Arginine-glycine-aspartic acid (RGD)–mimetic platelet inhibitors act by occupying the RGD recognition site of αIIb/β3 integrin (GPIIb/IIIa), thereby preventing the activated integrin from reacting with fibrinogen. Thrombocytopenia is a well-known side effect of treatment with this class of drugs and is caused by Abs, often naturally occurring, that recognize αIIb/β3 in a complex with the drug being administered. RGD peptide and RGD-mimetic drugs are known to induce epitopes (ligand-induced binding sites [LIBS]) in αIIb/β3 that are recognized by certain mAbs. It has been speculated, but not shown experimentally, that Abs from patients who develop thrombocytopenia when treated with an RGD-mimetic inhibitor similarly recognize LIBS determinants. We addressed this question by comparing the reactions of patient Abs and LIBS-specific mAbs against αIIb/β3 in a complex with RGD and RGD-mimetic drugs, and by examining the ability of selected non-LIBS mAbs to block binding of patient Abs to the liganded integrin. Findings made provide evidence that the patient Abs recognize subtle, drug-induced structural changes in the integrin head region that are clustered about the RGD recognition site. The target epitopes differ from classic LIBS determinants, however, both in their location and by virtue of being largely drug-specific.
Introduction
Ligand-mimetic platelet inhibitors bind specifically to the arginine-glycine-aspartic acid (RGD) recognition site of αIIb/β3 integrin (GPIIb/IIIa), thereby preventing the activated integrin from reacting with fibrinogen and participating in platelet thrombus formation.1,2 Two such drugs, tirofiban and eptifibatide, have been shown to reduce adverse complications in patients treated with percutaneous transluminal coronary angioplasty3,4 and are in widespread clinical use. Between 0.1% and 2.0% of patients treated with tirofiban, eptifibatide, and other drugs of this class evaluated in clinical trials experienced acute thrombocytopenia, often severe, within a few hours of starting treatment,5 a complication now known to be caused by Abs that recognize αIIb/β3 in a complex with the ligand mimetic drug being administered.6,,,–10 A unique feature of such Abs is that they can occur naturally in persons never previously exposed to one of these drugs, enabling thrombocytopenia to develop within a few hours of starting treatment.7,11
Various mechanisms have been identified by which drug-induced Abs cause thrombocytopenia.11,12 In patients sensitive to drugs like quinine, certain antibiotics, anticonvulsants, and many other medications, a soluble drug promotes binding of an otherwise nonreactive Ig to a platelet membrane glycoprotein by a mechanism that is not fully understood but does not appear to involve a preferred docking site for a drug on the target glycoprotein.11,13,,–16 In contrast, RGD-mimetic drugs bind to a well-defined recognition site at the junction of the αIIb β-propeller and the β3-β A domain (also designated β I) of αIIb /β317,,–20 and induce structural changes in the integrin. Numerous murine mAbs have been described that recognize conformational changes (ligand-induced binding sites [LIBS]) induced in αIIb/β3 by RGD peptide and by RGD-mimetic platelet inhibitors.21,,–24 By analogy, it has been proposed that Abs causing thrombocytopenia in patients treated with ligand-mimetic inhibitors likewise recognize structural changes (mimetic-induced binding site [MIBS]) induced in the integrin by drug5,7,25 but this has not been confirmed by experiment. In this report, we present evidence that Abs causing thrombocytopenia in patients treated with eptifibatide or tirofiban do recognize structural changes (neoepitopes) induced in αIIb/β3 by these drugs. However, the human Abs differ from classic LIBS-specific monoclonals in that they are largely drug-specific and appear to recognize subtle, drug-induced structural rearrangements, MIBS, in the integrin head region rather than the more widely dispersed LIBS epitopes.
Methods
Patient Abs
Abs from 43 patients who developed thrombocytopenia after treatment with eptifibatide or tirofiban were initially detected in testing done by the Platelet and Neutrophil Immunology Laboratory (BloodCenter of Wisconsin). Platelet nadirs in the affected patients averaged 19 000/μL (median 10 000/μL, range 1000-102 000/μL). Bleeding symptoms, consisting in most cases of petechial hemorrhages and ecchymoses, were observed in most patients who had severe thrombocytopenia (platelets < 20 000/μL) and 12 were given platelet transfusions. Twenty (47%) of 43 patients had no bleeding symptoms. Nineteen of 34 eptifibatide and 5 of 5 tirofiban samples (or 24 [62%] of 39 samples) were from patients with no known prior exposure to these drugs.
Drug-dependent Ab detection
Reactions of patient Abs with platelets pretreated with ligand-mimetic drugs or RGDW peptide were characterized by flow cytometry using an LSRII flow cytometer (BD Biosciences) as previously described.7,26 In brief, 5.0 × 106 platelets isolated from citrated blood were combined with test serum and eptifibatide 2.4μM, tirofiban 2.0μM, xemilofiban 2.7μM, orbofiban 3.0μM, or RGDW peptide 1.0mM in a 50-μL volume in HEPES buffer (137mM NaCl, 2mM KCl, 12mM NaHCO3, 0.3mM NaH2PO4, 5mM HEPES, 0.65mM CaCl2, pH 7.4), and incubated at room temperature for 60 minutes. Platelets were then washed twice in HEPES-buffered saline containing drug at the same concentration as in the primary reaction mixture and bound Ab was detected with FITC-labeled Fab′2 goat anti–human IgG (H + L) Ab (Jackson ImmunoResearch Laboratories). A reaction was considered to be positive if the signal (median fluorescence intensity) obtained with drug-treated platelets was at least twice the signal obtained with patient serum in the absence of drug and with normal serum in the presence of drug (ratio test serum/control serum > 2.0). Reactions positive by this criterion invariably exceeded the median signal obtained with randomly selected normal sera (N = 994) by > 3.0 SD.
Reagents
Unless otherwise stated, reagents were purchased from Sigma-Aldrich. Protein G sepharose was obtained from GE Healthcare; FBS was obtained from Hyclone; F12K media, PBS, G418, and gentamycin were obtained from Mediatech; Fugene 6 was obtained from Roche; sulfo-NHS LC-biotin was obtained from Pierce Biotechnology; and FITC goat (Fab′)2 anti–mouse IgG (H + L), FITC goat (Fab′)2 anti–human IgG (H + L), FITC goat (Fab′)2 anti–human IgG (Fc), and PE-labeled streptavidin were obtained from Jackson ImmunoResearch Laboratories. Eptifibatide and tirofiban were purchased from a local pharmacy. Xemilofiban and orbofiban (active forms) were gifts from G. D. Searle when that company was an independent entity. RGDW peptide was synthesized and purified by the Peptide Core Laboratory (BloodCenter of Wisconsin).
mAbs
Murine mAbs specific for αIIb/β3 were produced in BALB/c mice immunized with αIIb/β3 purified from human platelets.27 Details of the immunization protocol and selection for hybridomas secreting αIIb/β3-specific mAbs have been described previously.28 Other mAbs used were 10E5 (anti-αIIb) from B. S. Coller (Rockefeller University), 7E3 (anti-β3) from Centocor Inc, AP3 (anti-β3) and AP2 (anti-αIIb/β3) from the Hybridoma Core Laboratory (Blood Research Institute). mAbs were biotinylated with sulfo-NHS-LC-Biotin (Pierce Biotechnology) according to the manufacturer's protocol. In preliminary studies, it was found that the biotinylation process itself did not affect the ability of mAbs used in the study to react with αIIb/β3.
LIBS-specific monoclonals
mAbs specific for LIBS in αIIb/β3 were identified by screening integrin-specific mAbs produced as described above at a concentration of 10.0 μg/mL against untreated platelets and platelets pretreated with 1.0mM RGDW peptide24 and selecting those that gave a median fluorescence intensity signal in the presence of peptide at least 3 times greater than the signal obtained in the absence of peptide. The 14 LIBS-specific mAbs thus selected produced ratios (signal obtained with RGD-treated platelets to that obtained with untreated platelets) ranging from 3.0 to 43.7 (average 11.9, median 6.9). Other LIBS-specific mAbs used were AP5 specific for the N terminus of β324 , D3 specific for the β3 hybrid/EGF1 domain29 (from L. Jennings, University of Tennessee), LIBS-6 specific for a β3 EGF domain,21 and PMI-1 specific for the αIIb calf-2 domain30 (both from M. Ginsburg, University of California, San Diego). Reactions of the latter 4 mAbs with RGD-treated and untreated platelets were similar to those of our 14 newly produced LIBS-specific mAbs (average ratio RGD/no RGD 14.3, median 14.2, range 5.2-23.8).
Mapping of mAb-binding sites to specific domains of αIIb and β3
Stably transfected CHO cell lines expressing mixed αIIb/β3 integrins (rat/human) were described previously.28 mAbs were mapped to the α or β subunit of αIIb/β3 on the basis of their reactions with CHO cell lines expressing human αIIb paired with rat β3 or vice versa. After assignment to an α or β subunit, the mAbs were mapped to the β propeller domain of αIIb or the β A domain of β3 on the basis of their reactions with CHO cells expressing αIIb/β3 that was human except for substitution of a rat sequence in one of these domains.28 For example, mAb 290.5 reacted with a mixed integrin consisting of human αIIb and rat β3 and was therefore αIIb-specific. Its binding site was then localized to the β propeller domain of αIIb by showing that it failed to react when this domain alone consisted of a rat sequence but reacted with the reciprocal construct containing only a human β propeller sequence.
Further characterization of mAb-binding sites
Sites recognized by mAbs were further defined by investigating their ability to compete with one another for binding. For these studies, human platelets were isolated from whole blood anticoagulated with acid-citrate-dextrose (ACD) and were suspended in PBS containing 1.0% albumin. In preliminary studies, the quantity of each mAb required to saturate about 90% of the available platelet αIIb/β3 receptors was determined using flow cytometry. Platelets were then incubated with twice this amount of mAb for 30 minutes at room temperature to block binding sites. Biotinylated mAbs (0.5 μg) were then added to the mixture and incubated for an additional 30 minutes. Platelets were then washed twice, suspended in 50 μL of PE-labeled streptavidin (1:200) for 15 minutes and diluted to 0.2 mL. Platelet-bound PE was measured by flow cytometry. Binding of biotinylated mAb was expressed as a percentage of the signal (median fluorescence intensity) obtained in the absence of a blocking Ab.
Inhibition of patient Ab binding by selected mAbs
Platelets were suspended in 1% BSA in Tyrode/HEPES buffer (137mM NaCl, 2mM KCl, 12mM NaHCO3, 0.3mM NaH2PO4, 5mM HEPES, 0.65mM CaCl2, pH 7.4) and were first treated with the drug for which each Ab was specific and then with selected mAbs at twice the amounts needed to produce 90% saturation of their targets. Human Abs were then added in amounts that produced a signal ∼ 90% of the signal obtained with unblocked, drug-treated platelets. After incubation for 30 minutes at room temperature, platelets were washed once and bound Ab was detected with FITC-labeled goat (Fab′)2 anti–human IgG (H + L chain–specific). When abciximab was used for blocking, human Ab binding was detected with FITC-labeled goat (Fab′)2 anti–human IgG (Fc). Results were expressed as a percentage of the signal (median fluorescence intensity) obtained when platelets were treated with drug and an irrelevant mAb.
Research approvals
Human studies were approved by the Institutional Review Board of the BloodCenter of Wisconsin. Murine studies were approved by the Institutional Animal Care and Use Committee of the Medical College of Wisconsin.
Results
Patient Abs are mainly specific for the drug that caused thrombocytopenia
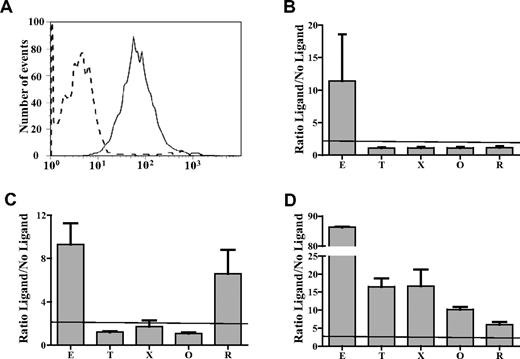
Abs from 43 patients who developed thrombocytopenia within 24 hours of treatment with eptifibatide (38 cases) or tirofiban (5 cases) were tested for reactions with intact platelets pretreated with eptifibatide, tirofiban, the nonapproved RGD-mimetic drugs xemilofiban and orbofiban, and RGDW peptide. As expected, all patient samples reacted with platelets pretreated with the drug that caused thrombocytopenia (Table 1). As illustrated in Figure 1A and B for a typical patient with eptifibatide-induced thrombocytopenia, this was the only reaction observed with 27 (72%) of 38 eptifibatide-dependent Abs and 3 (60%) of 5 tirofiban-dependent Abs. However, Abs from 11 of 38 cases of eptifibatide-induced thrombocytopenia and 2 of 5 cases of tirofiban-induced thrombocytopenia also recognized platelets pretreated with one or more of the other agents (Table 1). Five of 38 sera from patients sensitive to eptifibatide cross-reacted with tirofiban, 8 with xemilofiban, 3 with orbofiban, and 7 with RGDW; 2 of 5 sera from patients with tirofiban-induced thrombocytopenia cross-reacted with xemilofiban and one of these also recognized eptifibatide-treated platelets. The cross-reaction of serum from a patient with eptifibatide-induced thrombocytopenia against platelets pretreated with RGDW is shown in Figure 1C. A unique eptifibatide-dependent Ab reacted with platelets treated with all 5 agents (Figure 1D). With only 2 exceptions, reactions of the 13 cross-reacting sera were significantly stronger against platelets treated with the drug that caused thrombocytopenia than against platelets treated with any of the other 4 ligands.
Reactions of patient Abs against platelets pretreated with eptifibatide, tirofiban, and other ligands
| Cause of TP . | N . | Ligand . | ||||
|---|---|---|---|---|---|---|
| Epti . | Tiro . | Xemi . | Orbo . | RGDW . | ||
| Eptifibatide | 38 | 38 | 5 | 8 | 3 | 7 |
| Tirofiban | 5 | 1 | 5 | 2 | 2 | 0 |
| Cause of TP . | N . | Ligand . | ||||
|---|---|---|---|---|---|---|
| Epti . | Tiro . | Xemi . | Orbo . | RGDW . | ||
| Eptifibatide | 38 | 38 | 5 | 8 | 3 | 7 |
| Tirofiban | 5 | 1 | 5 | 2 | 2 | 0 |
Values shown indicate number of samples that gave positive reactions against platelets treated with the indicated ligands.
Epti indicates eptifibatide; Tiro, tirofiban; Xemi, xemilofiban; Orbo, orbofiban; and TP, thrombocytopenia.
Representative reactions of patient Abs. Patient samples were tested against platelets pretreated with eptifibatide (E), tirofiban (T), xemilofiban (X), orbofiban (O), and RGDW peptide (R). (A) Serum from a typical patient with eptifibatide-induced thrombocytopenia reacted with platelets pretreated with eptifibatide (solid tracing) but not with untreated platelets (dashed tracing) using flow cytometry. (B) The same serum failed to recognize platelets pretreated with T, X, O, and R. (C) A second patient sample reacted with platelets pretreated with E or R. (D) A third Ab recognized platelets pretreated with each of the 5 ligands. Reactions of patient serum with untreated platelets and normal serum with treated platelets were negative (not shown). Horizontal dashed line indicates upper limit of normal (mean + 3.0 SD). (B-D) Values on the ordinate indicate ratio of median fluorescence intensity value obtained with ligand-treated platelets to value obtained with untreated platelets.
Representative reactions of patient Abs. Patient samples were tested against platelets pretreated with eptifibatide (E), tirofiban (T), xemilofiban (X), orbofiban (O), and RGDW peptide (R). (A) Serum from a typical patient with eptifibatide-induced thrombocytopenia reacted with platelets pretreated with eptifibatide (solid tracing) but not with untreated platelets (dashed tracing) using flow cytometry. (B) The same serum failed to recognize platelets pretreated with T, X, O, and R. (C) A second patient sample reacted with platelets pretreated with E or R. (D) A third Ab recognized platelets pretreated with each of the 5 ligands. Reactions of patient serum with untreated platelets and normal serum with treated platelets were negative (not shown). Horizontal dashed line indicates upper limit of normal (mean + 3.0 SD). (B-D) Values on the ordinate indicate ratio of median fluorescence intensity value obtained with ligand-treated platelets to value obtained with untreated platelets.
In contrast to patient Abs, LIBS-specific mAbs tended not to discriminate between ligands
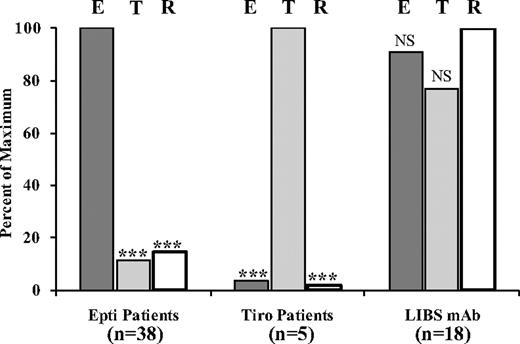
Figure 2 summarizes reactions of the 38 epifibatide-dependent Abs, the 5 tirofiban-dependent Abs, and 18 LIBS-specific mAbs against platelets pretreated with eptifibatide, tirofiban, and RGDW peptide. To facilitate comparisons, the reaction of each drug-dependent Ab against platelets pretreated with the drug that caused thrombocytopenia was assigned a value of 100 and reactions of the same Ab against platelets treated with the other ligands was expressed as a percentage of that value. Similarly, the signal obtained with each LIBS mAb against RGDW-treated platelets was assigned a value of 100, and results obtained with eptifibatide- and tirofiban-treated platelets were compared with that value. As a group, the LIBS-specific mAbs differed significantly from the patient Abs in that their reactions were relatively independent of the ligand that occupied the RGD recognition site under our experimental conditions. In contrast, reactions of the patient Abs were largely specific for the drug that caused thrombocytopenia.
Reactions of patient Abs and LIBS-specific mAbs against platelets pretreated with saturating concentrations of eptifibatide (E), tirofiban (T), and RGDW peptide (R; flow cytometry). For each Ab group, signals (median fluorescence intensity) obtained with (left) E, (center) T, and (right) R were expressed as “100%” and signals obtained with other ligands were expressed as a percentage of this value. Bars denote averages for 38 eptifibatide, 5 tirofiban, and 18 LIBS Abs. ***P < .001 compared with signal obtained with (left) E, (center) T, and (right) R. NS indicates not significant.
Reactions of patient Abs and LIBS-specific mAbs against platelets pretreated with saturating concentrations of eptifibatide (E), tirofiban (T), and RGDW peptide (R; flow cytometry). For each Ab group, signals (median fluorescence intensity) obtained with (left) E, (center) T, and (right) R were expressed as “100%” and signals obtained with other ligands were expressed as a percentage of this value. Bars denote averages for 38 eptifibatide, 5 tirofiban, and 18 LIBS Abs. ***P < .001 compared with signal obtained with (left) E, (center) T, and (right) R. NS indicates not significant.
Binding sites for 10 mAbs were mapped to 5 subdomains in the αIIb/β3 head region
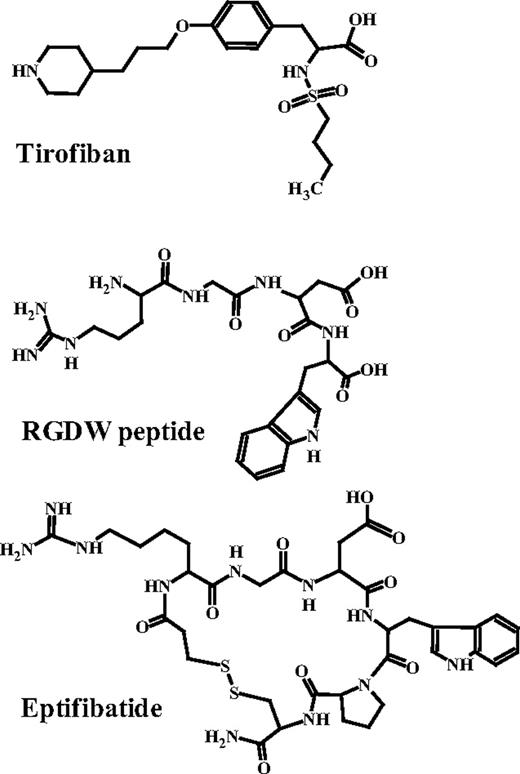
Eptifibatide, tirofiban, and RGDW have distinctly different structures (Figure 3), making it extremely unlikely that cross-reactions observed with 13 patient Abs (Figure 1, Table 1) are due to direct recognition of some common element of drug structure. An alternative possibility is that the patient Abs recognize epitopes created by structural changes induced in αIIb/β3 by the ligand-mimetic agents. If this is the case, failure of most eptifibatide-dependent Abs to cross-react with tirofiban and vice versa (Figure 1, Table 1) suggests that these 2 agents, even though they both bind to αIIb/β3 by mimicking the structure of RGD, induce different sets of epitopes in the integrin that can be distinguished by the patient Abs. We examined this possibility by determining whether mAbs specific for different epitopes in the “head” domains of the integrin might produce different patterns of inhibition when their ability to block binding of the patient Abs to their targets was determined.
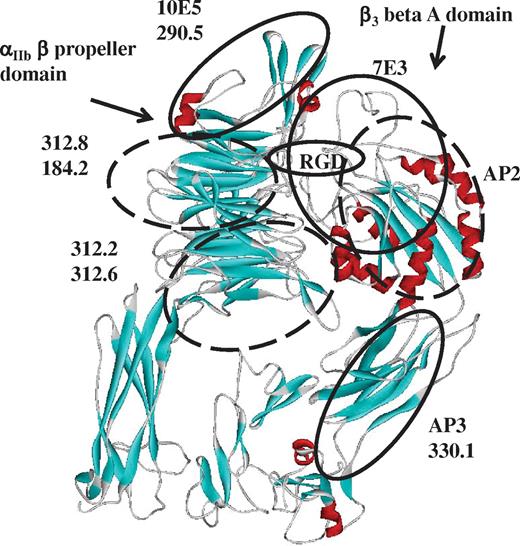
Ten mAbs specific for epitopes expressed on N-terminal domains of αIIb or β3 (Table 2) were used for these studies. Specific amino acid residues on αIIb recognized by mAb 10E5 were previously defined by x-ray crystallography17 and approximate binding sites for 4 others (7E3, AP3, 312.6, and 312.8) were identified on the basis of their reactions with selectively mutated αIIb/β3.28,31,32 Binding sites for the remaining 5 mAbs were localized to the αIIb β propeller (290.5, 312.2, and 184.2) or the β3-β A (AP2) or hybrid (330.1) domains on the basis of their reactions with CHO cells expressing chimeric forms of αIIb/β3 in which a rat domain (αIIb β propeller or β3 β A) was substituted for the human domain and vice versa as described in “Mapping of mAb-binding sites to specific domains of αIIb and β3.” Each of the 10 mAbs was tested for its ability to block binding of the others labeled with biotin to platelet αIIb/β3. As expected, each mAb completely blocked its biotinylated counterpart (Table 3 diagonal). In addition, 5 pairs of mAbs (the αIIb-specific pairs 10E5/290.5, 184.2/312.8, and 312.2/312.6, and the β3-specific pairs 7E3/AP2 and AP3/330.1) completely blocked each other but had only partial or no effect on binding of the remaining 8 (Table 3). Possible binding sites for the 5 pairs of mAbs on N-terminal domains of αIIb and β3 based on these patterns of inhibition and prior knowledge of specific amino acid residues recognized by mAbs 10E5, 7E3, AP3, 312.6, and 312.817,28,31,–33 are illustrated in Figure 4, where ovals representing Ab footprints are constructed so that, when projected onto the integrin, they occupy a surface area of ∼ 1000 sq Å, consistent with structural studies showing that the total solvent-accessible surface area buried in typical mAb-Ag complexes ranges from 1300 to 2300 sq Å (650-1150 sq Å for each component of the complex).34,–36
Characteristics of mAbs mapped to subdomains of the αIIb/β3 head structure
| Designation . | Specificity . | Reference . | ||
|---|---|---|---|---|
| Glycoprotein . | Domain . | Amino acids . | ||
| 10E5 | αIIb | Beta propeller | 77-82, 206, 208, 213-215 | 17 |
| 290.5 | αIIb | Beta propeller | ||
| 184.2 | αIIb | Beta propeller | ||
| 312.8 | αIIb | Beta propeller | 28 | |
| 312.2 | αIIb | Beta propeller | ||
| 312.6 | αIIb | Beta propeller | 28 | |
| PMI-1 | LIBS-αIIb | Calf-2 | 844-859 | 30 |
| 7E3 | β3 | Beta A | 129, 177-184 | 31 |
| AP2 | β3 | Beta A | ||
| AP3 | β3 | PSI/hybrid | 50-62 | 32 |
| 330.1 | β3 | PSI/hybrid | ||
| AP5 | LIBS-β3 | PSI | 1-6 | 24 |
| D3 | LIBS-β3 | Hybrid/EGF | 29 | |
| LIBS-6 | LIBS-β3 | EGF | 21 | |
| Designation . | Specificity . | Reference . | ||
|---|---|---|---|---|
| Glycoprotein . | Domain . | Amino acids . | ||
| 10E5 | αIIb | Beta propeller | 77-82, 206, 208, 213-215 | 17 |
| 290.5 | αIIb | Beta propeller | ||
| 184.2 | αIIb | Beta propeller | ||
| 312.8 | αIIb | Beta propeller | 28 | |
| 312.2 | αIIb | Beta propeller | ||
| 312.6 | αIIb | Beta propeller | 28 | |
| PMI-1 | LIBS-αIIb | Calf-2 | 844-859 | 30 |
| 7E3 | β3 | Beta A | 129, 177-184 | 31 |
| AP2 | β3 | Beta A | ||
| AP3 | β3 | PSI/hybrid | 50-62 | 32 |
| 330.1 | β3 | PSI/hybrid | ||
| AP5 | LIBS-β3 | PSI | 1-6 | 24 |
| D3 | LIBS-β3 | Hybrid/EGF | 29 | |
| LIBS-6 | LIBS-β3 | EGF | 21 | |
LIBS indicates ligand-induced binding site; PSI, plexin-semaphorin-integrin; and EGF, epidermal growth factor.
Reciprocal blocking studies performed with 10 αIIb/β3–specific mAbs
| . | Blocking (unlabeled) monoclonal . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 10E5 . | 290.5 . | 184.2 . | 312.8 . | 312.2 . | 312.6 . | 7E3 . | AP2 . | AP3 . | 330.1 . | |
| 10E5 | 100.0 | 99.6 | 24.8 | 37.2 | 10.9 | 4.5 | 3.7 | 23.9 | 5.7 | 8.6 |
| 290.5 | 99.6 | 100.0 | 12.5 | 14.1 | 5.3 | 0.5 | 8.6 | 25.7 | 4.0 | 15.4 |
| 184.2 | 29.7 | 24.9 | 100.0 | 99.0 | 23.4 | 14.6 | 28.9 | 36.8 | 5.2 | 33.4 |
| 312.8 | 43.4 | 37.1 | 99.7 | 100.0 | 16.4 | 15.6 | 17.9 | 52.2 | 21.7 | 0.0 |
| 312.2 | 60.7 | 74.2 | 40.7 | 20.8 | 100.0 | 100.0 | 43.2 | 3.1 | 41.6 | 18.4 |
| 312.6 | 40.5 | 43.0 | 12.4 | 5.3 | 100.0 | 100.0 | 7.0 | 20.3 | 20.8 | 3.7 |
| 7E3 | 40.6 | 42.7 | 14.3 | 16.3 | 13.6 | 18.6 | 100.0 | 99.4 | 22.1 | 8.2 |
| AP2 | 18.1 | 34.8 | 29.4 | 62.8 | 16.8 | 7.7 | 98.2 | 100.0 | 7.7 | 0.0 |
| AP3 | 10.1 | 19.2 | 25.1 | 43.7 | 14.5 | 26.1 | 34.6 | 12.4 | 100.0 | 100.0 |
| 330.1 | 6.9 | 4.5 | 7.8 | 24.0 | 10.0 | 14.1 | 25.1 | 10.3 | 99.6 | 100.0 |
| . | Blocking (unlabeled) monoclonal . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 10E5 . | 290.5 . | 184.2 . | 312.8 . | 312.2 . | 312.6 . | 7E3 . | AP2 . | AP3 . | 330.1 . | |
| 10E5 | 100.0 | 99.6 | 24.8 | 37.2 | 10.9 | 4.5 | 3.7 | 23.9 | 5.7 | 8.6 |
| 290.5 | 99.6 | 100.0 | 12.5 | 14.1 | 5.3 | 0.5 | 8.6 | 25.7 | 4.0 | 15.4 |
| 184.2 | 29.7 | 24.9 | 100.0 | 99.0 | 23.4 | 14.6 | 28.9 | 36.8 | 5.2 | 33.4 |
| 312.8 | 43.4 | 37.1 | 99.7 | 100.0 | 16.4 | 15.6 | 17.9 | 52.2 | 21.7 | 0.0 |
| 312.2 | 60.7 | 74.2 | 40.7 | 20.8 | 100.0 | 100.0 | 43.2 | 3.1 | 41.6 | 18.4 |
| 312.6 | 40.5 | 43.0 | 12.4 | 5.3 | 100.0 | 100.0 | 7.0 | 20.3 | 20.8 | 3.7 |
| 7E3 | 40.6 | 42.7 | 14.3 | 16.3 | 13.6 | 18.6 | 100.0 | 99.4 | 22.1 | 8.2 |
| AP2 | 18.1 | 34.8 | 29.4 | 62.8 | 16.8 | 7.7 | 98.2 | 100.0 | 7.7 | 0.0 |
| AP3 | 10.1 | 19.2 | 25.1 | 43.7 | 14.5 | 26.1 | 34.6 | 12.4 | 100.0 | 100.0 |
| 330.1 | 6.9 | 4.5 | 7.8 | 24.0 | 10.0 | 14.1 | 25.1 | 10.3 | 99.6 | 100.0 |
Platelets were incubated with saturating quantities of the unlabeled (blocking) mAbs listed in the first horizontal row or with buffer alone. After a single wash with buffer solution, biotinylated mAbs listed in the first column were added and bound Ab was detected with PE-labeled streptavidin using flow cytometry. Values shown (average of triplicate determinations) indicate the extent to which “blocking” mAbs reduced the signal obtained with unblocked platelets (expressed as percentage of inhibition).
Bold indicates reactions inhibited by 90% or more; and underlined values, reactions inhibited by 45% or more.
Ribbon diagram of the αIIb/β3 head region indicating possible binding footprints of 5 mAb pairs. Structural coordinates were from Protein Data Bank (3FCS)19 viewed with WebLab viewer Pro 3.7 (Molecular Simulations Inc). Amino acid residues 606-959 of αIIb and 561-690 of β3 were omitted for the sake of clarity. Alpha helical structures are red, and β sheets are turquoise. Solid ovals represent likely footprints of mAbs 10E5, 7E3, AP3, and RGD peptide based on prior crystallographic17 or mutagenic31,32 studies. Dashed ovals represent likely binding footprints of 7 other mAbs inferred from their reactions with chimeric αIIb/β3 constructs and from blocking studies summarized in Table 3.
Ribbon diagram of the αIIb/β3 head region indicating possible binding footprints of 5 mAb pairs. Structural coordinates were from Protein Data Bank (3FCS)19 viewed with WebLab viewer Pro 3.7 (Molecular Simulations Inc). Amino acid residues 606-959 of αIIb and 561-690 of β3 were omitted for the sake of clarity. Alpha helical structures are red, and β sheets are turquoise. Solid ovals represent likely footprints of mAbs 10E5, 7E3, AP3, and RGD peptide based on prior crystallographic17 or mutagenic31,32 studies. Dashed ovals represent likely binding footprints of 7 other mAbs inferred from their reactions with chimeric αIIb/β3 constructs and from blocking studies summarized in Table 3.
The 5 pairs of mAbs differed from one another in their ability to block binding of eptifibatide and tirofiban-dependent human Abs to αIIb/β3
We then examined the ability of the 10 mAbs to inhibit drug-dependent binding to αIIb/β3 of 6 human drug-dependent Abs, 3 specific for eptifibatide (E1, E2, E3), and 3 specific for tirofiban (T1, T2, T3) that were available in quantities sufficient for these studies. Results are shown in Table 4. mAb 7E3, which recognizes a peptide loop in β3 very close to the RGD-binding site,31 completely blocked each human Ab. The same pattern of inhibition was obtained with the Fab fragment, abciximab, derived from 7E331 (data not shown). mAb AP2, which competes with 7E3 for binding (Table 3), also inhibited each Ab by 34%-96%. However, patterns of inhibition distinctly different from that of mAbs 7E3 and AP2 were obtained with the other 8 mAbs. For example, the mAb pair 10E5/290.5 inhibited Abs E1 and E2 almost totally, and inhibited E3 by 50%-60% but had little effect on the tirofiban-dependent Abs T1, T2, and T3. In contrast, the 312.2/312.6 pair inhibited E2 and E3 by 68%-87% and inhibited T1 by 41%-45% but had little effect on E1, T2, or T3. Several contrasting patterns of inhibition are illustrated in Figure 5.
Inhibition of eptifibatide- and tirofiban-dependent patient Abs by GPIIb- and GPIIIa-specific monoclonals
| Patient Abs . | mAbs . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GPIIb-specific . | GPIIIa-specific . | |||||||||
| 10E5 . | 290.5 . | 184.2 . | 312.8 . | 312.2 . | 312.6 . | 7E3 . | AP2 . | AP3 . | 330.1 . | |
| E1 | 97.0 | 96.6 | 19.9 | 17.5 | 25.9 | 27.5 | 98.6 | 48.0 | 40.0 | 49.7 |
| E2 | 94.1 | 94.5 | 14.5 | 11.2 | 82.8 | 87.4 | 99.4 | 42.3 | 46.7 | 49.9 |
| E3 | 59.8 | 50.3 | 49.5 | 47.5 | 68.3 | 74.6 | 99.5 | 75.4 | 80.9 | 75.0 |
| T1 | 26.8 | 24.6 | 6.5 | 45.2 | 41.1 | 44.6 | 93.2 | 44.9 | 40.9 | 30.0 |
| T2 | 18.7 | 0.0 | 34.2 | 3.1 | 17.6 | 25.6 | 97.5 | 95.8 | 17.8 | 4.5 |
| T3 | 16.1 | 11.1 | 5.7 | 22.9 | 17.4 | 29.3 | 93.7 | 34.4 | 44.8 | 58.2 |
| Patient Abs . | mAbs . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GPIIb-specific . | GPIIIa-specific . | |||||||||
| 10E5 . | 290.5 . | 184.2 . | 312.8 . | 312.2 . | 312.6 . | 7E3 . | AP2 . | AP3 . | 330.1 . | |
| E1 | 97.0 | 96.6 | 19.9 | 17.5 | 25.9 | 27.5 | 98.6 | 48.0 | 40.0 | 49.7 |
| E2 | 94.1 | 94.5 | 14.5 | 11.2 | 82.8 | 87.4 | 99.4 | 42.3 | 46.7 | 49.9 |
| E3 | 59.8 | 50.3 | 49.5 | 47.5 | 68.3 | 74.6 | 99.5 | 75.4 | 80.9 | 75.0 |
| T1 | 26.8 | 24.6 | 6.5 | 45.2 | 41.1 | 44.6 | 93.2 | 44.9 | 40.9 | 30.0 |
| T2 | 18.7 | 0.0 | 34.2 | 3.1 | 17.6 | 25.6 | 97.5 | 95.8 | 17.8 | 4.5 |
| T3 | 16.1 | 11.1 | 5.7 | 22.9 | 17.4 | 29.3 | 93.7 | 34.4 | 44.8 | 58.2 |
Platelets were treated with tirofiban or eptifibatide and were then incubated with saturating quantities of the indicated mAbs or with buffer alone. Abs from patients who experienced thrombocytopenia after treatment with eptifibatide (E1, E2, E3) or tirofiban (T1, T2, T3) were then added, and platelet-bound human IgG was measured by flow cytometry. Values shown (average of triplicate determinations) indicate the extent to which each mAb reduced the signal obtained with platelets treated with eptifibatide or tirofiban alone (expressed as percentage of inhibition).
Bold values indicate reactions inhibited by 90% or more; and underlined values, reactions inhibited by 45% or more.
mAb pairs produce different patterns of inhibition when tested for their ability to block binding of patient Abs. Platelets pretreated with eptifibatide or tirofiban were incubated with saturating quantities of one of 10 “blocking” mAbs. The signal (median fluorescence intensity) obtained with a patient Ab was then determined by flow cytometry, and was expressed as a percentage of the signal obtained in the absence of a blocking mAb (percentage of relative binding). (Top panel) The eptifibatide-dependent Ab E2 was blocked almost completely by mAb pairs 10E5/290.5 and 312.2/312.8 specific for the αIIb β propeller and by 7E3, but was relatively unaffected by the other 5 mAbs. (Bottom panel) In contrast, the tirofiban-dependent Ab T2 was completely blocked by 7E3/AP2 but not by the other 4 pairs of monoclonals. Values shown are the average of triplicate measurements. Brackets indicate ± 1.0 SD.
mAb pairs produce different patterns of inhibition when tested for their ability to block binding of patient Abs. Platelets pretreated with eptifibatide or tirofiban were incubated with saturating quantities of one of 10 “blocking” mAbs. The signal (median fluorescence intensity) obtained with a patient Ab was then determined by flow cytometry, and was expressed as a percentage of the signal obtained in the absence of a blocking mAb (percentage of relative binding). (Top panel) The eptifibatide-dependent Ab E2 was blocked almost completely by mAb pairs 10E5/290.5 and 312.2/312.8 specific for the αIIb β propeller and by 7E3, but was relatively unaffected by the other 5 mAbs. (Bottom panel) In contrast, the tirofiban-dependent Ab T2 was completely blocked by 7E3/AP2 but not by the other 4 pairs of monoclonals. Values shown are the average of triplicate measurements. Brackets indicate ± 1.0 SD.
Discussion
Acute, severe thrombocytopenia was recognized in patients treated with tirofiban, eptifibatide, and other RGD-mimetic platelet inhibitors soon after this class of drugs was introduced.5 When serologic studies showed that this complication is caused by Abs specific for αIIb/β3 in a complex with drug,7,9,10,26 it was suggested that the human Abs, like LIBS-specific mAbs, might recognize epitopes created by conformational changes induced in the integrin by ligand.5,7,25 Our studies were designed to examine this possibility.
Of 43 patient Abs studied, 13 reacted with platelets pretreated with a ligand-mimetic drug in addition to the one that caused thrombocytopenia and/or with RGD-treated platelets (Table 1), consistent with the possibility that this subset of Abs is specific for LIBS-like epitopes induced in common by various ligands. However, 30 patient Abs recognized the integrin only in a complex with the drug that caused thrombocytopenia. This behavior differed from that of 18 different LIBS-specific mAbs, which reacted equally well with αIIb/β3 in a complex with eptifibatide, tirofiban, and RGD peptide under our experimental conditions (Figure 2). In its resting state, αIIb/β3 assumes a bent configuration in which the head region is in close proximity to the lower leg pieces17,37 and epitopes recognized by LIBS mAbs are relatively inaccessible.17,19,38 LIBS Ab binding is favored by ligand-induced rearrangements leading to exposure of these sites.19,39,–41 In general, LIBS-specific mAbs tend to recognize epitopes in the αIIb calf domains, the β3 hybrid domain, and the more distal PSI, epidermal growth factor–like, and cystatin domains of β3 (Table 2) that are totally or partially sequestered in the resting state.17,24 The 18 LIBS mAbs we studied fit this pattern. The contrasting reaction patterns of LIBS mAbs and patient Abs against αIIb/β3 occupied by different ligands (Figure 2) suggested that the latter were not specific for classic LIBS determinants.
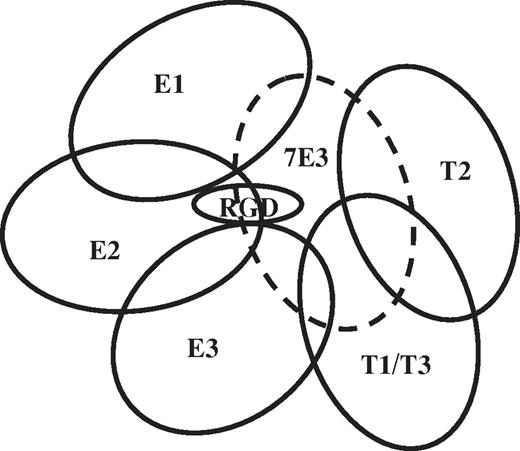
Drug-dependent binding of each of the patient Abs was blocked by mAb 7E3 and by abciximab, a Fab fragment derived from 7E3, which recognizes the specificity-determining loop of β3-β A and adjacent amino acid residues close to the metal ion-dependent adhesion site and the RGD recognition site.32,33 A trivial explanation for this finding would be that abciximab and 7E3 dislodge drug from its binding site. However, abciximab has been shown not to compete with low-molecular-weight RGD-mimetic inhibitors for binding to the integrin42 or to displace bound inhibitor.43 Moreover, we found that RGD peptide induces the epitopes recognized by LIBS mAbs PMI-1, LIBS6, 319.1, 322.5, and AP5 even when abciximab is bound to the integrin (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). To characterize the binding sites for patient Abs more fully, we defined the ability of mAbs specific for subdomains of the αIIb/β3 head structure distinct from the abciximab-binding site to interfere with drug-dependent binding of 6 representative patient Abs, 3 eptifibatide- and 3 tirofiban-specific. As shown in Table 4 and Figure 5, distinctly different patterns of inhibition were observed. For example, Ab E1 was completely blocked by the 10E5/290.5 mAb pair specific for the cap subdomain of the αIIb β propeller17,28 but was relatively unaffected by 2 other mAb pairs (184.2/312.8 and 312.2/312.6) specific for other sites on the β propeller. In contrast, Ab E2 was blocked by both the 10E5/290.5 and 312.2/312.6 mAb pairs. A plausible interpretation for these findings is that Abs E1 and E2 recognize sites on the β propeller located close to the cap subdomain but that their footprints are slightly different, enabling E2 to be blocked by both of the mAb pairs and E1 to be blocked only by 10E5/290.5. In contrast to Abs E1 and E2, the tirofiban-dependent Abs T1 and T3 were blocked completely only by 7E3 and a third tirofiban-dependent Ab, T2, was blocked by both 7E3 and AP2. These findings suggest that T1, T2, and T3 occupy at least 2 distinct footprints on the β3-β A domain. A schematic representation of the likely spatial relationships of binding sites for the 6 human Abs is shown in Figure 6.
Schematic representation of likely “footprints” for patient Abs E1, E2, and E3 and T1, T2, and T3 based on inhibition of Ab binding by mAbs specific for subdomains of the αIIb and β3 head regions. The footprint for mAb 7E3 (and abciximab) is shown by the dashed oval. “RGD” indicates approximate location of the binding site for RGD peptide, eptifibatide, and tirofiban.
Schematic representation of likely “footprints” for patient Abs E1, E2, and E3 and T1, T2, and T3 based on inhibition of Ab binding by mAbs specific for subdomains of the αIIb and β3 head regions. The footprint for mAb 7E3 (and abciximab) is shown by the dashed oval. “RGD” indicates approximate location of the binding site for RGD peptide, eptifibatide, and tirofiban.
Evidence suggests that immune thrombocytopenia associated with drugs like quinine and many others is caused by Abs that directly recognize some element of drug structure and bind to their target in such a way that the drug is trapped at the Ab-Ag interface.11,13,16 The finding that mAb 7E3 and its Fab fragment abciximab, which bind to an epitope immediately adjacent to the RGD recognition site,33 prevented each of 6 patient Abs from binding to αIIb/β3 occupied by ligand (Table 4) is consistent with the possibility that the patient Abs studied, like quinine-dependent Abs, recognize bound drug and adjacent amino acid residues. However, other findings argue against this possibility. As shown in Table 1, 13 of 43 patient Abs recognized αIIb/β3 occupied by a ligand other than the drug that caused thrombocytopenia. Cross-reactions of 5 Abs from patients with eptifibatide-induced thrombocytopenia against αIIb/β3 occupied by tirofiban are especially noteworthy because these 2 drugs have very different structures (Figure 3), making it very unlikely that direct contact between Ab and drug is involved in Ab recognition. Moreover, most of the patients had no prior exposure to the drug that caused thrombocytopenia. Substances like tirofiban and eptifibatide are not found in nature and it is extremely unlikely that patients would have potent, naturally occurring Abs that recognize these structures. Finally, because RGD-mimetic drugs all occupy the same binding pocket on the integrin,19 footprints of the patient Abs although not necessarily identical, would be tightly localized by a requirement that they contact drug itself. However, blocking studies with various mAbs (Table 4) indicate that the Ab footprints are distinctly different and that not all are overlapping despite being clustered about the 7E3-binding site. Finally, RGD and RGD-mimetic drugs are known to induce significant structural changes in αIIb/β3 that can be immunogenic,21,24,29 whereas drugs like quinine appear to have no preferred binding site15,20,44 and are not known to induce structural changes. These considerations favor the possibility that Abs causing thrombocytopenia in patients treated with ligand-mimetic drugs are distinctly different from those found in patients with quinine-associated thrombocytopenia, being specific for structural changes (neoepitopes, MIBS) created adjacent to the RGD recognition site when ligand binds. Findings shown in Figure 2 and the blocking studies summarized in Table 4 indicate that the epitopes recognized are distinct from classic LIBS determinants, both in their locations and in being largely drug-specific.
Other possible interpretations require discussion. Intact mAbs were used for the blocking studies; the Fc component of an intact mAb, although flexible, might interfere with binding of another Ab (mAb or patient) some distance away from its binding site. This seems unlikely to have influenced our findings because the 5 pairs of mAbs used for blocking interfered only with each other and not with any of the other 4 pairs (Table 3). A second concern is that the blocking mAbs could, like LIBS-specific mAbs, have induced conformational changes in the integrin that modified epitopes recognized by one or more patient Abs. LIBS mAbs do induce conformational changes in αIIb/β3 by shifting its equilibrium toward an “active” conformation.23,24,29 However, none of the blocking mAbs or 7E3/abciximab had LIBS activity (Table 2) and, as noted, each affected the binding of only one other mAb (Table 3). Third, ligand-induced structural change in integrins is a dynamic process, possibly involving several intermediate conformations,17,18 any one of which might conceivably be recognized preferentially by some patient Abs. This is unlikely to have influenced our findings because saturating quantities of ligand-mimetic drugs were maintained in the reaction mixtures throughout the assays to maximize activation. Moreover, we performed a study in which platelets were treated with tirofiban and eptifibatide at concentrations ranging from subthreshold (0.005 μg/mL) to supermaximal (4.0 μg/mL) and found that patient Ab specificities were uniform throughout the entire concentration range (data not shown). Finally, it is possible that more than one ligand-dependent Ab was present in some patient samples, which could explain why patient Ab E3 was at least partially inhibited by each of the blocking mAbs (Table 4). Even if this were the case, the distinctly different inhibitory patterns obtained with the panel of blocking mAbs (Table 4, Figure 5) indicate that the individual patient Abs are recognizing distinctly different epitopes in the integrin head region.
Further studies are needed to define the structural basis for drug-induced epitopes in the αIIb/β3 head region recognized by eptifibatide- and tirofiban-dependent patient Abs. RGD peptide and RGD-mimetic drugs bind to a defined pocket in the head region of αIIb/β3 where a basic nitrogen of the ligand forms a hydrogen bond with Asp224 of αIIb and an acidic group contacts the Mg++ in the β3 metal ion-dependent adhesion site.17,20 Eptifibatide, tirofiban, and RGDW peptide differ slightly in the distance between their positive and negative charges,17,20 and mimetic compounds differ in the flexibility of covalent bonds that connect the Arg-mimetic group and Asp-mimetic groups. For example, tirofiban has a more flexible alkyl-oxy chain connecting to its piperidine moiety than the homoarginine connecting to the guanidinium group in eptifibatide. Varying degrees of flexibility among ligand mimetics could influence allosteric effects on integrin conformation around the ligand-binding pocket, leading to subtle ligand-mimetic–specific structural changes in the surrounding regions of the integrin and creating ligand mimetic-specific epitopes (MIBS) recognized by patient Abs. Xiao et al resolved the crystal structures of constructs comprising αIIb residues 1-452 and β3 residues 1-440 in a complex with eptifibatide and tirofiban17 and compared these structures with that of unliganded αv/β317,45 . Movements up to 5.3 Å in magnitude resulting from ligation were observed in various β strands and helical structures of the β3-β A domain but ligand-specific structural differences were not described.17 A possible explanation for this is that patient Abs might recognize (and perhaps stabilize) drug-specific structural isomers that are present in aqueous solution but are lost on crystallization. We recently succeeded in producing a murine mAb that recognizes αIIb/β3 only in a complex with eptifibatide and thus closely mimics the behavior of Abs that cause thrombocytopenia in patients treated with this drug. Further studies with this mAb may enable identification of a ligand-specific epitope at the structural level.
Thrombocytopenia in patients treated with RGD ligand-mimetic platelet inhibitors can be life-threatening,5,7,46,47 and drugs that are equally effective but do not cause thrombocytopenia would be desirable. Zhu and coworkers recently described an inhibitor designated RUC-1, which like eptifibatide and tirofiban, blocks binding of fibrinogen to the activated integrin.20 However, RUC-1 contacts only Asp224 and adjacent αIIb amino acid residues and fails to induce the “open” (activated) integrin configuration or the epitope recognized by the LIBS mAb AP5.20 It seems possible that drugs of this type may be incapable of inducing structural changes recognized by ligand-dependent human Abs and will therefore be much less likely to trigger thrombocytopenia.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Platelet and Neutrophil Immunology Laboratory (BloodCenter of Wisconsin, Brian Curtis, PhD, Director) for patient materials, Trudy Holyst (Peptide Core Laboratory) for peptide synthesis, and Renee Bordeaux and Erin Yttre (Hybridoma Core Laboratory) for production of mAbs.
This work was supported by grants HL-13629 and HL-44612 from the National Heart, Lung, and Blood Institute.
National Institutes of Health
Authorship
Contribution: D.W.B. designed the study, provided oversight of experiments, carried out selected studies, and wrote the manuscript; M.R. performed laboratory studies and contributed to experimental design; J.Z. aided in the interpretation of results and in the writing of the manuscript; and R.H.A. designed the study, provided oversight of experimental work, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel W. Bougie, BloodCenter of Wisconsin, Blood Research Institute, PO Box 2178, Milwaukee WI 53201; e-mail: dan.bougie@bcw.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal