Abstract
MHC I–associated peptides (MIPs) play an essential role in normal homeostasis and diverse pathologic conditions. MIPs derive mainly from defective ribosomal products (DRiPs), a subset of nascent proteins that fail to achieve a proper conformation and the physical nature of which remains elusive. In the present study, we used high-throughput proteomic and transcriptomic methods to unravel the structure and biogenesis of MIPs presented by HLA-A and HLA-B molecules on human EBV-infected B lymphocytes from 4 patients. We found that although HLA-different subjects present distinctive MIPs derived from different proteins, these MIPs originate from proteins that are functionally interconnected and implicated in similar biologic pathways. Secondly, the MIP repertoire of human B cells showed no bias toward conserved versus polymorphic genomic sequences, were derived preferentially from abundant transcripts, and conveyed to the cell surface a cell-type–specific signature. Finally, we discovered that MIPs derive preferentially from transcripts bearing miRNA response elements. Furthermore, whereas MIPs of HLA-disparate subjects are coded by different sets of transcripts, these transcripts are regulated by mostly similar miRNAs. Our data support an emerging model in which the generation of MIPs by a transcript depends on its abundance and DRiP rate, which is regulated to a large extent by miRNAs.
Introduction
All nucleated cell types constitutively express MHC class I molecules that present self-peptides to CD8 T cells. MHC I–associated peptides (MIPs) play crucial roles in many processes, including the development and homeostasis of CD8 T cells, self/nonself discrimination, tumor immunosurveillance, tissue rejection, GVHD, and odorant-based mate selection.1–6 Classic MHC I molecules are encoded by 3 genes in humans, HLA-A, HLA-B, and HLA-C, which have an astounding allelic diversity.7 Genetic polymorphisms that distinguish HLA allomorphs affect mainly their peptide-binding groove. Accordingly, studies on the presentation of various peptides suggest that a minority of peptides may bind to several MHC I molecules, but that different HLA allomorphs present largely nonoverlapping sets of peptides.8,9 The generation of MIPs is tightly linked to protein economy because it depends on protein synthesis and degradation.10,11 Nevertheless, the most striking (and intriguing) feature of MIPs is that they derive mainly from defective ribosomal products (DRiPs).12,13 DRiPs are a subset of nascent proteins that fail to achieve a proper conformation and are therefore rapidly degraded by proteasomes. A key unresolved issue is the physical nature of DRiPs. In theory, DRiPs could originate from a variety of processes that impinge on transcription, translation, posttranslational modifications, and protein folding.12,13
No algorithm can predict the amount of DRiPs produced by a gene.14 Furthermore, the composition of the self-MIP repertoire (or immunopeptidome) is not correlated with the abundance of MIP source proteins.15 Finally, studies on the relationship between the transcriptome and the immunopeptidome have yielded seemingly conflicting results.9,16 Therefore, large-scale mass spectrometry (MS) studies are the only approach that can yield a global appraisal of the MIP landscape. MS studies have highlighted the complexity of the MIP repertoire and shown that MIPs derive from all cell compartments.5,16–19 They also revealed that the immunopeptidome is plastic, conveys to the cell surface an integrative view of cellular regulation, and can be affected by cell-intrinsic and cell-extrinsic factors.17,18
Given the quintessential importance of self-MIPs, it is imperative to develop and exploit systems-level quantitative methods that will yield insights into MIP biogenesis and enable modeling of the immunopeptidome. In the present study, we used high-throughput proteomic and transcriptomic methods to unravel the structure and biogenesis of MIPs presented by HLA-A and HLA-B molecules on human EBV-infected B lymphocytes. Our results show that different HLA allomorphs present MIPs derived from distinct but functionally related source proteins. We further demonstrate that 2 features determine whether a transcript will generate MIPs: the transcript abundance and whether it contains miRNA response elements (MREs).
Methods
Cell culture and HLA typing
This study was approved by the Comité Éthique Recherche de l'Hôpital Maisonneuve-Rosemont, and all subjects provided written informed consent in accordance with the Declaration of Helsinki. PBMCs were isolated from blood samples of 4 subjects (2 pairs of HLA-identical siblings). EBV-transformed B-lymphoblastoid cell lines (B-LCLs) were derived from PBMCs with Ficoll-Paque Plus (Amersham) as described previously.20 Established B-LCLs were maintained in RPMI 1640 medium supplemented with 10% FBS, 25mM HEPES, 2mM l-glutamine, and penicillin-streptomycin (all from Invitrogen). HLA genotyping was performed at the Maisonneuve-Rosemont Hospital (Montréal, QC). Siblings of sibship 1 were: HLA-A*0101/*0205, HLA-B*1501/*5001, HLA-C*0602/*0701, and DRB1*0101/*1104. Siblings of sibship 2 were: HLA-A*0301/2902, HLA-B*0801/*4403, HLA-C*0701/*1601, and DRB1*0301/*0701. HeLa and HEK293 cell lines were maintained in DMEM medium supplemented with 10% FBS, 2mM l-glutamine, and penicillin-streptomycin (all from Invitrogen).
Mass spectrometry and peptide sequencing
Three to 4 biologic replicates of 4 × 108 exponentially growing B-LCLs were prepared from each subject. MIPs were released by mild acid treatment and separated by cation exchange chromatography using an offline 1100 series binary LC system (Agilent Technologies) as described previously.16 MIP fractions were analyzed by LC-MS/MS using an Eksigent LC system coupled to a LTQ-Orbitrap mass spectrometer (Thermo Electron).16–18 Additional details are provided in supplemental Methods (see the Supplemental Materials link at the top of the article). MS/MS of all peptide identifications are available through ProteoConnections21 at http://www.thibault.iric.ca/proteoconnections.
MIPs selection, predicted binding affinity, and identification of MIP source proteins
We filtered peptides by their length and kept those peptides with the canonical MIP length (8-11 mers) and predicted binding affinity (IC50 < 500nM. In addition, only MIPs detected in at least 3 replicates of both siblings per sibship (ie, MIPs detected in 6-8 samples derived from 2 different HLA-identical siblings) were considered for further analyses. The predicted binding affinity of peptides to the allelic products was obtained using NetMHC Version 3.2 (http://www.cbs.dtu.dk/services/NetMHC/)22 for the frequent alleles HLA-A*0101/*0301/*2902 and HLA-B*0801/*1501/*4403) or NetMHCpan Version 2.2 (http://www.cbs.dtu.dk/services/NetMHCpan-2.2/)23 for the rare alleles HLA-A*0205 and HLA-B*5001. MIPs were further inspected for mass accuracy and MS/MS spectra were validated manually. The list of MIPs reported in the present study has been provided to The Immune Epitope Database and Analysis Resource (IEDB; http://www.immuneepitope.org/)24 under the reference ID 1022782. MIP source proteins were identified using the Sidekick resource (http://www.bioinfo.iric.ca/sidekick/) and only MIPs unambiguously assigned to one source gene were analyzed further.
Analysis of enriched pathways and functional categories
Ingenuity Pathway Analysis Version 5.5.1 software (Ingenuity Systems, http://www.ingenuity.com/) was used to identify overrepresented canonical pathways and functional categories for MIP source proteins. The right-tailed Fisher exact test was used to calculate a P value determining the probability that each overrepresented pathway or biologic function was due to chance alone.
Functional connectivity score
An all-pairs-shortest-path matrix developed in-house18 was used to calculate the functional association between 2 lists of nodes (transcripts or proteins), in this case: (1) MIP source proteins of sibship 1 and MIPs source proteins of sibship 2, and (2) all MIP source proteins and all protein-coding transcripts expressed in B-LCLs. The functional association corresponds to a connectivity score. Details on the calculation of the score are provided in supplemental Methods. A bootstrapping procedure was used as a statistical sampling method to calculate control connectivity scores from 105 sets of randomly selected human protein coding transcripts (1165 transcripts are shown in Figure 2C and 12 384 in Figure 4B). The P value corresponds to the number of times that the score of the bootstrap is smaller than the score of the sample/number of bootstrap iterations (105).
Analysis of transcripts containing miRNA-binding sites
Transcripts containing MREs were obtained from 2 sources: TargetScan Version 5.1 (http://www.targetscan.org/),25 which includes both validated and predicted miRNA targets, and the Molecular Signature Database Version 3.0 (http://www.broadinstitute.org/gsea/msigdb/index.jsp),26 which includes gene sets with a known 3′-untranslated region (3′-UTR) miRNA-binding motif. The MIP datasets used in the analyses were obtained from MS-based studies by us,16,17 by others,19,27 or were available at IEDB (accessed on May 2011).24 The human and mouse proteome were downloaded from the National Center for Biotechnology Information by excluding entries with a gene type for noncoding RNA, “transposon,” or “pseudogene.” The number of transcripts containing MREs in the different datasets was compared with the 2-tailed Fisher exact test.
miRNA enrichment analysis
The miRNA target analysis module of the Web-based Gene Set Analysis Toolkit (WebGestalt; http://bioinfo.vanderbilt.edu/webgestalt/)28 was used to identify transcripts containing MREs that are overrepresented among MIP source transcripts in B-LCLs. This tool also allowed us to predict miRNAs recognizing MREs found in enriched MIP source transcripts. A hypergeometric test and a Benjamini and Hochberg correction were used to identify enriched miRNAs and target transcripts.
RNA extraction and cell sorting
PBMCs (5 × 106) were labeled with FITC-conjugated anti-CD19 (clone HIB19), allophycocyanin-conjugated anti-CD20 (clone 2H7), and propidium iodide to exclude cells in the later apoptotic stages (all from BD Biosciences). CD19+CD20+ B cells were sorted on a FACSAria cell sorter (BD Biosciences) before RNA extraction. Cells (0.5-1.0 × 106) were lysed in QIAzol (QIAGEN) and total RNA was isolated with the miRNeasy Mini Kit including DNase I treatment (QIAGEN) according to the manufacturer's instructions. Total RNA was quantified using the NanoDrop 2000 (Thermo Scientific) and RNA quality was assessed with the 2100 Bioanalyzer (Agilent Technologies).
miRNA profiling
miRNA labeling, hybridization, and washing steps were performed with the miRNA Complete Labeling and Hybridization Kit (Agilent Technologies) according to the manufacturer's instructions. A total of 100 ng of each RNA sample was hybridized to Agilent Human miRNA Microarray Release Version 16.0 (G4872A-031181; Agilent Technologies) containing 60K probes for 1205 human and 144 human viral miRNAs (including 25 specific for EBV). Images were acquired with a GenePix 4000B scanner (Molecular Devices) at a 5 μM/pixel resolution, and features were extracted with GenePix Version 6.1 software. Analyses were performed using BRB-ArrayTools Version 4.2.0 Stable Release developed by Dr Richard Simon and the BRB-ArrayTools Development Team (http://en.bio-soft.net/chip/BRBArrayTools.html). The data were background subtracted and quantile normalized. To estimate a single intensity measure for each miRNA, we calculated the mean of its different probes. We applied the Self-Organizing Maps method, followed by hierarchical clustering analysis using the average linkage method and uncentered correlation as similarity metric. Clustering analyses were performed with the Cluster Version 3.0 program (http://bonsai.hgc.jp/∼mdehoon/software/cluster/)29 and visualized with Java TreeView Version 1.1.6r2 (http://jtreeview.sourceforge.net/).30 The miRNA array data have been deposited into the Gene Expression Omnibus database under accession number GSE35319.
Results
High-throughput MS-based identification of the MIP repertoire of B-LCLs
To characterize the MIP repertoire of B-LCLs, we performed mild acid elution of peptides from 4 B-LCLs derived from PBMCs of 4 different subjects. The 4 subjects belonged to 2 HLA-disparate sibships, each composed of 1 pair of HLA-identical siblings. Eluted peptides were fractionated by ion exchange chromatography and analyzed by nano-LC-MS/MS.16–18 To identify MIPs consistently expressed by B-LCLs, we selected those MIPs detected in at least 3 replicates of both siblings in each sibship (ie, MIPs constantly detected in 6-8 samples derived from 2 different HLA-identical siblings). We further filtered peptides according to the canonical MIP length (8-11 mers) and kept only those that were predicted to bind to their corresponding HLA-A and B allelic products for further analysis. To evaluate the HLA-binding affinity of our peptides, we used NetMHC and NetMHCpan,22,23 which are the best-performing MHC-binding predictors.31,32 HLA-C–associated peptides were not included in this study because predictors for HLA-C allomorphs are not fully developed.
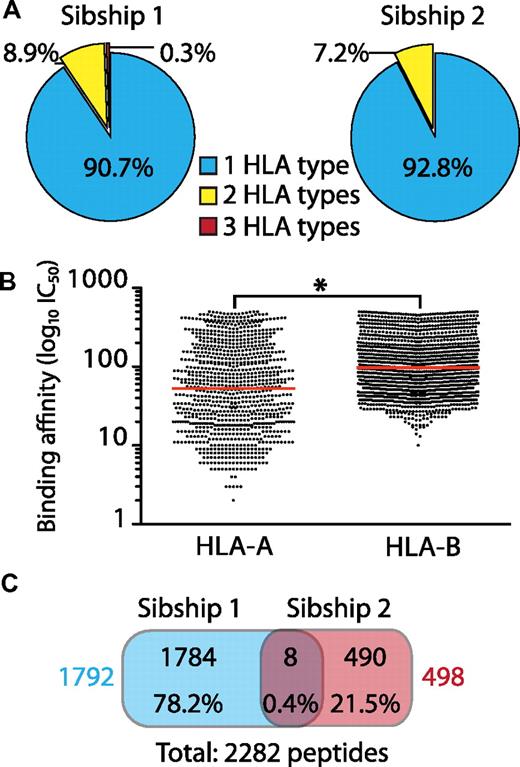
We identified a total of 2375 unique 8-11 mers associated with 8 HLA-A and B allelic products: HLA-A*0101/*0205/*0301/*2902 and HLA-B*0801/*1501/*4403/*5001 (supplemental Table 1). Of the identified MIPs, 29 are listed in the IEDB and have been identified previously by various techniques, including MHC ligand elution, MHC binding, and T cell–response assays. Most of the peptides identified herein are therefore novel and enlarge the list of MIPs associated with MHC allomorphs significantly, especially those for which fewer than 10 human peptides are currently known (eg, HLA-A*0205 and HLA-B*0801/*5001). We found that a large number of MIPs were specific to one HLA (Figure 1A). Less than 10% of MIPs were predicted to bind to more than 1 HLA (Figure 1A), although a stronger binding preference was found for a single HLA allelic product (supplemental Table 1). Accordingly, the overlap between MIPs from HLA-disparate subjects was minimal (0.4%; Figure 1C). We observed locus-dependent differences in the predicted binding affinity of our MIPs. On average, HLA-A–associated peptides had a significantly stronger binding affinity than HLA-B–associated peptides (Figure 1B and supplemental Figure 1). We conclude that HLA allomorphs present essentially distinct MIP repertoires and that, at the organismal level, the HLA genotype ultimately defines the MIP repertoire of an individual.
General features of MIPs eluted from B-LCLs from 2 HLA-disparate sibships. There were 2 HLA-identical siblings per sibship and each MIP included in the analysis was found in both siblings. (A) Number of MIPs associated with 1, 2, or more different HLA allelic products based on bioinformatic predictions with NetMHC/NetMHCpan. Most MIPs are associated with a single allelic product (HLA type). (B) Predicted MHC I–binding affinity (IC50) of eluted peptides shows that HLA-B–associated peptides are weaker binders than HLA-A–associated peptides (*P < .0001 by 2-tailed Mann-Whitney test). Each dot represents a peptide and the red line corresponds to the mean binding affinity. (C) Venn diagram showing minimal overlap between MIPs from HLA-disparate sibships. Peptide numbers and percentages are depicted for each category. Total number of MIPs for each sibship are shown in colors.
General features of MIPs eluted from B-LCLs from 2 HLA-disparate sibships. There were 2 HLA-identical siblings per sibship and each MIP included in the analysis was found in both siblings. (A) Number of MIPs associated with 1, 2, or more different HLA allelic products based on bioinformatic predictions with NetMHC/NetMHCpan. Most MIPs are associated with a single allelic product (HLA type). (B) Predicted MHC I–binding affinity (IC50) of eluted peptides shows that HLA-B–associated peptides are weaker binders than HLA-A–associated peptides (*P < .0001 by 2-tailed Mann-Whitney test). Each dot represents a peptide and the red line corresponds to the mean binding affinity. (C) Venn diagram showing minimal overlap between MIPs from HLA-disparate sibships. Peptide numbers and percentages are depicted for each category. Total number of MIPs for each sibship are shown in colors.
MIPs of HLA-disparate individuals derive from different sets of source proteins that are functionally interconnected and implicated in similar biologic pathways
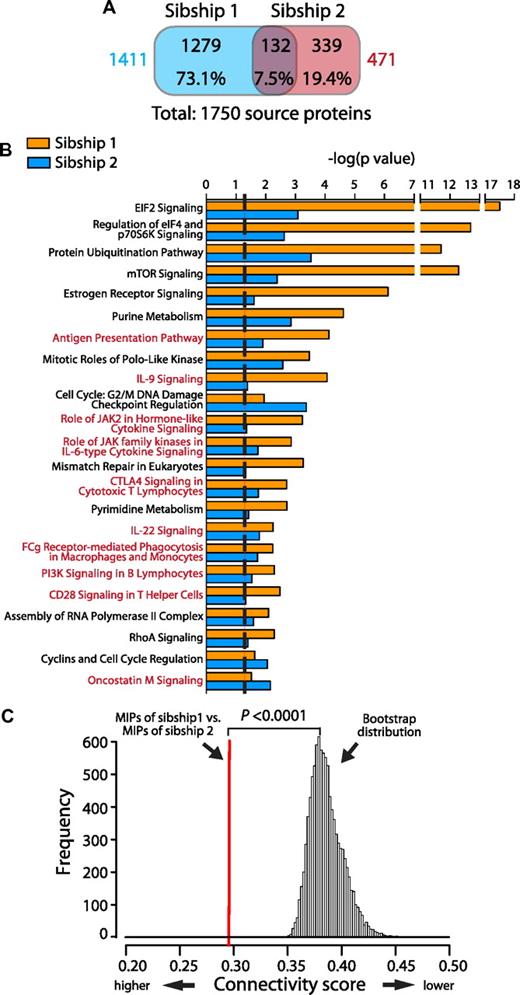
We identified unambiguously 1750 proteins source of 2290 peptides (supplemental Table 1), indicating that most MIP source proteins are represented by a single MIP. We investigated whether distinct sets of MIPs associated with nonoverlapping sets of HLA molecules originated from the same or from different sets of proteins and found that the overlap between MIP source proteins of sibships 1 and 2 was minimal (7.5%; Figure 2A). We then performed an enrichment analysis to identify pathways overrepresented in the protein source of MIPs in each sibship, which revealed that most of the overrepresented pathways were common to both sibships (P < .05 by Fisher exact test; Figure 2B). Therefore, although MIPs of HLA-disparate subjects originated mostly from different proteins, a significant number of those proteins are implicated in the same biologic pathways. We confirmed this result by analyzing the functional connectivity between the sets of proteins giving rise to MIPs in sibships 1 and 2 using an all-pairs-shortest-path matrix developed in-house (Figure 2C). This analysis is based on the fact that proteins acting in the same biologic pathway are closer in an interaction network (ie, they are more functionally connected). Functional association was measured as a connectivity score, which corresponds to the mean of the shortest path distance between every pair of proteins (one protein from each sibship) in an interaction network. We found that the set of MIP source proteins of sibship 1 was highly connected to the set of proteins of sibship 2. A bootstrap procedure (100 000 iterations) failed to reveal a single randomly selected set of proteins in the human proteome that was so tightly connected to the MIP source proteins of our subjects (P < .0001). These results show that B-LCLs from subjects with no shared HLA alleles present different immunopeptidomes that originate from different sets of proteins, which are nevertheless interconnected functionally and implicated in the same biologic pathways.
MIP source proteins are very different in the 2 sibships, but are implicated in similar biologic pathways and are interconnected functionally. (A) Venn diagram showing the minimal overlap between MIP source proteins from sibships 1 versus 2. Protein numbers and percentages are depicted for each category. Total numbers of MIP source proteins for each sibship are shown in colors. (B) The Ingenuity Pathway Analysis resource was used to identify overrepresented biologic pathways for MIPs source proteins in sibships 1 and 2. Pathways are sorted by their statistical significance as follows: −log(P value calculated with the right-tailed Fisher exact test). Higher scores indicate an increased association between the MIP source proteins and a given pathway. The dotted line represents the threshold for statistical significance (P < .05). Immune-associated pathways are highlighted in red. (C) MIP source proteins from HLA-disparate sibships are interconnected functionally. An all-pairs-shortest-path matrix was used to calculate the functional association between MIP source proteins of sibship 1 and MIPs source proteins of sibship 2 (see “Functional connectivity score” for details). The functional association was measured as a connectivity score, which corresponds to the mean of the shortest path distance between every pair of proteins (1 protein from each sibship) in an interaction network. Lower scores indicate increased connectivity. The red line represents the connectivity score between the MIP source proteins of sibship 1 and the MIP source proteins of sibship 2. A bootstrap procedure, represented by the Gaussian distribution, was used to calculate control connectivity scores between MIP source proteins (of sibships 1 or 2) and random sets of proteins from the human proteome (*P < .0001, calculated as the number of times that the score of the bootstrap is smaller than the score of the sample/number of bootstraps).
MIP source proteins are very different in the 2 sibships, but are implicated in similar biologic pathways and are interconnected functionally. (A) Venn diagram showing the minimal overlap between MIP source proteins from sibships 1 versus 2. Protein numbers and percentages are depicted for each category. Total numbers of MIP source proteins for each sibship are shown in colors. (B) The Ingenuity Pathway Analysis resource was used to identify overrepresented biologic pathways for MIPs source proteins in sibships 1 and 2. Pathways are sorted by their statistical significance as follows: −log(P value calculated with the right-tailed Fisher exact test). Higher scores indicate an increased association between the MIP source proteins and a given pathway. The dotted line represents the threshold for statistical significance (P < .05). Immune-associated pathways are highlighted in red. (C) MIP source proteins from HLA-disparate sibships are interconnected functionally. An all-pairs-shortest-path matrix was used to calculate the functional association between MIP source proteins of sibship 1 and MIPs source proteins of sibship 2 (see “Functional connectivity score” for details). The functional association was measured as a connectivity score, which corresponds to the mean of the shortest path distance between every pair of proteins (1 protein from each sibship) in an interaction network. Lower scores indicate increased connectivity. The red line represents the connectivity score between the MIP source proteins of sibship 1 and the MIP source proteins of sibship 2. A bootstrap procedure, represented by the Gaussian distribution, was used to calculate control connectivity scores between MIP source proteins (of sibships 1 or 2) and random sets of proteins from the human proteome (*P < .0001, calculated as the number of times that the score of the bootstrap is smaller than the score of the sample/number of bootstraps).
MIPs encoded by conserved and polymorphic genomic sequences
One interesting, yet unexplored, question is whether MIPs derive preferentially from conserved versus polymorphic genomic regions. Polymorphic MIPs, commonly referred to as minor histocompatibility antigens, are important in hematology because they elicit GVHD and the GVL effect after allogeneic hematopoietic cell transplantation.33–36 The most common form of polymorphism is the single nucleotide polymorphism (SNP). We used software developed in-house (pyGeno) to estimate the frequency of SNPs (SNPs/bps) in the MIP-coding DNA sequences and compared it with the SNP frequency in the human exome (Table 1). Our analysis took into account all validated synonymous and nonsynonymous SNPs reported in the Single Nucleotide Polymorphism database (http://www.ncbi.nlm.nih.gov/projects/SNP/). We found no significant differences between the frequency of synonymous or nonsynonymous SNPs within MIP-coding sequences versus the rest of the human exome. This result suggests that the MIP repertoire shows no bias toward polymorphic or invariant genomic sequences.
SNP frequency in MIPs and in the human exome
| Dataset . | Length, bp . | Nonsynonymous SNPs . | Synonymous SNPs . | Total SNPs . | |||
|---|---|---|---|---|---|---|---|
| n . | SNPs/bp* . | n . | SNPs/bp† . | n . | SNPs/bp‡ . | ||
| MIPs | 59994 | 230 | 0.0038 | 216 | 0.0036 | 446 | 0.0074 |
| Exome | 35201356 | 150158 | 0.0043 | 115376 | 0.0033 | 265534 | 0.0075 |
| Dataset . | Length, bp . | Nonsynonymous SNPs . | Synonymous SNPs . | Total SNPs . | |||
|---|---|---|---|---|---|---|---|
| n . | SNPs/bp* . | n . | SNPs/bp† . | n . | SNPs/bp‡ . | ||
| MIPs | 59994 | 230 | 0.0038 | 216 | 0.0036 | 446 | 0.0074 |
| Exome | 35201356 | 150158 | 0.0043 | 115376 | 0.0033 | 265534 | 0.0075 |
MIPs correspond to 2282 unique peptides eluted from B-LCLs. The human exome corresponds to 264 401 CDS's extracted from Ensembl (GRCh37.65). dbSNP (Build 135) was used to calculate SNP frequency (SNPs/bp). P values were calculated using the 2-tailed Fisher exact test comparing MIP-coding sequences with the exome not encoding MIPs.
P = .12.
P = .16.
P = .50.
MIPs derive preferentially from abundant transcripts
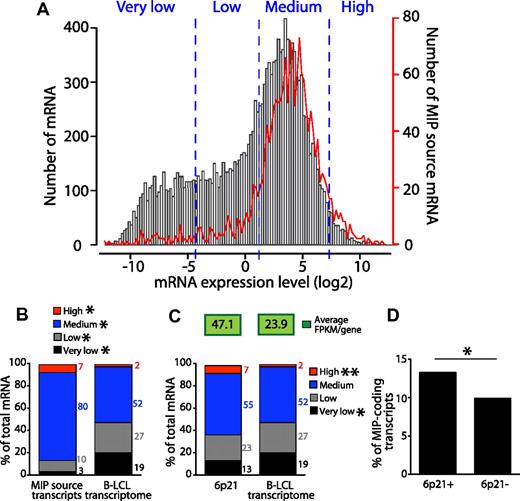
We reported previously that in primary mouse thymocytes, MIPs derived preferentially from highly abundant mRNAs.16 However, studies on human cells have cast some doubts on the correlation between the immunopeptidome and the transcriptome.9 To evaluate this relationship directly, we compared the expression level of 15 737 protein-coding transcripts expressed by B-LCLs37 with the expression level of 1707 MIP-coding transcripts (Figure 3A-B). We set 4 expression categories based on Toung et al,37 very low, low, medium, and high, and compared the number of transcripts belonging to each category in the 2 sets of transcripts. We found that MIP-coding transcripts were skewed toward higher expression categories (Figure 3B). Whereas 54% of all transcripts were expressed at a medium to high level, 87% of those coding for MIPs were. Furthermore, the number of highly abundant transcripts was enriched by 3.5 times in MIP-coding transcripts relative to the entire protein-coding transcriptome. Consistent with this, whereas 46% of transcripts were expressed at low or very low levels, only 13% of MIP-coding transcripts fell into these categories (Figure 3B). We further demonstrated that changing the thresholds that defined the various categories did not affect the differential expression of MIP-coding transcripts relative to all transcripts expressed in B-LCLs (supplemental Figure 2).
MIPs derive preferentially from abundant transcripts. (A) MIP source transcripts are expressed at a wide range of levels. Expression levels were obtained from Toung et al37 and measured in FPKMs by RNA sequencing. Frequency distribution of expression levels of 15 737 protein-coding transcripts expressed by B-LCLs (black bars) and 1707 (of 1750) transcripts source of MIPs (red line) for which an entry was found in the RNA-seq data. The frequencies of mRNAs and of MIPs source mRNAs are displayed on the left and right y-axes, respectively. Frequencies were calculated for 0.2 FPKM-bin increments. Expression categories (very low, low, medium, and high) were set based on Toung et al37 and confirmed to be unbiased (supplemental Figure 2). (B) The number of transcripts belonging to the expression categories shown in panel A among the 2 sets of transcripts indicate that MIPS derive preferentially from transcripts expressed at moderate to high levels as opposed to transcripts expressed at very low to low levels (*P < 2.2 × 10−6 by Fisher exact test). (C) The 6p21 chromosomal region is a transcriptional hot spot in B-LCLs. Graph shows the number of transcripts belonging to the expression categories shown in panel A among 316 protein-coding transcripts located in the 6p21 chromosomal region and the protein-coding transcriptome of B-LCLs (*P < .008 and **P < 2.05 × 10−7 by Fisher exact test). The average expression level (FPKMs/gene) is shown on the top for both datasets. The 6p21 region analyzed was located between positions 30400001 and 46200000 (UCSC Genome Browser, http://genome.ucsc.edu/; and NCBI Map viewer, http://www.ncbi.nlm.nih.gov/projects/mapview/). (D) The 6p21 transcriptional hot spot is a preferential source of MIPs. Comparison of the number of MIP-coding transcripts located in 6p21 (6p21+) and in the rest of the genome (6p21−; *P = .013 × 10−6 by Fisher exact test).
MIPs derive preferentially from abundant transcripts. (A) MIP source transcripts are expressed at a wide range of levels. Expression levels were obtained from Toung et al37 and measured in FPKMs by RNA sequencing. Frequency distribution of expression levels of 15 737 protein-coding transcripts expressed by B-LCLs (black bars) and 1707 (of 1750) transcripts source of MIPs (red line) for which an entry was found in the RNA-seq data. The frequencies of mRNAs and of MIPs source mRNAs are displayed on the left and right y-axes, respectively. Frequencies were calculated for 0.2 FPKM-bin increments. Expression categories (very low, low, medium, and high) were set based on Toung et al37 and confirmed to be unbiased (supplemental Figure 2). (B) The number of transcripts belonging to the expression categories shown in panel A among the 2 sets of transcripts indicate that MIPS derive preferentially from transcripts expressed at moderate to high levels as opposed to transcripts expressed at very low to low levels (*P < 2.2 × 10−6 by Fisher exact test). (C) The 6p21 chromosomal region is a transcriptional hot spot in B-LCLs. Graph shows the number of transcripts belonging to the expression categories shown in panel A among 316 protein-coding transcripts located in the 6p21 chromosomal region and the protein-coding transcriptome of B-LCLs (*P < .008 and **P < 2.05 × 10−7 by Fisher exact test). The average expression level (FPKMs/gene) is shown on the top for both datasets. The 6p21 region analyzed was located between positions 30400001 and 46200000 (UCSC Genome Browser, http://genome.ucsc.edu/; and NCBI Map viewer, http://www.ncbi.nlm.nih.gov/projects/mapview/). (D) The 6p21 transcriptional hot spot is a preferential source of MIPs. Comparison of the number of MIP-coding transcripts located in 6p21 (6p21+) and in the rest of the genome (6p21−; *P = .013 × 10−6 by Fisher exact test).
Because MIPs derived preferentially from abundant transcripts, we investigated whether the number of MIP-coding transcripts was increased in a region with high transcriptional activity. The 6p21 chromosomal region containing the extended human MHC is recognized as a canonical transcriptional hot spot.7 We first confirmed that this region generates an increased number of highly abundant transcripts and then calculated the average expression of transcripts derived from 6p21 versus the whole transcriptome of B-LCLs (Figure 3C). We found that the average gene-expression level (measured in fragments per kilobase of exon per million fragments mapped [FPKMs]) was 2 times greater for 6p21 than for all expressed protein-coding transcripts. We then compared the number of MIP-coding transcripts located in 6p21 versus in the rest of the genome (Figure 3D) and found that the number of MIP-coding transcripts was significantly higher for transcripts located in 6p21 than elsewhere in the genome. These results show that the 6p21 transcriptional hot spot is a preferential source of MIPs.
The MIP repertoire is connected functionally to the transcriptome
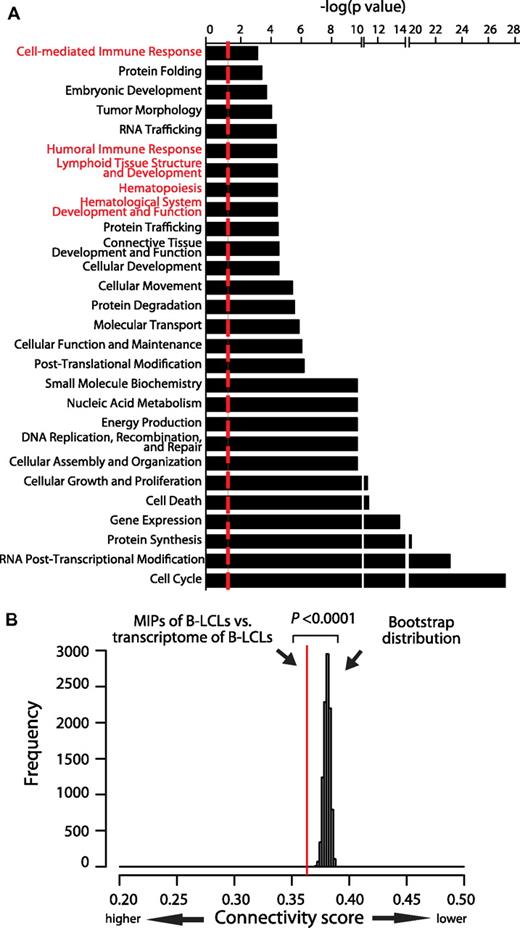
In mouse thymocytes and dendritic cells (DCs), we found that the MIP repertoire conceals a cell-type–specific signature.16,17 To determine whether this is also true in human cells, we first used the Ingenuity Pathway Analysis resource to identify major functional categories that were overrepresented in the set of proteins encoding MIPs in B-LCLs (Figure 4A). Some overrepresented categories were associated with basic cellular biologic processes such as cell cycling, protein synthesis, and gene expression, reflecting intracellular events occurring in any proliferating cell. The salient finding is that several immune-specific functional categories were also overrepresented, including hematopoiesis, lymphoid tissue structure and development, and humoral and cell-mediated immune responses (Figure 4A categories in red). Furthermore, these major immune categories included B cell–specific functions such as proliferation, development, and differentiation of B lymphocytes; class switching; and development of pre-B and pro-B lymphocytes (supplemental Table 2). Moreover, the MIP repertoire reflected immune-associated and intracellular pathways important for B-cell biology (Figure 2B pathways in red). Among the overrepresented pathways, we found antigen presentation, IL-6 signaling (required for B-cell maturation), PI3K signaling in B lymphocytes (crucial in B-cell development), and JAK2 in cytokine signaling (very active in stimulated B cells).
The MIP repertoire is connected functionally to the transcriptome. (A) Graph showing the major functional categories that are overrepresented in the MIP repertoire of B-LCLs. Overrepresented functional categories were identified with the Ingenuity Pathway Analysis resource and are sorted by their statistical significance as follows: −log (P value calculated with the right-tailed Fisher exact test). Higher values indicate an increased association between the MIP source proteins and a given category. The dotted line represents the threshold for statistical significance (P < .05). Immune-associated categories are highlighted in red. More details on functional annotations represented in immune-associated categories are provided in supplemental Table 2. (B) The immunopeptidome of B-LCLs is connected functionally to the transcriptome of B-LCLs. An all-pairs-shortest-path matrix was used to calculate the functional association between MIP source proteins and the protein-coding transcriptome of B-LCLs (see “Functional connectivity score” for details). The functional association was measured as a connectivity score corresponding to the mean of the shortest path distance between every pair of proteins in an interaction network (ie, 1 MIP-source protein and 1 expressed transcript of B-LCLs). The red line represents the connectivity score between MIP source proteins and transcripts expressed by B-LCLs. A bootstrap procedure, represented by the Gaussian distribution, was used to calculate the control connectivity scores between MIP source proteins and random sets of the human transcripts. (*P < .0001).
The MIP repertoire is connected functionally to the transcriptome. (A) Graph showing the major functional categories that are overrepresented in the MIP repertoire of B-LCLs. Overrepresented functional categories were identified with the Ingenuity Pathway Analysis resource and are sorted by their statistical significance as follows: −log (P value calculated with the right-tailed Fisher exact test). Higher values indicate an increased association between the MIP source proteins and a given category. The dotted line represents the threshold for statistical significance (P < .05). Immune-associated categories are highlighted in red. More details on functional annotations represented in immune-associated categories are provided in supplemental Table 2. (B) The immunopeptidome of B-LCLs is connected functionally to the transcriptome of B-LCLs. An all-pairs-shortest-path matrix was used to calculate the functional association between MIP source proteins and the protein-coding transcriptome of B-LCLs (see “Functional connectivity score” for details). The functional association was measured as a connectivity score corresponding to the mean of the shortest path distance between every pair of proteins in an interaction network (ie, 1 MIP-source protein and 1 expressed transcript of B-LCLs). The red line represents the connectivity score between MIP source proteins and transcripts expressed by B-LCLs. A bootstrap procedure, represented by the Gaussian distribution, was used to calculate the control connectivity scores between MIP source proteins and random sets of the human transcripts. (*P < .0001).
If the MIP repertoire is molded by the cell-type–specific transcriptome, then the set of MIP-encoding proteins in B-LCLs should be connected functionally to the B-LCL transcriptome. To test this hypothesis, we used our all-pairs-shortest-path matrix method18 to evaluate the functional connectivity between the set of MIP source proteins and the protein-coding transcriptome of B-LCLs and to compare this connectivity with the connectivity scores of 105 random human transcriptome sets (Figure 4B). The functional connectivity found between MIP-coding transcripts and the transcriptome of B-LCLs was significantly greater than the MIP connectivity to control transcriptomes (P < .0001 by bootstrapping). Interestingly, the MIP-transcriptome connectivity increased as a function of the expression level of MIP-coding transcripts (supplemental Figure 3). Therefore, the connectivity of MIP-encoding transcripts to the transcriptome reached its maximum value when only highly abundant transcripts were considered. We conclude that the MIP repertoire of human cells is connected functionally to the cell transcriptome in a transcript-level–dependent manner.
MIPs derive preferentially from transcripts containing miRNA-binding sites
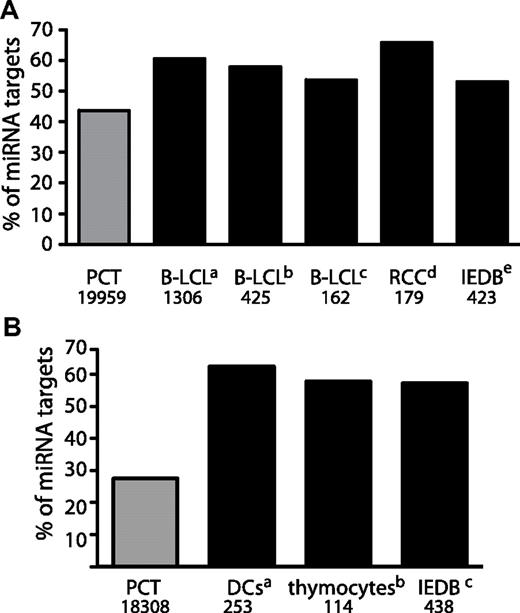
According to the dominant paradigm, MIPs derive primarily from DRiPs generated by an as-yet elusive biochemical processes.10,13 Theoretically, the number of MIPs generated by a transcript should be dictated by 2 factors: the transcript abundance and the transcript DRiP rate. In accordance with this assumption, medium and highly abundant transcripts were a preferential source of MIPs (Figure 3A), However, some MIPs derived from transcripts expressed at low or very low levels (Figure 3A). We therefore speculated that low abundance transcripts/proteins that generate MIPs do so because they have a high DRiP rate. miRNAs are a class of non-protein-coding RNAs that bind to MREs on target transcripts, causing destabilization of the target transcript or inhibition of its translation.38 Destabilization of target mRNAs by miRNAs could constitute a possible mechanism through which DRiPs are generated. To determine whether miRNA targets are a preferential source of MIPs, we first calculated the number of transcripts containing MREs in 3 datasets: MIP source transcripts in our B-LCLs, all human-protein–coding transcripts (Figure 5A and supplemental Figure 4A), and protein-coding transcripts expressed in our B-LCLs (supplemental Figure 4C-D). We used 2 different databases to identify validated and predicted transcripts recognized by miRNAs, TargetScan Version 5.125 and the Molecular Signature Database Version 3.0,26 and performed independent analyses with both databases (Figure 5A and supplemental Figure 4). These analyses revealed that, compared with the B-LCL transcriptome (supplemental Figure 4C-D) with to the human-protein–coding transcriptome (Figure 5A), the set of transcripts encoding MIPs identified in B-LCLs from each sibship was enriched significantly in transcripts containing MREs.
The MIP-coding transcriptome is enriched in transcripts containing miRNA-binding sites. (A) Human transcripts containing miRNA-binding sites are a preferential source of MIPs. The number of MIPs that derive from miRNA targets was calculated for various datasets: B-LCLs from sibships 1 and 2 (this study), B-LCLs,19 renal cell carcinomas27 and all human peptides listed in the IEDB.24 A list of 8725 human-protein–coding miRNA targets was extracted from TargetScan.25 Numbers on the bottom indicate the number of entries for each dataset. The number of MIPs that derive from miRNA targets for all datasets was significantly higher than the number of miRNA targets in the human protein coding transcriptome (PCT; aP < 2.2 × 10−16, bP < 3.3 × 10−9, cP < .01, dP < 2.12 × 10−9, and eP < 1.38 × 10−4 by Fisher exact test). (B) Mouse transcripts containing MREs are preferential sources of MIPs. The number of MIPs that derive from miRNA targets was calculated for various datasets: DCs,17 thymocytes,16 and all mouse peptides listed in the IEDB.24 A list of 504 mouse protein-coding miRNA targets was extracted from TargetScan.25 Numbers on the bottom indicate the number of entries for each dataset. The number of MIPs that derive from miRNA targets for all MIP datasets was significantly higher than the number of miRNA targets in the mouse protein coding transcriptome (aP < 1.0 × 10−11 and b,cP < 2.2 × 10−16 by Fisher exact test).
The MIP-coding transcriptome is enriched in transcripts containing miRNA-binding sites. (A) Human transcripts containing miRNA-binding sites are a preferential source of MIPs. The number of MIPs that derive from miRNA targets was calculated for various datasets: B-LCLs from sibships 1 and 2 (this study), B-LCLs,19 renal cell carcinomas27 and all human peptides listed in the IEDB.24 A list of 8725 human-protein–coding miRNA targets was extracted from TargetScan.25 Numbers on the bottom indicate the number of entries for each dataset. The number of MIPs that derive from miRNA targets for all datasets was significantly higher than the number of miRNA targets in the human protein coding transcriptome (PCT; aP < 2.2 × 10−16, bP < 3.3 × 10−9, cP < .01, dP < 2.12 × 10−9, and eP < 1.38 × 10−4 by Fisher exact test). (B) Mouse transcripts containing MREs are preferential sources of MIPs. The number of MIPs that derive from miRNA targets was calculated for various datasets: DCs,17 thymocytes,16 and all mouse peptides listed in the IEDB.24 A list of 504 mouse protein-coding miRNA targets was extracted from TargetScan.25 Numbers on the bottom indicate the number of entries for each dataset. The number of MIPs that derive from miRNA targets for all MIP datasets was significantly higher than the number of miRNA targets in the mouse protein coding transcriptome (aP < 1.0 × 10−11 and b,cP < 2.2 × 10−16 by Fisher exact test).
To determine whether this feature of the MIP-coding transcriptome was cell-type specific, we performed the same analysis on publicly available datasets of human MIPs: B-LCLs expressing HLA-B*1801,19 renal cell carcinomas,27 and a list of all human peptides identified in at least 20 different cell types and extracted from the IEDB.24 In all cases, the number of MIP source transcripts bearing MREs was higher than expected (Figure 5A and supplemental Figure 4A). We then investigated whether this was also true for mouse MIPs (Figure 5B and supplemental Figure 4B). Analysis of mouse MIPs identified previously by our group in DCs17 and thymocytes,16 as well as MIPs from different cell lines listed in the IEDB,24 showed that the mouse MIP-encoding transcriptome was also enriched significantly in miRNA targets. In summary, these results show that, irrespective of cell type, both mouse and human MIP-coding transcripts are enriched in miRNA targets.
MIPs from HLA-disparate sibships derive from different sets of transcripts regulated by similar miRNomes
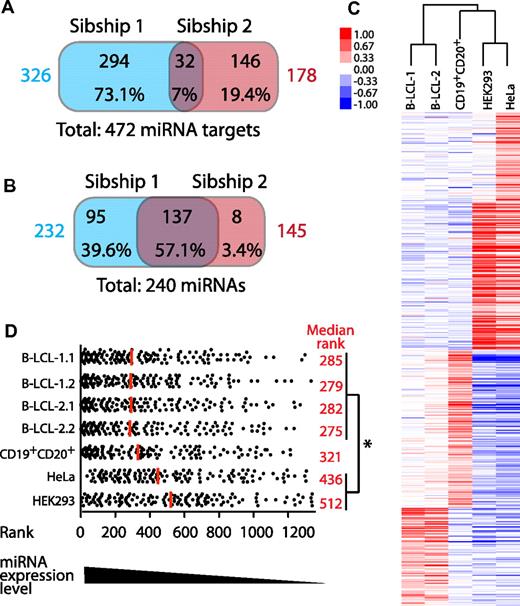
We used the WebGestalt analysis program28 to identify the specific MREs enriched in the 3′-UTR of MIP-coding transcripts identified in B-LCLs from our 2 HLA-disparate sibships (Figure 6A). A representative set of enriched MREs is shown in Table 2, and the complete list can be found in supplemental Table 3. We then investigated whether the MIP repertoire would reflect the miRNome. Consistent with the observation that MIPs of HLA-disparate sibships derive from different sets of proteins (Figure 2A), the sets of MIP source transcripts bearing enriched MREs in the 2 sibships displayed minimal overlap (7%; Figure 6A). Having compared miRNA targets, we next compared the miRNAs recognizing MREs in MIP source transcripts. Enriched miRNA targets were classified according to their specific 3′-UTR MREs. We then compared the miRNAs that were predicted to regulate MIP source transcripts in the 2 sibships and found a 57% overlap among enriched miRNAs regulating MIP source transcripts in the 2 sibships (Figure 6B). These results suggest that MIPs from HLA-disparate subjects derive from different sets of transcripts regulated by mostly similar miRNAs.
MIPs from HLA-disparate sibships derive from different sets of transcripts regulated by similar miRNomes. (A) Venn diagram showing that MIPs of HLA-disparate sibships derive from different sets of miRNA targets. The WebGestalt analysis program28 was used to identify transcripts significantly enriched in 3′-UTR–binding sites of specific miRNAs among all MIP source transcripts identified in sibships 1 and 2 (P < .05 by hypergeometric test). The total number of MIP source transcripts that are miRNA targets for each sibship are shown in colors. (B) Different sets of MIP source transcripts are predicted to be regulated by mostly the same miRNAs. The WebGestalt analysis program28 was used to identify miRNAs that recognize the 3′-UTR–binding site present in enriched MIP source transcripts. The numbers and percentages of miRNAs are depicted for each category. Total number of miRNAs that target MIP source transcripts in each sibship are shown in colors. (C) miRNA profiling showing that B-LCLs from HLA-disparate sibships have similar miRNA-expression profiles. Human miRNA microarrays were used to assess the expression of 1205 human and 144 viral miRNAs in B-LCLs (2 subjects from each sibship), CD19+CD20+ cells isolated from PBMCs of one member of sibship 2, and 2 nonlymphoid cell lines (HEK293 and HeLa). Hierarchical clustering was used to calculate the correlation between various profiles (represented by a dendrogram). (D) miRNAs that target MIP source transcripts in B-LCLs are expressed at higher levels in B-LCLs than in other cell types. All miRNAs studied were ranked from high to low according to their expression level in each cell line. Among them, the ranking of each of the 133 miRNAs predicted to regulate MIP source transcripts in B-LCLs was determined in B-LCLs, primary CD19+CD20+ cells, HEK293 cells, and HeLa cells. Each dot corresponds to 1 miRNA. The median rank is shown for each cell population (red line and values). On average, miRNAs predicted to recognize MIP source transcripts were ranked significantly higher (ie, were more expressed) in B-LCLs than in nonlymphoid cell lines (*P < .05 by 1-way ANOVA and the Bonferroni multiple comparison test). B-LCL-1.1 indicates B-LCLs from subject 1 of sibship 1; B-LCL-1.2, from subject 2 of sibship 1; B-LCL-2.1, from subject 1 of sibship 2; and B-LCL-2.2, from subject 2 of sibship 2.
MIPs from HLA-disparate sibships derive from different sets of transcripts regulated by similar miRNomes. (A) Venn diagram showing that MIPs of HLA-disparate sibships derive from different sets of miRNA targets. The WebGestalt analysis program28 was used to identify transcripts significantly enriched in 3′-UTR–binding sites of specific miRNAs among all MIP source transcripts identified in sibships 1 and 2 (P < .05 by hypergeometric test). The total number of MIP source transcripts that are miRNA targets for each sibship are shown in colors. (B) Different sets of MIP source transcripts are predicted to be regulated by mostly the same miRNAs. The WebGestalt analysis program28 was used to identify miRNAs that recognize the 3′-UTR–binding site present in enriched MIP source transcripts. The numbers and percentages of miRNAs are depicted for each category. Total number of miRNAs that target MIP source transcripts in each sibship are shown in colors. (C) miRNA profiling showing that B-LCLs from HLA-disparate sibships have similar miRNA-expression profiles. Human miRNA microarrays were used to assess the expression of 1205 human and 144 viral miRNAs in B-LCLs (2 subjects from each sibship), CD19+CD20+ cells isolated from PBMCs of one member of sibship 2, and 2 nonlymphoid cell lines (HEK293 and HeLa). Hierarchical clustering was used to calculate the correlation between various profiles (represented by a dendrogram). (D) miRNAs that target MIP source transcripts in B-LCLs are expressed at higher levels in B-LCLs than in other cell types. All miRNAs studied were ranked from high to low according to their expression level in each cell line. Among them, the ranking of each of the 133 miRNAs predicted to regulate MIP source transcripts in B-LCLs was determined in B-LCLs, primary CD19+CD20+ cells, HEK293 cells, and HeLa cells. Each dot corresponds to 1 miRNA. The median rank is shown for each cell population (red line and values). On average, miRNAs predicted to recognize MIP source transcripts were ranked significantly higher (ie, were more expressed) in B-LCLs than in nonlymphoid cell lines (*P < .05 by 1-way ANOVA and the Bonferroni multiple comparison test). B-LCL-1.1 indicates B-LCLs from subject 1 of sibship 1; B-LCL-1.2, from subject 2 of sibship 1; B-LCL-2.1, from subject 1 of sibship 2; and B-LCL-2.2, from subject 2 of sibship 2.
Representative set of enriched miRNA-binding sites present in transcripts that are a source of MIPs in B-LCLs from both sibships
| 3′-UTR–binding site . | miRNA . | Sibship 1 . | Sibship 2 . | ||||
|---|---|---|---|---|---|---|---|
| n . | RE . | P . | n . | RE . | P . | ||
| TGCACTT | MIR-519C MIR-519B MIR-519A | 51 | 4.21 | 5.93 × 10−18 | 9 | 2.23 | 2.15 × 10−02 |
| ACATTCC | MIR-1 MIR-206 | 40 | 4.94 | 8.63 × 10−17 | 11 | 4.07 | 9.95 × 10−05 |
| TGCCTTA | MIR-124A | 53 | 3.53 | 2.87 × 10−15 | 11 | 5.02 | 1.31 × 10−02 |
| CTTGTAT | MIR-381 | 30 | 5.64 | 1.66 × 10−14 | 5 | 2.81 | 3.36 × 10−02 |
| ACCAAAG | MIR-9 | 48 | 3.48 | 9.38 × 10−14 | 16 | 3.48 | 2.01 × 10−05 |
| TAGCTTT | MIR-9 | 48 | 3.48 | 9.38 × 10−14 | 16 | 3.48 | 1.95 × 10−02 |
| TGAATGT | MIR-181A MIR-181B MIR-181C MIR-181D | 47 | 3.51 | 1.23 × 10−13 | 9 | 2.01 | 3.72 × 10−02 |
| GTGCCTT | MIR-506 | 57 | 2.93 | 7.65 × 10−13 | 20 | 3.08 | 1.16 × 10−05 |
| ACTTTAT | MIR-142-5P | 34 | 4.28 | 1.18 × 10−12 | 11 | 4.15 | 8.38 × 10−05 |
| TTTGCAC | MIR-19A MIR-19B | 46 | 3.28 | 2.41 × 10−12 | 14 | 2.99 | 3.00 × 10−04 |
| TTTGTAG | MIR-520D | 35 | 3.93 | 6.75 × 10−12 | 7 | 2.36 | 3.08 × 10−02 |
| GTATTAT | MIR-369-3P | 25 | 4.51 | 3.62 × 10−10 | 6 | 3.24 | 1.11 × 10−02 |
| GTTTGTT | MIR-495 | 27 | 4.05 | 8.41 × 10−10 | 8 | 3.6 | 1.90 × 10−03 |
| TTTGCAG | MIR-518A-2 | 24 | 4.33 | 1.88 × 10−09 | 7 | 3.79 | 2.70 × 10−03 |
| CTTTGTA | MIR-524 | 36 | 3.01 | 5.82 × 10−09 | 13 | 3.26 | 2.00 × 10−04 |
| CAGTGTT | MIR-141 MIR-200A | 28 | 3.33 | 3.32 × 10−08 | 10 | 3.57 | 6.00 × 10−04 |
| TATTATA | MIR-374 | 26 | 3.44 | 5.41 × 10−08 | 8 | 3.17 | 4.10 × 10−03 |
| TTTTGAG | MIR-373 | 23 | 3.77 | 5.75 × 10−08 | 8 | 3.93 | 1.10 × 10−03 |
| CTCAGGG | MIR-125B MIR-125A | 28 | 3.20 | 7.76 × 10−08 | 7 | 2.4 | 2.84 × 10−02 |
| TGTTTAC | MIR-30A-5P MIR-30C MIR-30D MIR-30B MIR-30E-5P | 40 | 2.54 | 9.80 × 10−08 | 11 | 2.09 | 1.78 × 10−02 |
| TGGTGCT | MIR-29A MIR-29B MIR-29C | 37 | 2.65 | 9.83 × 10−08 | 12 | 2.58 | 2.80 × 10−03 |
| GCAAAAA | MIR-129 | 20 | 4.09 | 1.07 × 10−07 | 6 | 3.68 | 6.20 × 10−03 |
| ATTCTTT | MIR-186 | 25 | 3.35 | 1.63 × 10−07 | 6 | 2.41 | 4.03 × 10−02 |
| ATACTGT | MIR-144 | 21 | 3.81 | 1.77 × 10−07 | 10 | 5.44 | 1.83 × 10−05 |
| 3′-UTR–binding site . | miRNA . | Sibship 1 . | Sibship 2 . | ||||
|---|---|---|---|---|---|---|---|
| n . | RE . | P . | n . | RE . | P . | ||
| TGCACTT | MIR-519C MIR-519B MIR-519A | 51 | 4.21 | 5.93 × 10−18 | 9 | 2.23 | 2.15 × 10−02 |
| ACATTCC | MIR-1 MIR-206 | 40 | 4.94 | 8.63 × 10−17 | 11 | 4.07 | 9.95 × 10−05 |
| TGCCTTA | MIR-124A | 53 | 3.53 | 2.87 × 10−15 | 11 | 5.02 | 1.31 × 10−02 |
| CTTGTAT | MIR-381 | 30 | 5.64 | 1.66 × 10−14 | 5 | 2.81 | 3.36 × 10−02 |
| ACCAAAG | MIR-9 | 48 | 3.48 | 9.38 × 10−14 | 16 | 3.48 | 2.01 × 10−05 |
| TAGCTTT | MIR-9 | 48 | 3.48 | 9.38 × 10−14 | 16 | 3.48 | 1.95 × 10−02 |
| TGAATGT | MIR-181A MIR-181B MIR-181C MIR-181D | 47 | 3.51 | 1.23 × 10−13 | 9 | 2.01 | 3.72 × 10−02 |
| GTGCCTT | MIR-506 | 57 | 2.93 | 7.65 × 10−13 | 20 | 3.08 | 1.16 × 10−05 |
| ACTTTAT | MIR-142-5P | 34 | 4.28 | 1.18 × 10−12 | 11 | 4.15 | 8.38 × 10−05 |
| TTTGCAC | MIR-19A MIR-19B | 46 | 3.28 | 2.41 × 10−12 | 14 | 2.99 | 3.00 × 10−04 |
| TTTGTAG | MIR-520D | 35 | 3.93 | 6.75 × 10−12 | 7 | 2.36 | 3.08 × 10−02 |
| GTATTAT | MIR-369-3P | 25 | 4.51 | 3.62 × 10−10 | 6 | 3.24 | 1.11 × 10−02 |
| GTTTGTT | MIR-495 | 27 | 4.05 | 8.41 × 10−10 | 8 | 3.6 | 1.90 × 10−03 |
| TTTGCAG | MIR-518A-2 | 24 | 4.33 | 1.88 × 10−09 | 7 | 3.79 | 2.70 × 10−03 |
| CTTTGTA | MIR-524 | 36 | 3.01 | 5.82 × 10−09 | 13 | 3.26 | 2.00 × 10−04 |
| CAGTGTT | MIR-141 MIR-200A | 28 | 3.33 | 3.32 × 10−08 | 10 | 3.57 | 6.00 × 10−04 |
| TATTATA | MIR-374 | 26 | 3.44 | 5.41 × 10−08 | 8 | 3.17 | 4.10 × 10−03 |
| TTTTGAG | MIR-373 | 23 | 3.77 | 5.75 × 10−08 | 8 | 3.93 | 1.10 × 10−03 |
| CTCAGGG | MIR-125B MIR-125A | 28 | 3.20 | 7.76 × 10−08 | 7 | 2.4 | 2.84 × 10−02 |
| TGTTTAC | MIR-30A-5P MIR-30C MIR-30D MIR-30B MIR-30E-5P | 40 | 2.54 | 9.80 × 10−08 | 11 | 2.09 | 1.78 × 10−02 |
| TGGTGCT | MIR-29A MIR-29B MIR-29C | 37 | 2.65 | 9.83 × 10−08 | 12 | 2.58 | 2.80 × 10−03 |
| GCAAAAA | MIR-129 | 20 | 4.09 | 1.07 × 10−07 | 6 | 3.68 | 6.20 × 10−03 |
| ATTCTTT | MIR-186 | 25 | 3.35 | 1.63 × 10−07 | 6 | 2.41 | 4.03 × 10−02 |
| ATACTGT | MIR-144 | 21 | 3.81 | 1.77 × 10−07 | 10 | 5.44 | 1.83 × 10−05 |
The WebGestalt analysis tool was used to calculate the enrichment. P values were calculated using the hypergeometric test.
n indicates the number of transcripts that were a source of MIPs containing the 3′-UTR–binding site recognized by the specified miRNAs; and RE, ratio of enrichment (observed/expected number of transcripts).
To confirm that the miRNome of our 2 sibships was indeed similar, we performed genome-wide miRNA profiling in 8 different cell lines: B-LCLs from the 4 subjects (2 siblings from each sibship), primary CD19+CD20+ B cells from 1 of the subjects, and 2 nonlymphoid human cell lines, HEK293 and HeLa (Figure 6C). Hierarchical clustering analysis revealed that the highest similarity was found between B-LCLs from the 2 sibships, which formed a separate cluster. Therefore, as expected, B-LCLs express a cell-type–specific miRNome that is independent of the HLA genotype or other interindividual genetic differences. We then investigated whether enriched miRNAs that were predicted to regulate MIP source transcripts in the 2 sibships (Table 2 and supplemental Table 3) were indeed expressed in the B-LCLs of our 4 subjects and, if so, at what level. All 1349 profiled miRNAs were ranked from high to low according to their expression level in each cell line. Among them, the ranking of each of the 133 miRNAs predicted to regulate MIP source transcripts in B-LCLs was determined in B-LCLs, primary CD19+CD20+ cells, HEK293 cells, and HeLa cells (Figure 6D). Enriched MREs can be recognized by more than one miRNA (Table 2), and we could not identify which of the predicted miRNAs was indeed acting on the transcript source of MIPs. Despite this source of uncertainty, we found that, on average, miRNAs predicted to recognize MIP source transcripts ranked significantly higher in B-LCLs than in control cell lines. Therefore, miRNAs predicted to recognize MIP source transcripts in B-LCLs were expressed at higher levels in B-LCLs than in other cell types. This supports the concept that the immunopeptidome of a cell is molded by its miRNome.
Discussion
Self-MIPs regulate all key events in the life of CD8 T cells. Indeed, CD8 T cells are selected on, sustained by, and activated in the presence of self-MIPs.39 Moreover, MIPs are the targets of several immune processes, including autoimmunity, graft rejection, GVHD, and the GVL effect.5,6,35 Estimates suggest that the immunopeptidome comprises approximately 0.1% of the nonapeptide sequences (the typical length of MIPs) found in the proteome.5 Despite the important role of MIPs in health and disease, we know little about their biogenesis except for the fact that they derive from DRiPs, the physical nature of which remains ill-defined.13 In the present study, we sought to characterize the global landscape of MIPs on human cells and to gain insights into the biogenesis of the human immunopeptidome. We used B-LCLs because they can be obtained from nearly any subject, proliferate extensively in vitro, express high levels of MHC I molecules at the surface, and have been shown to be a reliable tool for high-throughput genomic studies.40 We found that most MIPs bind one unique HLA among all available MHC allelic products and do so with different predicted binding affinities. In general, HLA-A–associated peptides had a significantly higher binding affinity than HLA-B–associated peptide, probably resulting from differences in the permissiveness of the HLA-binding motifs. We also observed that subjects with no shared HLA-A and HLA-B alleles presented different MIP repertoires, showing that the immunopeptidome is ultimately determined by the combination of HLA allelic products. This result was expected based on the specificities of the various HLA-binding motifs,9 and is consistent with a recent MS-based study showing minimal overlap in the identity of soluble plasma MIPs found in 2 HLA-different individuals.
Three salient findings emerged from the present study. First, even though HLA-different subjects presented different MIPs derived from different proteins, these MIPs originated from proteins that are interconnected functionally and implicated in the same biologic pathways. In our B-LCLs, many MIP source proteins were associated with functional categories or involved in signaling pathways characteristic of the immune system or specific to B-cell biology. Accordingly, we found that the MIP repertoire of B-LCLs was linked intimately to the transcriptome of B-LCLs. This supports our previous results on mouse MIPs identified on thymocytes and DCs showing that the MIP repertoire conceals a tissue/cell-specific signature.16,17 This means that the immunopeptidome is not monopolized by MIPs derived from ubiquitous housekeeping genes. The notion that the MIP repertoire is cell-type specific has to be taken into account in immunotherapeutic interventions such as leukemia immunotherapy.41
Second, our results show that under basal conditions, the MIP repertoire of human B-LCLs derived preferentially from abundant transcripts. This was true whether taking into consideration the whole exome (Figure 3A-B) or a single transcriptional hot spot (the 6p21 chromosomal region; Figure 3C-D). We used as a reference the average mRNA expression estimated by RNA-seq in B-LCLs from 20 unrelated individuals,37 which provides an accurate estimation of transcript abundance in this cell type. A correlation between transcript abundance and MIP presentation was also observed in MS studies on primary mouse thymocytes.16 However, this correlation was absent in studies analyzing changes in MIP abundance induced by neoplastic transformation or metabolic stress.16,18,27 All of these results can be reconciled with a parsimonious explanation: (1) in healthy normal cells, MIPs derive preferentially from the most abundant transcripts, and (2) in stressed cells, changes in MIP abundance result to a large extent from co- or posttranslational processes that regulate the DRiP rate.
Finally, perhaps the most exciting finding reported herein is that MIPs derive preferentially from transcripts bearing MREs. miRNA-profiling experiments revealed that the miRNAs that recognized MIP source transcripts in our B-LCLs were expressed at higher levels in B-LCLs than in other cell types. Furthermore, even though MIPs from HLA-disparate subjects derived from different transcripts, miRNA-profiling experiments confirmed that these sets of transcripts were regulated by similar miRNomes, as was predicted by bioinformatic analysis. We found that transcripts containing MREs were a preferential source of MIPs, not only in our B-LCLs but also in various mouse and human cell types analyzed by us and others. The relationship between MREs and MIPs is therefore very robust, because it holds true across species and cell types. Although we still have to investigate how miRNAs could enhance the generation of MIPs, our working hypothesis is that destabilization of mRNAs by miRNAs generates DRiPs. Consistent with this idea, studies on cells transfected with shRNA or mRNAs carrying premature stop codons have revealed that the nonsense-mediated decay pathway can generate MIPs, presumably as a result of DRiP formation.42,43 Indeed, miRNAs can decrease the levels of the targeted protein via various mechanisms, including slowing of elongation, ribosome drop-off, and nascent polypeptide degradation.44,45 We therefore postulate that miRNAs are major regulators of the DRiP rate. Given the pervasive influence of miRNAs on all cellular processes, including normal and neoplastic lymphohematopoiesis,46–49 further studies are warranted to investigate how miRNA may mold the immunopeptidome of normal and neoplastic cells. In addition, we propose that the engineering of the MIP repertoire via the miRNome might be relevant in immunotherapy.
There is an Inside Blood commentary on this article in this issue.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Simon Drouin, Raphaëlle Lambert, and Pierre Chagnon at the Genomics facility; Olivier Caron Lizotte and Eric Bonneil at the proteomics facility; and Danièle Gagné and Gaël Dulude at the flow cytometry facility of the Institute for Research in Immunology and Cancer (IRIC); and Véronique Lisi, Moutih Rafei, Chantal Baron, and Marie-Christine Meunier for advice and thoughtful comments.
This work was supported by a grant from the Fonds d'Innovation Pfizer–Fonds de la Recherche en Santé du Québec. D.P.G. is supported by a studentship from the Canadian Institutes for Health Research. C.L. is supported by the Perseverance Program of IRIC. C.P. and P.T. are supported by the Canada Research Chairs Program. IRIC is supported in part by the Canadian Center of Excellence in Commercialization and Research, the Canada Foundation for Innovation, and the FRSQ.
Authorship
Contribution: D.P.G. designed the study, performed the experiments, analyzed the data, prepared the figures, and wrote the first draft of the manuscript; W.Y. and T.L.M.-S. performed the MS experiments and analysis; C.M.L. analyzed the data and prepared the figures; T.D. and J.-P.L. performed the bioinformatics analyses; C.C. performed experiments; S.L. designed the study, performed the statistical analyses, and discussed the results; P.T. and C.P. designed the study, discussed the results, and wrote the manuscript; and all authors edited and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Claude Perreault or Pierre Thibault, Institute for Research in Immunology and Cancer, PO Box 6128, Station Centre-Ville, Montreal, QC, Canada H3C 3J7; e-mail: claude.perreault@umontreal.ca or pierre.thibault@umontreal.ca.
References
Author notes
P.T. and C.P. contributed equally to this work and share senior authorship.