Abstract
Regulation of V(D)J recombination events at immunoglobulin (Ig) and T-cell receptor loci in lymphoid cells is complex and achieved via changes in substrate accessibility. Various studies over the last year have identified the DNA-binding zinc-finger protein CCCTC-binding factor (CTCF) as a crucial regulator of long-range chromatin interactions. CTCF often controls specific interactions by preventing inappropriate communication between neighboring regulatory elements or independent chromatin domains. Although recent gene targeting experiments demonstrated that the presence of the CTCF protein is not required for the process of V(D)J recombination per se, CTCF turned out to be essential to control order, lineage specificity and to balance the Ig V gene repertoire. Moreover, CTCF was shown to restrict activity of κ enhancer elements to the Ig κ locus. In this review, we discuss CTCF function in the regulation of V(D)J recombination on the basis of established knowledge on CTCF-mediated chromatin loop domains in various other loci, including the imprinted H19-Igf2 locus as well as the complex β-globin, MHC class II and IFN-γ loci. Moreover, we discuss that loss of CTCF-mediated restriction of enhancer activity may well contribute to oncogenic activation, when in chromosomal translocations Ig enhancer elements and oncogenes appear in a novel genomic context.
Introduction
Development of B and T lymphocytes requires proper spatial and temporal control of gene expression. This is achieved through the coordinate binding of lineage-specific transcription factors to cis-regulatory elements. In many developmentally regulated gene clusters (eg, the antigen receptor and cytokine loci), gene regulation is particularly complex and involves cell type-specific interactions over large genomic distances between gene promoters and remote enhancers, silencers, or locus control regions (LCRs).1,2 Regulation of higher-order chromatin structures is mediated by protein complexes composed of lineage-specific and ubiquitously expressed nuclear factors. It has recently become apparent that the DNA-binding protein CCCTC-binding factor (CTCF) is a leading candidate for the regulation of gene expression at complex loci during lymphoid differentiation.
General introduction into CTCF biology
CTCF is a ubiquitously expressed and highly conserved 11-zinc finger protein that was originally identified as a negative transcriptional regulator of the c-Myc oncogene.3 CTCF has since been implicated in a variety of regulatory functions, including transcriptional activation and repression, enhancer blocking, gene insulation, gene silencing, genomic imprinting, X chromosome inactivation, and long-range chromatin interactions.4
CTCF occupancy studies in various human and murine cell types, including embryonic stem cells and T lymphocytes, revealed approximately 14 000 to approximately 40 000 CTCF-binding sites, sharing a 15- to 20-bp core consensus sequence.5–8 Although the genome-wide distribution of these sites strongly correlates with gene density, it suggests a role for CTCF distinct from that of canonical transcription factors: binding patterns do not correlate with transcriptional start sites and do not appear to predict cell-specific gene expression levels. Domains with few or no CTCF-binding sites tend to include clusters of transcriptionally coregulated genes that are often flanked by CTCF-binding sites. Conversely, CTCF-rich regions contain genes displaying extensive alternative promoter usage, including the T-cell receptor (TCR) and immunoglobulin (Ig) gene loci.4
Many studies have suggested that CTCF is the archetypal vertebrate protein that binds insulator elements. Insulators affect gene expression by blocking inappropriate communication between neighboring regulatory elements (enhancer-blocking activity, when positioned between enhancer and promoter) and by preventing the spread of repressive heterochromatin (barrier function).9 Based on studies showing that a 5′ boundary element of the chicken β-globin locus serves as an insulator in erythroid cells and protects against position effect in Drosophila, Chung et al were the first to hypothesize that insulator function may depend on interactions between distant elements that result in looping out of intervening sequences.10 The first association of CTCF with enhancer blocking activity was proposed on the basis of CTCF-binding to the DNAse hypersensitive site (5′HS4) upstream of the chicken β-globin locus and another insulator sequence at the 3′ end.11 Based on functional analysis in transgenic systems, CTCF has been demonstrated to mediate insulation at several model loci discussed below in detail. In addition, computational analyses showing CTCF binding to highly conserved noncoding elements across 12 mammalian species provided evidence supporting a global role for CTCF as an insulator protein.12
CTCF-mediated gene insulation via chromatin loops in model loci
The H19/Igf2 locus
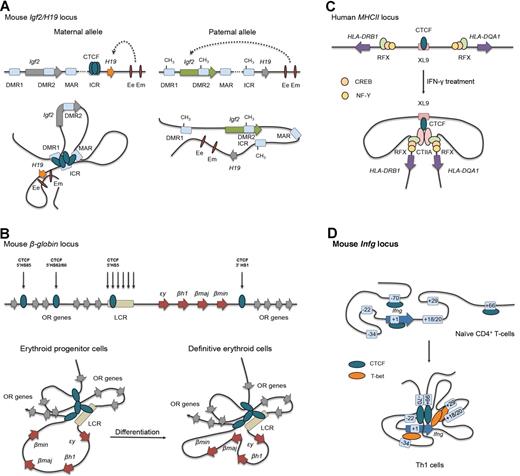
CTCF has been implicated in genomic imprinting, the epigenetically regulated process that causes genes to be expressed in a parental-origin–specific manner rather than from both chromosomes.14 In the H19/Igf2 locus, an imprinting control region, containing 4 CTCF-binding sites, is located immediately upstream of the H19 gene and is essential for regulation of imprinted expression of the H19 and Igf2 genes (Figure 1A). On the maternally inherited allele, the imprinting control region is unmethylated, allowing for CTCF binding and the formation of a tightly coiled chromatin loop insulating the Igf2 gene and preventing its promoter to access the enhancers downstream of H19. On the paternal allele, however, the imprinting control region is methylated, which abrogates CTCF binding, resulting in functional interactions between the Igf2 promoters and the enhancer elements (Figure 1A). Moreover, loss of CTCF binding is also associated with H19 promoter methylation leading to H19 gene silencing on the paternal allele.
CTCF-mediated gene insulation via chromatin looping at several model loci. (A) Allele-specific, DNA methylation-sensitive binding of CTCF and chromatin looping account for parent-of-origin H19 and Igf2 gene expression patterns during mouse development. DMR indicates differentially methylated region; Ee, endodermal tissue enhancer; ICR, imprinting control region; and Em, mesodermal tissue enhancer. (B) In progenitor cells, mouse globin genes embedded within the olfactory receptor (OR) gene cluster are looped out from the CTCF-mediated chromatin hub, and ϵy and βh1 are expressed. On differentiation, dynamic interactions between the definitive erythroid genes β-major (βmaj) and β-minor (βmin) and the LCR are observed. HS indicates hypersensitive site. (C) IFN-γ treatment induces cotranscription of the human HLA-DRB1 and HLA-DQA1 genes in nonexpressing cell types by up-regulating the coactivator CIITA. Gene activation results from heterodimer formation between CIITA and the transcription factors RFX, CREB, and NF-Y bound to the inactive promoter of the MHC class II genes and the CTCF-bound XL9 enhancer. (D) On mouse Th1 cell differentiation, CTCF cooperates with T-bet to juxtapose distal regulatory elements, including the conserved noncoding sequence (CNS)–34 enhancer, to the Ifng gene. This results in a Th1-cell–specific chromatin structure at the Ifng locus for efficient IFN-γ transcription.
CTCF-mediated gene insulation via chromatin looping at several model loci. (A) Allele-specific, DNA methylation-sensitive binding of CTCF and chromatin looping account for parent-of-origin H19 and Igf2 gene expression patterns during mouse development. DMR indicates differentially methylated region; Ee, endodermal tissue enhancer; ICR, imprinting control region; and Em, mesodermal tissue enhancer. (B) In progenitor cells, mouse globin genes embedded within the olfactory receptor (OR) gene cluster are looped out from the CTCF-mediated chromatin hub, and ϵy and βh1 are expressed. On differentiation, dynamic interactions between the definitive erythroid genes β-major (βmaj) and β-minor (βmin) and the LCR are observed. HS indicates hypersensitive site. (C) IFN-γ treatment induces cotranscription of the human HLA-DRB1 and HLA-DQA1 genes in nonexpressing cell types by up-regulating the coactivator CIITA. Gene activation results from heterodimer formation between CIITA and the transcription factors RFX, CREB, and NF-Y bound to the inactive promoter of the MHC class II genes and the CTCF-bound XL9 enhancer. (D) On mouse Th1 cell differentiation, CTCF cooperates with T-bet to juxtapose distal regulatory elements, including the conserved noncoding sequence (CNS)–34 enhancer, to the Ifng gene. This results in a Th1-cell–specific chromatin structure at the Ifng locus for efficient IFN-γ transcription.
The β-globin locus
CTCF-mediated contacts are associated with transcriptional activation of genes in the extensively characterized β-globin locus in erythroid cells.15 Developmentally regulated expression of individual globin genes is controlled by many cis-acting elements, the most important of which are located in the upstream LCR (Figure 1B). CTCF binds to 3 DNAse hypersensitive sites upstream in the LCR and 1 downstream (3′HS1) of the mouse β-globin locus in a cell type–specific manner.16 During erythroid differentiation, CTCF-bound regulatory sequences throughout the locus come into close spatial proximity to form an “active chromatin hub,” as was shown by chromosome conformation capture (3C) technology detecting long-range DNA interactions.15–17 Formation of this “active chromatin hub” allows stage-specific interaction of active globin genes with the LCR and concomitant looping-out of transcriptionally silent isoforms (Figure 1B). Conditional CTCF deletion or mutation of the 3′HS1 CTCF-binding site resulted in disruption of CTCF contacts in erythroid progenitors but surprisingly had no effects on kinetics or levels of globin expression during erythroid differentiation.16 Because of the high density of CTCF-binding sites in the β-globin locus, this finding is explained by redundancy of chromatin contacts, which may not be abrogated by mutations in a single regulatory element. Genetic disruption of CTCF-binding to 3′HS1 also did not affect transcription of the neighboring olfactory receptor genes.16 Collectively, these findings indicate that correct globin gene expression can be achieved by different CTCF-dependent loops and thus by different chromatin conformations.
The MHC class II locus
CTCF organizes the MHC class II locus into a conformation that rearranges in the presence of the crucial non-DNA–binding transcriptional coactivator CIITA. In the human locus, CTCF binds to the intergenic enhancer XL9 that coregulates expression of the HLA-DRB1 and HLA-DQA1 genes18,19 (Figure 1C). Induction of CIITA in non-MHC class II–expressing cells results in the initiation of interactions between XL9 and the promoters upstream of HLA-DRB1 and HLA-DQA. Hereby chromatin loop formation depends on XL9-bound CTCF, CIITA, as well as the transcription factor RFX, which is constitutively bound to regulatory sequences within the HLA-DRB1 and HLA-DQA1 proximal promoters. Therefore, in the human MHC class II locus, CTCF is involved in the control of gene expression through loop formation via hetero-multimerization.
The Ifng and Th2 cytokine loci
The key transcription factor of T helper-1 (Th1) cells, T-bet, promotes the expression of IFN-γ in part by facilitating CTCF binding in the Ifng locus.20 CTCF acts to establish a Th1 cell–specific Ifng locus architecture, which helps to drive the juxtaposition of T-bet–binding enhancers, the flanking CTCF-binding elements, and CTCF sites in the first exon of the Ifng gene (Figure 1D). Cooperation between CTCF and T-bet is required for robust Th1 cell–specific IFN-γ expression,20 which is also supported by our finding of an approximately 50% reduction in IFN-γ–producing cells when naive CTCF-deficient CD4+ T cells were cultured under Th1-polarized conditions.21 CTCF may act in a similar fashion in the Th2 cytokine locus containing the genes for IL-4, IL-5, and IL-13. Indeed, Th2 cytokines were strongly reduced in CTCF-deficient T lymphocytes cultured under Th2-polarized conditions, despite proper induction of the key Th2 transcriptional regulators Gata3 and Satb1.21
CTCF as a genome-wide organizer of chromatin architecture
Its capacity to form dimers and to participate in multiprotein complexes allows CTCF to organize long-range 3-dimensional looping of genomic regions. Interestingly, genome-wide identification of binding sites for CTCF and for cohesin, the protein complex responsible for sister chromatid cohesion, showed that a large proportion of these sites are shared.22,23 CTCF enables cohesin recruitment to specific sites, and cohesin is required for CTCF-mediated long-range intrachromosomal interaction (eg, at the H19/Igf2, Ifng, or MHC class II loci).18,22,24 Because cohesin forms a large ring structure that holds sister chromatids together during mitosis, it is conceivable that cohesin may function to stabilize loops that have been initiated by CTCF.
Several lines of evidence support a role for CTCF as a crucial genome-wide organizer of chromatin architecture: (1) the high number of CTCF-binding sites in the genome, both near transcription start sites, in exons, in introns, and in intergenic regions4 ; (2) the essential function of CTCF in the orchestration of long-range chromatin interaction at model loci; (3) CTCF has many binding partners, including transcription factors, chromatin-modifying proteins, the nucleolar protein nucleophosmin, RNA polymerases, and Smad signaling proteins4,25 ; and (4) the recent genome-wide identification of CTCF-mediated chromatin interactions implicates CTCF in the global organization of chromatin.26,27 Based on combining CTCF binding and mapping of histone modifications, CTCF often demarcates the boundaries of repressed chromatin associated with nuclear lamina domains.26 Recent 3C-based analyses showed that indeed CTCF-mediated loops separate H3K4 active domains from H3K27me3 repressive domains, which coincide with lamina domains.27 These findings demonstrate that CTCF is involved in the division of the mammalian genome into large, discrete domains that are units of chromosome organization within the nucleus.
Regulation of gene rearrangements in antigen receptor loci
Lymphocyte development is a stepwise process involving ordered rearrangement of variable (V), diversity (D), and joining (J) antigen receptor gene segments, also known as V(D)J recombination.1,28 Because these gene segments are spread over several megabases of the genome, it is obvious that locus compaction and looping are essential for the site-specific DNA recombination reactions that create a diverse repertoire of Ig and TCRs. The question how V(D)J recombination is regulated over great distances is, however, largely unanswered. Various studies over the last year have identified CTCF as a crucial regulator of long-range chromatin interactions in antigen receptor loci.
The lymphoid-specific recombination activation gene endonucleases Rag1 and Rag2 cooperate to induce double-strand DNA breaks at recombination signal sequences flanking the V, D, and J gene segments, which are subsequently joined by the nonhomologous end-joining DNA-repair machinery.1,28 Cell cycle–dependent regulation of Rag expression during lymphocyte development is critical for preventing Rag-mediated DNA breaks leading to translocation in lymphocytes undergoing active V(D)J recombination.29 B-cell development is initiated in pro-B cells in the bone marrow by sequential D-to-JH and V-to-DJH joining. Successful rearrangement of the Ig heavy chain (Igh) locus results in IgH μ protein deposition on the surface of pre-B cells, together with the preexisting surrogate light chain proteins λ5 and VpreB, as the pre-B cell receptor (pre-BCR) complex. Pre-BCR signaling controls the selective expansion of cells that have a functionally rearranged IgH chain and redirects V(D) recombination activity from the Igh to the Igκ or Igλ light chain loci.30,31 Successful Igκ/Igλ chain rearrangement results in surface expression of the BCR and additional checkpoints follow to limit autoreactivity.32 Likewise, in the thymus, V(D)J recombination events in the TCR loci accompany the development of mature T cells, expressing an αβ TCR or, in a minor subset, a γδ TCR on the cell surface.32
Given that all Ig and TCR loci are rearranged by a common recombination machinery, an essential level of regulation of the V(D)J recombination process relies on lineage and developmental stage-specific changes in the accessibility of antigen receptor loci for recombination. The “accessibility hypothesis” was proposed based on the initial observation that rearranging gene segments are concomitantly undergoing germline transcription.28,33 It has been demonstrated that transcription itself can direct recombinase targeting: blockade of transcriptional elongation through the mouse TCR-α locus suppressed Vα to Jα recombination and chromatin remodeling of Jα segments.34 Germline transcription increases recombination of Jα segments located close to an upstream crucial promoter, called TEA, and prevents activation of downstream promoters through transcriptional interference.34 Accessibility is controlled by local cis-regulatory elements (promoters and enhancers) and involves subnuclear relocation, histone acetylation and/or methylation, DNA demethylation, and locus compaction.1,28 These processes are dependent on various lineage-specific transcription factors that bind to enhancer elements present in the Ig and TCR loci.35–37 Among these transcription factors, Pax5, EBF, and E2A play a central role in B-cell differentiation, as they also establish a hierarchical regulatory network for specification and commitment to the B-cell fate.38 Next to CTCF, only a few general nuclear factors that modulate antigen receptor loci topology have been identified to date, including YY1 and Ezh2.35,37,39,40 However, the precise mechanism by which these factors act in concert with lineage-specific transcription factors to regulate chromatin structure at the Ig loci remains unclear.
CTCF function in IgH locus gene rearrangement
The Igh locus in the mouse spans an approximately 2.7-Mbp region and is composed of more than 100 VH gene segments and DH, JH, and CH clusters located downstream. The locus contains 2 important regulatory elements: the intronic enhancer (iEμ), located 3′ of the JH genes, and the 3′ regulator region (3′RR; Figure 2A).28 The stepwise activation of the locus, reflected by DH-to-JH preceding V-to-DHJH rearrangement, is paralleled by germline transcription. First, Iμ transcripts are generated from iEμ, μ0 transcripts from the promoter of the most 3′ DH segments, and antisense intergenic transcripts throughout the DH and JH domains.28,41,42 After DH-to-JH recombination, noncoding sense and antisense transcripts are generated from the VH genes.28,33,42,43 Three-dimensional FISH experiments have elucidated why pro-B cells deficient for Pax5, Yy1, or Ikaros only recombine D-proximal VH genes, suggesting that locus compaction mediated by these proteins is necessary to bring distal VH genes in close proximity to DH-JH.35,37,39 Spatial distance measurements of genomic markers spanning the entire Igh locus have demonstrated that this locus is organized into compartments that contain clusters of loops or rosette-type structures separated by linkers.44 This topology compacts the VH region and should ensure that VH gene segments scattered over approximately 2.5 Mbp encounter DH-JH segments or enhancers with appropriate frequencies.
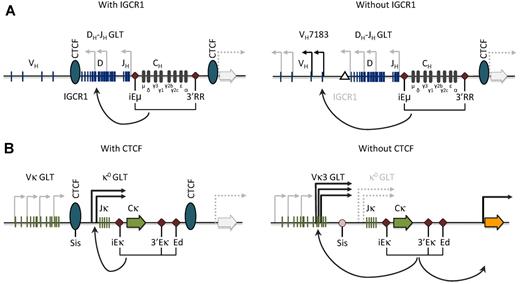
Model for CTCF function at the Igh and Igκ loci. (A) CTCF binding at the IGCR1 in the VH-D intergenic region in the Igh locus prevents inappropriate communication between the Igh enhancers and the proximal VH gene segments. Targeted IGCR1 deletion in the mouse (right) leads to antisense transcription extending into proximal VH segments of the VH7381 family and concomitant aberrant rearrangement.53,54 GLT indicates germline transcript; and IGCR1, intergenic control region 1. (B) Likewise, CTCF binding at the SIS element in the Vκ-Jκ intergenic region in the Igκ locus is important for proper germline transcription over the Jκ region, limits proximal Vκ recombination, and restricts κ enhancer interactions to the Igκ locus.49,60 Conditional deletion of Ctcf in the mouse (right) leads to increased proximal Vκ3-family transcription and rearrangement, and increased expression of the neighboring Rpia gene (orange arrow). GLT indicates germline transcript; and SIS, silencer of intergenic sequence.
Model for CTCF function at the Igh and Igκ loci. (A) CTCF binding at the IGCR1 in the VH-D intergenic region in the Igh locus prevents inappropriate communication between the Igh enhancers and the proximal VH gene segments. Targeted IGCR1 deletion in the mouse (right) leads to antisense transcription extending into proximal VH segments of the VH7381 family and concomitant aberrant rearrangement.53,54 GLT indicates germline transcript; and IGCR1, intergenic control region 1. (B) Likewise, CTCF binding at the SIS element in the Vκ-Jκ intergenic region in the Igκ locus is important for proper germline transcription over the Jκ region, limits proximal Vκ recombination, and restricts κ enhancer interactions to the Igκ locus.49,60 Conditional deletion of Ctcf in the mouse (right) leads to increased proximal Vκ3-family transcription and rearrangement, and increased expression of the neighboring Rpia gene (orange arrow). GLT indicates germline transcript; and SIS, silencer of intergenic sequence.
Emerging data have revealed CTCF as an attractive candidate for controlling Igh locus topology. CTCF and cohesin bind to multiple sites throughout the VH region,45 implicating CTCF in Igh chromatin loop formation and effective distal VH-DHJH recombination.46,47 However, CTCF occupancy in the VH region is largely unchanged between pro-B and pre-B cells,45 making it unlikely that CTCF alone accounts for Igh locus topology changes. Consistently, 3C-based experiments demonstrated that shRNA-mediated CTCF depletion only modestly affected Igh locus contraction.48 In agreement with these findings, on mb1-Cre–mediated conditional deletion of the Ctcf gene in the B-cell lineage, Igμ+ pre-B cells were still generated and both distal and proximal VH gene segments were used.49
Using chromatin immunoprecipitation coupled to microarray analysis (ChIP-chip) technology, Ebert et al have mapped Igh active histone modifications and transcription factor occupancy.50 They hereby identified 14 copies of a potential regulatory Pax5-activated intergenic repeat (PAIR) element in the distal Igh region, upstream of VH3609 genes and interspersed among VHJ558 genes. These PAIR elements contain Pax5-dependent active chromatin and binding sites for E2A, CTCF, and the cohesin component Rad21. Detailed analysis of 3 PAIR elements showed that immediately upstream of the CTCF-binding sites noncoding antisense transcription is initiated exclusively in pro-B cells. The authors hypothesize that PAIR elements mediate looping of distal VH genes in pro-B cells, whereby Pax5 orchestrates the formation of a bridging complex that mediates long-range interactions between distal and proximal VH gene regions. In contrast, on the basis of anti-CTCF Chip-loop assays, Guo et al51 presented evidence for a CTCF-mediated but iEμ-independent multiloop domain of approximately 500 kb in the 5′ VHJ558/VH3609 region containing the PAIR elements, whereby locus contraction may result from interaction between Eμ-bound39 Yy1 and Yy1 sites in the distal VH region. These interactions may be increased by heterotypic interactions with CTCF sites or by inter-PAIR interactions in the distal VH region.51 Future experiments should focus on functional characterization of the PAIR elements, which would require deletion of these proposed regulatory elements to test the effects on V(D)J recombination and VH repertoire in vivo.
In addition, CTCF-binding sites are present in the intergenic control region I (IGCR1), which is located 3 to 5 kb upstream of the most 5′ DH element (Figure 2A).45,52,53 These CTCF-binding sites have enhancer-blocking activity and mark a sharp decrease in Eμ-dependent antisense DH-JH transcription.52 Targeted IGCR1 deletion in the mouse leads to DH-JH antisense transcripts extending into proximal VH segments and concomitant aberrant rearrangement of the proximal VH gene segments in developing thymocytes.53,54 Importantly, IGCR1 balances proximal versus distal VH usage by inhibiting germline transcription and rearrangement of D-proximal VH7183 gene segments (Figure 2A). Moreover, IGCR1 maintains ordered V(D)J recombination by suppressing VH joining to D segments that are not joined to JH. Both FISH and 3C-based experiments showed that CTCF-binding sites in IGCR1 are in close spatial proximity with a region immediately downstream of 3′RR with a high density of CTCF-binding sites (Figure 2A).48,51,55 These CTCF-mediated interactions would create a domain that contains all of the DH, JH, and C region elements, as well as Eμ and 3′RR. It therefore seems probable that CTCF binding to IGCR1 prevents chromatin accessibility extending into the independently activated VH chromatin domains. This notion is supported by the findings that (1) insertion of a VH gene segment just 5′ of the DH cluster resulted in loss of ordered and lineage-specific Igh locus rearrangement,56 and (2) mice with an approximately 8-kb deletion encompassing the CTCF-binding sites downstream 3′RR showed a 2-fold increase in the usage of VH7183.57
In summary, by binding to IGCR1 and downstream of 3′RR, CTCF acts as an insulator that ensures ordered and lineage-specific V(D)J recombination at the Igh locus.
CTCF function in Igκ locus gene rearrangement
The Igκ locus in the mouse spans approximately 3.2 Mbp and contains approximately 100 Vκ gene segments, a Jκ cluster and a single Cκ region.28 Igκ locus rearrangement and expression are positively regulated by an intronic enhancer (iEκ) located between Jκand Cκ, and 2 enhancers downstream of Cκ (3′Eκ and Ed; Figure 2B). As D elements are lacking, κ light chains can be expressed on a single productive Vκ-to-Jκ recombination event and autoreactive BCR specificities can be replaced by “receptor editing,” the process of ongoing Ig light chain gene recombination.1,28 Germline κ0 transcripts from promoters located upstream of Jκ (κ0 1.1 and κ0 0.8) or Vκ gene segments indicate the accessibility over Jκ and Vκ regions, respectively.58 A recombination silencer element in the Vκ-Jκ region, called “silencer in intervening sequence” (SIS), negatively regulates Igκ rearrangement by targeting of the nonrearranging allele to centromeric heterochromatin (Figure 2B).59 Deletion of the SIS element resulted in decreased monoallelic Igκ heterochromatin localization, reduced κ0 germline transcription, and enhanced proximal Vκ usage.59,60
Next to strong CTCF binding at the 5′ and 3′ boundaries and the SIS element,49 we identified approximately 60 CTCF-binding sites in the Vκ region (in contrast to the previously reported low density of CTCF occupancy in the Igκ locus45 ). CTCF-binding sites were not evenly distributed over the Igκ locus. Detailed analysis of long-range interactions in pre-B cells revealed a correlation between Vκ usage, local density of CTCF-binding sites, and the frequencies of contacts with SIS, iEκ, or 3′Eκ.49 Using 3C-seq, we observed that long-range interactions between the SIS-, iEκ-, or 3′Eκ- and Vκ region were not uniformly affected by conditional deletion of the CTCF gene in the B-cell lineage. Loss of CTCF resulted in significantly increased interactions with the most proximal Vκ region containing the Vκ3 family. In line with these findings, CTCF-deficient pre-B cells showed significantly increased Vκ3 germline transcription and recombination.49 On the other hand, loss of CTCF resulted in reduced contacts with the 2 most distal Vκ genes, Vκ2-137 and Vκ1-135, which are often rearranged in control but not in CTCF-deficient (pre-)B cells.
Based on these studies,49,59,60 we proposed that CTCF binding in the SIS element provides enhancer-blocking activity that limits interactions between the κenhancers and the proximal Vκ promoters. CTCF, however, plays a dual role, as, next to the enhancer-blocking activity, it tethers remote Vκ gene promoters to the κenhancers.
Comparison of CTCF function in the Igh and Igκ locus
The enhancer blocking activity of CTCF at the SIS element in the Vκ-Jκ intergenic region in the Igκ locus49,60 remarkably parallels the role of CTCF at IGCR1 in the VH-D intergenic region in the Igh locus.53 Likewise, in thymocytes, CTCF colocalizes with cohesin at the TCR-α regulatory elements TEA just upstream of Jα. Loss of cohesion in double-positive thymocytes led to reduced long-range TEA-Eα interactions and significantly decreased germline transcription and rearrangement over the 3′Jα gene segments.61 Cohesin and CTCF also colocalize at 5′ boundary sites just downstream of the Eα enhancer, demarcating the TCR-α locus from the neighboring gene Dad1,61 a region with CTCF-dependent insulator activity.62
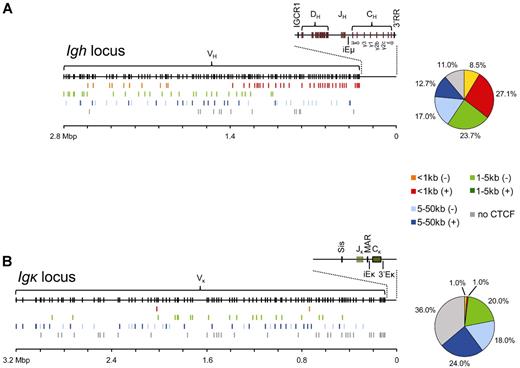
On virtually complete deletion of CTCF protein in the B-cell lineage in our conditionally targeted mice, we found that productive Igh and Igκ rearrangements were still generated, demonstrating that CTCF is not required for V(D)J recombination per se. This is also consistent with the presence of TCR-α– or TCR-β–chain rearrangement in CTCF-deficient thymocytes in vivo.63 The Igh and Igκ loci both contain approximately 60 CTCF-binding sites that influence the genomic architecture of these loci. The identification in the Igh locus of PAIR elements just upstream of distal VH3609 genes,50 together with the observation that many proximal VH gene segments are located within 100 bp of CTCF-binding sites,64 suggests that proximity of a CTCF-binding site may affect the probabilities of individual VH genes to encounter a DH-JH element for recombination. In this aspect, the Igh and Igκ loci are remarkably different: whereas approximately 27% of VH genes have a CTCF-binding site within 1-kb distance, this is the case for only 2 Vκ genes (Figure 3). Moreover, only approximately 11% of VH genes but approximately 36% Vκ genes do not have a CTCF-binding site in the intervening sequences up to their 5′ or 3′ neighboring V genes. Thus, although CTCF binds at many sites throughout the Igκ locus, most CTCF occupancy is located between Vκ segments and not adjacent to the Vκ segments.1,49 It has been suggested that V regions are organized as rosettes by CTCF, whereby CTCF-binding adjacent to a V gene increases its recombination probability.64 Because the Igh and Igκ loci show large differences in the proximity of CTCF-binding sites to V genes, in such a model the 2 loci would be very differently organized in 3-dimensional space to provide appropriate access of individual V regions to the proposed recombination center.65 Interestingly, it was recently proposed that E2A proteins may modulate Igκ locus topology by acting as anchors in a mechanism similar to that put forward for CTCF in the Igh locus.1 This hypothesis still needs to be tested but is supported by the remarkable, nonrandom distribution of E2A binding sites across the Igκ locus, in particular within 200 bp of the 5′ or the 3′ end of Vκ regions.1,46,47
VH and Vκ gene positions relative to CTCF occupancy. (A) Overview of VH gene positions relative to the nearest CTCF binding site for the mouse Igh locus. The germline Igh locus is shown with the locations of VH, DH, JH, CH, and enhancer elements (top). VH genes were grouped according to the location of the nearest CTCF site and whether this site was located upstream (−) or downstream (+) relative to VH transcriptional orientation. Color-coded groups of VH genes were plotted at their respective locations below the germline VH genes. On the right, a pie-chart represents the distribution of the different VH gene groups expressed in percentages of the total number of VH genes. (B) Identical to panel A, but for the Vκ genes within the mouse Igκ locus.
VH and Vκ gene positions relative to CTCF occupancy. (A) Overview of VH gene positions relative to the nearest CTCF binding site for the mouse Igh locus. The germline Igh locus is shown with the locations of VH, DH, JH, CH, and enhancer elements (top). VH genes were grouped according to the location of the nearest CTCF site and whether this site was located upstream (−) or downstream (+) relative to VH transcriptional orientation. Color-coded groups of VH genes were plotted at their respective locations below the germline VH genes. On the right, a pie-chart represents the distribution of the different VH gene groups expressed in percentages of the total number of VH genes. (B) Identical to panel A, but for the Vκ genes within the mouse Igκ locus.
CTCF and long-range gene interactions in Ig enhancer-mediated oncogene activation
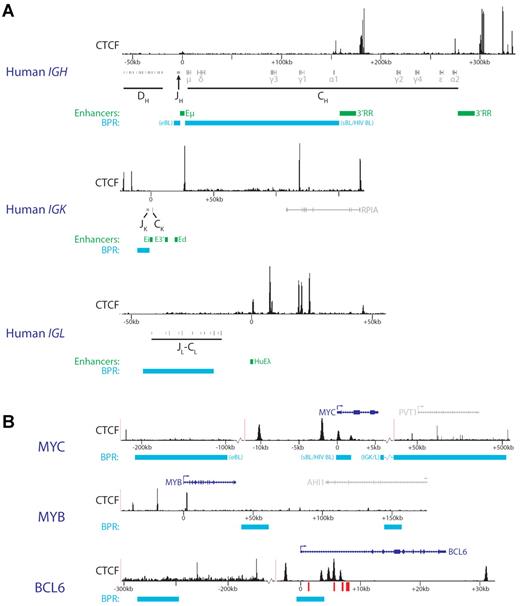
Many leukemias an lymphomas contain chromosomal translocation involving Ig or TCR loci whereby the strong enhancers in these loci are placed in a new genomic context. Because CTCF and cohesin have been implicated in interchromosomal interactions, they probably contribute to physical proximity of particular translocation targets in lymphocytes. For example, in many Burkitt B-cell lymphomas, translocations place the MYC oncogene up to approximately 200 kb upstream of the IgH 3′RR (Figure 4A). In normal B cells, IgH and MYC are preferentially positioned in close spatial proximity relative to each other and are present in the same transcription factories.66–68 Recently developed new genome-wide methods that measure primary chromosomal rearrangements in the absence of growth selection have shown that translocations between IGH and MYC and all other genes are indeed directly related to their contact frequencies.69–72 However, 3C-based measurements showed that approximately 30% of all genes interact with IGH at equal or higher frequency than MYC.71 Therefore, growth selection and AID-mediated DNA damage rather than a high contact frequency between IGH and the translocations partners account for their high rate of translocation.
CTCF distribution and chromosomal breakpoint regions (BPR) around Ig enhancers and oncogenes involved in translocation with antigen receptor loci. (A) Genomic regions surrounding the human IgH enhancers. CTCF ChIP-Seq data (from the human GM12878 B lymphoblastoid cell line81) is shown above the localization of DH, JH, and CH fragments. Green represents enhancers; and cyan, most common chromosomal BPRs. For the IGH locus, 2 common BPRs are annotated. eBL indicates endemic Burkitt lymphoma; sBL, sporadic Burkitt lymphoma; and HIV BL, HIV-induced Burkitt lymphoma. For scaling purposes, the position of Eμ (IGH), Ei (IGK), or HuEλ (IGL) enhancers was set to 0. (B) CTCF ChIP-Seq data (from the human GM12878 B lymphoblastoid cell line81) are shown for 3 genomic regions around oncogenes involved in translocations with antigen receptor loci. Locations of genes are shown on top, with the relevant oncogenes in blue and other genes in gray. The oncogene transcriptional start site was set to 0 for scaling purposes. Cyan indicates the most common chromosomal BPRs. BPRs linked to distinct B-cell malignancies are indicated as in panel A. For MYC, breakpoints found specifically in translocations with Ig light chain loci are annotated (IGK/L). Red bars represent the BCL6 CpG islands located in intron 1.
CTCF distribution and chromosomal breakpoint regions (BPR) around Ig enhancers and oncogenes involved in translocation with antigen receptor loci. (A) Genomic regions surrounding the human IgH enhancers. CTCF ChIP-Seq data (from the human GM12878 B lymphoblastoid cell line81) is shown above the localization of DH, JH, and CH fragments. Green represents enhancers; and cyan, most common chromosomal BPRs. For the IGH locus, 2 common BPRs are annotated. eBL indicates endemic Burkitt lymphoma; sBL, sporadic Burkitt lymphoma; and HIV BL, HIV-induced Burkitt lymphoma. For scaling purposes, the position of Eμ (IGH), Ei (IGK), or HuEλ (IGL) enhancers was set to 0. (B) CTCF ChIP-Seq data (from the human GM12878 B lymphoblastoid cell line81) are shown for 3 genomic regions around oncogenes involved in translocations with antigen receptor loci. Locations of genes are shown on top, with the relevant oncogenes in blue and other genes in gray. The oncogene transcriptional start site was set to 0 for scaling purposes. Cyan indicates the most common chromosomal BPRs. BPRs linked to distinct B-cell malignancies are indicated as in panel A. For MYC, breakpoints found specifically in translocations with Ig light chain loci are annotated (IGK/L). Red bars represent the BCL6 CpG islands located in intron 1.
An oncogenic role of the IGH locus 3′RR has been defined based on long-range activation of translocated MYC genes.73 In this context, an important function of CTCF is to restrict enhancer interaction to Ig loci49 (Figure 4). Obviously, CTCF-mediated restriction might be lost when strong Ig enhancers appear in an aberrant genomic context in chromosomal translocations, which may then lead to activation of oncogenes. In addition, CTCF sites clustered around Ig enhancers may affect local chromatin conformation to promote novel promoter-enhancer interactions. The genes encoding c-Myb and c-Myc, 2 essential hematopoietic regulators and oncogenic transcription factors, harbor multiple CTCF-binding sites in their vicinity (Figure 4B). Indeed, CTCF was originally discovered as a transcriptional repressor of c-Myc.74 Both genes are regulated at the level of transcriptional elongation by attenuator elements located in their first introns,75,76 close to CTCF-binding sites77,78 (Figure 4B). The exact role played by CTCF at these sites is still unclear, but the recent demonstration that CTCF regulates RNA polymerase II processing79 suggests that CTCF could directly affect elongation. Loss of intronic CTCF binding resulting from translocations could disrupt normal locus structure and transcriptional control, allowing the juxtaposed IGH enhancers to drive MYC expression. Because MYB seems to be regulated by a similar mechanism, this may also occur within the MYB locus, when translocated to the TCR-β locus in acute T-cell leukemia.80
Interestingly, based on published CTCF-binding sites,81 translocation breakpoints at the CRLF2 and BCL3 loci found in several types of B-cell malignancies are both confined to an approximately 25-kb region between the transcriptional start site and an upstream CTCF element.82,83 Translocations therefore result in loss of the upstream CTCF-binding site, which may allow strong Ig enhancers to interact and activate the CRLF2 and BCL3 promoters. In contrast, many other loci frequently involved in leukemic translocations (eg, BCL2 and MYC),84,85 show more complex breakpoint patterns and harbor numerous CTCF-binding sites (Figure 4B).
Expression of BCL6 in diffuse large B cell lymphomas is maintained through hypermethylation of intragenic CpG islands in its first intron.86 Hypermethylation, which has been observed in a wide range of cancer cells, prevents CTCF-mediated silencing of BCL6 because CTCF binds these CpG islands in a methylation-sensitive fashion. Furthermore, long-range regulatory regions approximately 150 to 260 kb upstream have been implicated in BCL6 regulation,87 possibly representing another layer of control in which CTCF might play an important role (Figure 4B).
In conclusion, based on studies in the Igh and Igκ, it has become clear that CTCF-binding provides enhancer-blocking activity that restricts Ig enhancer activity to the Ig loci. Nevertheless, CTCF also serves to bring remote V gene promoters into close proximity of Ig enhancers. It will be challenging to identify the exact mechanism by which CTCF regulates V gene choice because individual developing B cells are expected to manifest distinct and highly dynamic interactions between V regions and Ig enhancers. Moreover, the high density of CTCF-binding sites and their anticipated partial redundancy may complicate the interpretation of in vivo targeted deletion experiments.
Chromosomal translocations present in lymphoid malignancies will (1) remove CTCF sites close to the oncogenes, (2) bring in enhancer-associated CTCF sites, or (3) dramatically alter the spacing between CTCF sites and their cognate genes. These events are likely to influence local communication between cis-regulatory elements by imposing new structural constraints and thereby increasing oncogene expression. Additional epigenetic changes, including DNA methylation and histone modifications, might also interfere with CTCF function. For example, the BCL6 oncogene is involved in chromosomal translocations through breakpoints frequently found near the intronic CpG islands where CTCF binds in a methylation-sensitive manner.86,88
In a normal context, oncogene expression is tightly regulated in response to extracellular signaling events. It is an intriguing question why these signals are no longer able to efficiently regulate expression of the translocation products. Trying to address this issue might reveal additional roles for CTCF in the integration of signaling to chromatin. Future research should clarify the complex roles played by CTCF in the wiring of cis-DNA regulatory elements and in organizing networks of genomic interactions. The field of spatial chromatin organization research is currently exploding with the development of new exciting technologies, allowing high-throughput analysis of chromatin organization of the whole genome.27,89,90 As the resolution of these technologies increases, they might rapidly become valuable tools to reveal the complex 3-dimensional changes in cancer genomes and unravel their relationships with critical regulatory factors, including CTCF.
Acknowledgments
This work was supported in part by Fundação para a Ciência e a Tecnologia (C.R.d.A.), Royal Netherlands Academy of Arts and Sciences (R.S.), the Center of Biomedical Genetics, the Cancer Genomics Center, and the EuTRACC Consortium (E.S. and R.S.).
Authorship
Contribution: C.R.d.A. wrote the manuscript and designed figures; R.S. analyzed and interpreted data, wrote the manuscript, and designed figures; S.T. analyzed data; E.S. wrote the manuscript; and R.W.H. designed figures and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for C.R.d.A. is Genome Function Group, Medical Research Council Clinical Sciences Centre, Imperial College School of Medicine, London, United Kingdom.
Correspondence: Rudi W. Hendriks, Department of Pulmonary Medicine, Room Ee2251a, Erasmus MC Rotterdam, PO Box 2040, NL 3000 CA Rotterdam, The Netherlands; e-mail: r.hendriks@erasmusmc.nl.
References
Author notes
C.R.d.A. and R.S. contributed equally to this study.