To the editor:
Bignami et al recently reported on miRNA profiles of treatment-naive HIV-1 patients (naive), elite suppressors (éLTNP or ES), and multiply-exposed, uninfected individuals (MEUs).1 The authors identified only 1 putative miRNA expression difference between naive and ES and did not confirm results of earlier studies of miRNAs in HIV-1 susceptible cells and infected patients.2,,–5 After re-analyzing the data (Gene Expression Omnibus series GSE33514), we suggest that technical and experimental design issues, including comparisons of patients with a single control sample, may have inappropriately affected the authors' conclusions. Our alternative examination tended to corroborate previous findings.
We posit that the original conclusions may derive from analysis issues and batch effects. There were significant differences in the number of miRNAs detected in MEU or control versus naive/ES (Figure 1A; P < .001). Further, when we used the specified U6 normalization and exclusion criteria, 70 miRNAs of < 150 detected consistently in MEU had significant apparent expression differences (P < .05 with multiple comparison correction). Each was rarer in MEUs than infected patients. A uniformly 1-way directionality of expression differences is unusual and inconsistent with previous work2 ; Figure 1B details additional issues. We conclude that technical factors of RNA integrity and processing (no quality control assessments were provided), measurement, and analysis likely affected results.
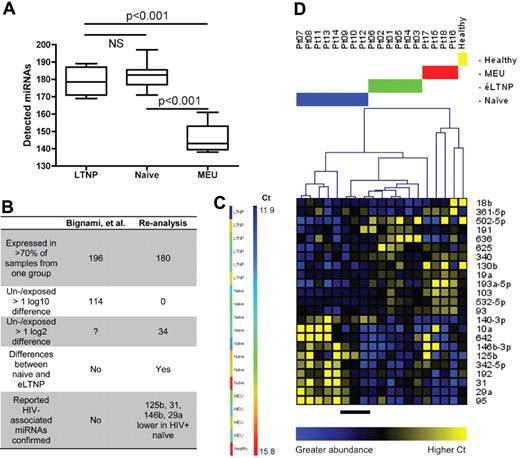
Data re-analysis. (A) There was a significant difference in the mean number of detected miRNAs (P < .0001, 1-way ANOVA), with significant individual differences between naive or éLTNP and MEU (P < .001, Bonferroni; NS indicates not significant). (B) Original/re-analysis differences using original exclusion criteria and normalization (first 3 rows) or stricter criteria (detected in > 80% of samples) and global normalization (bottom rows). No log10 changes were reported for 114 miRNAs, and only 34 log2 differences were seen. Naive versus éLTNP: the authors reported that miR-155 levels were significantly different, but in contrast with other studies, miR-155 was simply not detected in éLTNP or uninfected controls and was undetected or at low levels in naive. Bottom row: although Bignami et al provided values for some previously reported miRNAs,1 the assertion of changes > 1 log10 confuses interpretation. Re-analysis identified several previously reported miRNAs; miR-150 also appeared to be downmodulated in naive, but its abundance complicated interpretation of the quantile normalized data. Additional analyses (not shown) suggested differential expression of miR-150. (C) Across samples, U6 Cts varied from 11.9 (blue) to 15.8 (red), suggesting sample quality differences with implications for normalization choice. (D) Re-analysis using quantile normalized data. miRNAs differentially expressed in naive and éLTNP were used to cluster all samples (Pearson correlation, average linkage; patient numbers are at top and groups are color coded; for mean-centered data, blue represents greater abundance and yellow indicates low abundance/high Ct). Note that 3 naive samples (black underscore) diverge in expression of underexpressed miRNAs; disclosure of standard clinical parameters might reveal correlates of these differences.
Data re-analysis. (A) There was a significant difference in the mean number of detected miRNAs (P < .0001, 1-way ANOVA), with significant individual differences between naive or éLTNP and MEU (P < .001, Bonferroni; NS indicates not significant). (B) Original/re-analysis differences using original exclusion criteria and normalization (first 3 rows) or stricter criteria (detected in > 80% of samples) and global normalization (bottom rows). No log10 changes were reported for 114 miRNAs, and only 34 log2 differences were seen. Naive versus éLTNP: the authors reported that miR-155 levels were significantly different, but in contrast with other studies, miR-155 was simply not detected in éLTNP or uninfected controls and was undetected or at low levels in naive. Bottom row: although Bignami et al provided values for some previously reported miRNAs,1 the assertion of changes > 1 log10 confuses interpretation. Re-analysis identified several previously reported miRNAs; miR-150 also appeared to be downmodulated in naive, but its abundance complicated interpretation of the quantile normalized data. Additional analyses (not shown) suggested differential expression of miR-150. (C) Across samples, U6 Cts varied from 11.9 (blue) to 15.8 (red), suggesting sample quality differences with implications for normalization choice. (D) Re-analysis using quantile normalized data. miRNAs differentially expressed in naive and éLTNP were used to cluster all samples (Pearson correlation, average linkage; patient numbers are at top and groups are color coded; for mean-centered data, blue represents greater abundance and yellow indicates low abundance/high Ct). Note that 3 naive samples (black underscore) diverge in expression of underexpressed miRNAs; disclosure of standard clinical parameters might reveal correlates of these differences.
The titular case for a retroviral exposure signature is tenuous as presented. First, only unprotected sex suggested exposure of MEUs to HIV-1. The authors did not specify whether individuals had HIV-1–positive sex partners or if these partners were taking antiretrovirals. It seems unusual to define a patient class by conjecture. Second, the normalizing RNA, U6 was not “relatively stable” in these samples, but varied 14-fold. The highest threshold cycle (Ct) was in the healthy donor sample (Figure 1C), likely skewing results. Third, 5 members of the proposed octapartite exposure signature1 (Figure 2A) failed the authors' inclusion criterion (expression in > 70% of at least 1 patient group); each was undetected in 13-16 of 17 patient samples. The other 3 were considered up-regulated in patients only because they were undetected in the single control PCR well, insufficient to conclude absence or calculate accurate fold changes. Many such “undetected” miRNAs were found in previous studies of healthy CD4+ T cells.6,7 Finally, no meaningful conclusions can be drawn from comparisons made with a single control sample, pooled or not. The authors' gp120 experiments,1 (Figure 3) also lack biologic replicates. With an n = 1, assertions of a retroviral exposure signature and a gp120-mediated miRNA profile shift are preliminary.
Contrasting with the authors' conclusions, alternative analyses using quantile normalized data from miRNAs expressed in the majority of samples revealed multiple naive-to-ES differences. Although confidence in these results remains attenuated by the observations above and would be bolstered by verification with technical replicates and/or different measurement platforms, it is interesting that several identified miRNAs have been associated with HIV-12,3,5 or ES/naive differences,4 including miRs-29a, -31, -125b, and -146b (Figure 1D). We conclude that the data provide additional evidence for, not against, previously reported miRNAs in HIV-1 infection and elite suppression.
Authorship
Acknowledgments: This work was supported by the National Institutes of Health (grant NSO 76357, S.E.C.).
Contribution: K.W.W. and J.E.C. examined the data and designed the study; K.W.W. re-analyzed the data; and K.W.W. and J.E.C. wrote the letter.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kenneth W. Witwer, Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, 733 N Broadway, BRB Suite 831, Baltimore, MD 20125; e-mail: kwitwer1@jhmi.edu.
References
National Institutes of Health

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal