab 5E1 or ERK inhibitors. (A) Percentage of viable CLL cells compared with the DMSO control after 48 hours of treatment with the SMO inhibitor cyclopamine or the HH ligand–blocking Ab 5E1 or a combination of cyclopamine and the 5E1 Ab. The HH-blocking Ab 5E1 overcomes SMO inhibitor resistance induced by stromal cells (patient 1). (B) Percentage of viable cells after treatment of primary CLL cells with the SMO inhibitor cyclopamine and the HH-blocking Ab for 48 hours compared with control. Although in both karyotypes SMO inhibitor treatment with 10μM cyclopamine cannot significantly reduce viable cells (trisomy 12: n = 4; P = .1539, Student t test; del 13q14: n = 3; P = .0799, Student t test), the HH-blocking Ab 5E1 can significantly reduce the percentage of viable cells (trisomy 12: n = 4; 20 μg/mL 5E1 P = .0131, Student t test; Del 13q14: 40 μg/mL; n = 3; P = .0075, Student t test; patients 1, 7, 18, 50, 60, 66, 68, 69). (C) Numbers of viable CLL cells compared with the DMSO control in cells treated with cyclopamine or the ERK inhibitor U0126 or a combination of both for 48 hours and measured by flow cytometry after annexin V/7-AAD staining. Combination of SMO and ERK inhibitors shows a significant increase in apoptosis compared with single treatment alone (n = 7; P = .0007, Student t test). (D) Diagram shows the interaction between stromal cells and CLL cells and changes in canonical and noncanonical HH signaling on treatment with the HH ligand–blocking Ab 5E1 (left) or a combination of SMO and ERK inhibitors (right).
Stroma-induced SMO inhibitor resistance mediated by noncanonical HH signaling can be overcome by the HH-blocking Ab 5E1 or ERK inhibitors. (A) Percentage of viable CLL cells compared with the DMSO control after 48 hours of treatment with the SMO inhibitor cyclopamine or the HH ligand–blocking Ab 5E1 or a combination of cyclopamine and the 5E1 Ab. The HH-blocking Ab 5E1 overcomes SMO inhibitor resistance induced by stromal cells (patient 1). (B) Percentage of viable cells after treatment of primary CLL cells with the SMO inhibitor cyclopamine and the HH-blocking Ab for 48 hours compared with control. Although in both karyotypes SMO inhibitor treatment with 10μM cyclopamine cannot significantly reduce viable cells (trisomy 12: n = 4; P = .1539, Student t test; del 13q14: n = 3; P = .0799, Student t test), the HH-blocking Ab 5E1 can significantly reduce the percentage of viable cells (trisomy 12: n = 4; 20 μg/mL 5E1 P = .0131, Student t test; Del 13q14: 40 μg/mL; n = 3; P = .0075, Student t test; patients 1, 7, 18, 50, 60, 66, 68, 69). (C) Numbers of viable CLL cells compared with the DMSO control in cells treated with cyclopamine or the ERK inhibitor U0126 or a combination of both for 48 hours and measured by flow cytometry after annexin V/7-AAD staining. Combination of SMO and ERK inhibitors shows a significant increase in apoptosis compared with single treatment alone (n = 7; P = .0007, Student t test). (D) Diagram shows the interaction between stromal cells and CLL cells and changes in canonical and noncanonical HH signaling on treatment with the HH ligand–blocking Ab 5E1 (left) or a combination of SMO and ERK inhibitors (right).
Discussion
Previous publications showed contrary results about the role of HH signaling in CLL. Hedge et al reported dependency of all CLL cells on the presence of the HH-signaling pathway on the level of SMO.42 In contrast to those results, Desch et al described no dependency on the SMO receptor but dependency on the GLI1 transcription factor.43 Therefore, the aim of our study was to identify biomarkers or clinical parameters that can discriminate between SMO inhibitor responsive and nonresponsive CLLs. Because the SMO inhibitor cyclopamine is not completely specific for the HH-signaling pathway, we added 2 more specific and more potent SMO inhibitors, NVP-LDE22547 and IPI-92648,49 to our assays, which are already in clinical trials and therefore might be more relevant for future implications. We identified 3 different response groups in our CLL cohort with 40% of CLL cells showing no response to any of the inhibitors and 36.7% of the samples with good response to SMO inhibitors. The intermediate response group showed only responses to 1 or 2 of the SMO inhibitors, and, because this group does not have elevated GLI1 and PTCH1 transcript levels, this might be unspecific effects.
We identified several biomarkers, which can discriminate between the good response group and the intermediate/nonresponder group. An increase in GLI1 and PTCH1 transcript levels > 3-fold over the median of all samples is highly predictive for a good response to SMO inhibitors. Furthermore, all patients with trisomy 12 showed HH pathway activation measured by GLI1 transcript levels and showed high responsiveness to SMO inhibitor treatment. In addition, DHH was overexpressed in all patients with trisomy 12, another biomarker for SMO inhibitor responsiveness. Treatment of trisomy 12 patient samples in the absence of stromal cells with the HH ligand–blocking Ab 5E1 induced apoptosis rates up to 90%, indicating the importance of this autocrine HH pathway activation loop for the survival of this CLL subtype. However, treatment of del 13q14 patient samples with the 5E1 Ab did not induce any apoptosis in contrast to SMO inhibitor treatment, indicating a nonautocrine HH pathway activation mechanism in those samples. Interestingly, the gene locus for DHH and GLI1 is present on chromosome 12. The pathomechanism by which trisomy 12 influences CLL development is currently not understood, but according to our results overactivation of HH signaling might be one of the mechanisms involved in this process.
Another interesting finding is the SMO inhibitor protective effect of murine or human stromal cells on CLL cells (Figure 5A). In addition, stromal cells possess an intact HH-signaling pathway and SMO inhibitor treatment reduces stromal Gli1 levels but simultaneously increases Dhh levels. Therefore, high amounts of paracrine Dhh continue to stimulate the PTCH1 receptor in CLL cells and support noncanonical activation of ERK directly downstream of the PTCH1 receptor. The fact that SMO inhibitors cannot reduce the stroma-induced ERK activation indicates that ERK activation is not dependent on SMO signaling. In addition, the finding that the DHH-blocking Ab 5E1 can completely overcome stroma-mediated resistance to SMO inhibitors and abolishes stroma-induced ERK phosphorylation argues for the stroma-dependent Dhh production to be responsible for the resistance process. In addition, the fact that trisomy 12 patient samples with autocrine DHH secretion show a higher baseline ERK phosphorylation compared with patients with del 13q14 without autocrine DHH production indicate the ERK pathway to be downstream of the PTCH1 receptor.
Our results in CLL cells are the first to show the existence of noncanonical HH signaling in cancer cells. Furthermore, they show a potential resistance mechanism of cancer cells toward SMO inhibitors induced by stromal cells. The in vitro simulation of stroma-CLL interaction by cocultivation of high amounts of stromal cells with CLL cells might overestimate the protective effect of stromal cells on CLL cells, but still gives insights into important interaction mechanisms. The protective effect might not be relevant for circulating CLL cells, because they are not in direct contact with BM stromal cells and might therefore still be responsive to the treatment with SMO inhibitors alone.
In general, our results argue for further clinical development of HH ligand–blocking Abs, because they can effectively block canonical and noncanonical HH signaling and might therefore be more effective than SMO inhibitors for the treatment of human CLL or other cancer types with ligand-mediated HH pathway activation. Regarding CLL, our results argue for 2 different treatment strategies of patients with CLL according to their genetic background: patients with trisomy 12 with autocrine HH pathway activation should be treated with the HH ligand–blocking Abs to overcome both the autocrine- and stroma-mediated paracrine HH pathway activation. Other patients with CLL with high GLI1 and PTCH1 transcript levels, but intrinsic HH pathway activation, would need a combination treatment of HH-blocking Abs to overcome the stroma-protective effect and in addition SMO inhibitors for the intrinsic canonical HH pathway activation. Further experiments need to prove those concepts in xenograft models in vivo and could be tested in future clinical trials.
Taken together, our results show that the HH-signaling pathway is an interesting therapeutic target for a subset of patients with CLL, characterized by high GLI1 and PTCH1 transcript levels and/or the presence of trisomy 12 and indicate HH-blocking Abs to be favorable over SMO inhibitors in overcoming stroma-mediated protective effects.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof Roland Mertelsmann for helpful advice, Marie Follo for help with flow cytometry, Novartis for providing NVP-LDE225, and Infinity Pharmaceuticals for providing IPI-926.
The monoclonal Ab 5E1 developed by Thomas M. Jessell and Susan Brenner-Morton was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by The Department of Biology, University of Iowa.
This work was supported by the Deutsche Krebshilfe Verbund “Receptor signaling and comparative genomics in CLL” (grant 108935 TP03) and by the “BIOSS Center for Biologic Signaling Studies” at the University of Freiburg.
Authorship
Contribution: S.D. performed research and wrote the manuscript; K.Z., L.D., D. Hartmann, and D. Herchenbach performed experiments; G.I. performed the statistical analysis; M.W., H.J., A.S., and H.V. designed experiments; and C.D. performed and designed experiments and wrote the paper.
Conflict-of-interest disclosure: M.W. was employed by and owns stock in a company (Novartis) whose product was studied in the present work. The remaining authors declare no competing financial interests.
Correspondence: Christine Dierks, Department of Hematology/Oncology, University Medical Center Freiburg, Hugstetter Str 55, 79106 Freiburg, Germany; e-mail: christine.dierks@uniklinik-freiburg.de.
References
Author notes
S.D. and K.Z. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal