Candidate genetic associations with acute GVHD (aGVHD) were evaluated with the use of genotyped and imputed single-nucleotide polymorphism data from genome-wide scans of 1298 allogeneic hematopoietic cell transplantation (HCT) donors and recipients. Of 40 previously reported candidate SNPs, 6 were successfully genotyped, and 10 were imputed and passed criteria for analysis. Patient and donor genotypes were assessed for association with grades IIb-IV and III-IV aGVHD, stratified by donor type, in univariate and multivariate allelic, recessive and dominant models. Use of imputed genotypes to replicate previous IL10 associations was validated. Similar to previous publications, the IL6 donor genotype for rs1800795 was associated with a 20%-50% increased risk for grade IIb-IV aGVHD after unrelated HCT in the allelic (adjusted P = .011) and recessive (adjusted P = .0013) models. The donor genotype was associated with a 60% increase in risk for grade III-IV aGVHD after related HCT (adjusted P = .028). Other associations were found for IL2, CTLA4, HPSE, and MTHFR but were inconsistent with original publications. These results illustrate the advantages of using imputed single-nucleotide polymorphism data in genetic analyses and demonstrate the importance of validation in genetic association studies.
Introduction
During the past decade, many reports have identified genetic variants such as single-nucleotide polymorphisms (SNPs) and other polymorphisms that influence the risk of acute GVHD (aGVHD) after allogeneic hematopoietic cell transplantation (HCT). Many of these variants regulate the function of immune cells, their receptors, effector molecules, cytokines, and chemokines. Some results have led to the suggestion that pretransplantation assessment of these variants might help to assess the risk of adverse outcomes for each patient, guide the clinical management of patients who are at high risk, and ultimately serve as potential biologic targets for novel therapeutics.
Although these concepts have been greeted with some enthusiasm, the consistency of results leading to such suggestions has been questioned. Many of the reported results have been difficult to replicate in independent cohorts, and in no published studies have authors comprehensively evaluated these variants in the same cohort simultaneously. We recently had the opportunity to perform genome-wide scans of a cohort of patients and their donors with the use of an array that is informative for > 500 000 SNPs. In conjunction with the vast HapMap resource for SNP genotypes across the entire genome, these data provided a unique opportunity to use either the genotyped or imputed SNP data to determine how many of the previously published associations could be replicated.
Methods
Literature search
We performed a comprehensive PubMed search using the terms “acute GVHD” and “polymorphism” to identify all studies published by April 30, 2011, that reported an association between aGVHD and a genetic polymorphism at an α level < 0.05. Studies that did not meet this threshold were not included. Studies that reported associations with alternative genetic variants such as deletions, microsatellites, or VNTRs were excluded because our SNP genotyping array is not informative for such variants. Because our group previously published associations between SNPs in the IL10 and IL10RB genes and aGVHD using a cohort that overlaps with the current cohort,1,2 we used imputed genotypes for these SNPs as a quality control measure to evaluate whether previously identified associations could be replicated with imputed genotypes.
Study population
The source population included 1424 donor-recipient pairs randomly selected from among 3177 patients who received allogeneic HCT after myeloablative conditioning at the Fred Hutchinson Cancer Research Center and Seattle Cancer Care Alliance between 1992 and 2004. All recipient and donor samples were collected before HCT according to approved research protocols. Project-specific institutional research board approval from the Fred Hutchinson Cancer Research Center was obtained for the use of these samples. Recipient and donor demographic information collected at the time of pretransplantation evaluation, including sex, XY karyotype, ABO blood group, and race, was available through the FHCRC Clinical Research Division patient database (“Clinical Research Database”). All clinical data were prospectively collected and retrospectively reviewed.
Clinical characteristics of the patients and aGVHD prevalence data are summarized in Table 1. The cohort included recipients with either HLA-matched related donor (n = 612) or unrelated donors (n = 686) who underwent transplantation for a hematologic malignancy or myelodysplasia. Patients with related HLA-mismatched donors (n = 126) were excluded because of the limited number for meaningful stratified analysis. The pretransplantation disease risk category was determined on the basis of disease type and stage or remission/relapse status and classified as low, intermediate, or high. Patient-donor HLA matching was determined on the basis of genotyping for HLA-A, B, C, HLA-DRB1, and DQB1. All patients received T cell–replete BM or growth factor-mobilized blood cell grafts, and cyclosporine or tacrolimus plus methotrexate or mycophenylate for aGVHD prophylaxis. Conditioning regimens were categorized according to the use of total body irradiation. Peak severity of aGVHD after HCT, defined according to grade (0, I, IIa, IIb, III, IV), was recorded.
Summary demographic and outcome data for the GWAS cohort
| Variables . | Patient . |
|---|---|
| Number of patients | 1298 |
| Age, y | 38.2 ± 14 |
| Sex match (patient/donor), n (%) | |
| Male/male | 446 (34.3) |
| Male/female | 309 (23.8) |
| Female/male | 288 (22.2) |
| Female/female | 255 (19.7) |
| Race, n (%) | |
| White | 1116 (86.0) |
| Other | 182 (14.0) |
| Diagnosis, n (%) | |
| Acute leukemia | 465 (35.82) |
| CML | 486 (37.44) |
| MDS | 229 (17.64) |
| CLL, HD, and NHL | 79 (6.1) |
| Multiple myeloma | 39 (3.0) |
| Disease risk, n (%) | |
| Low | 361 (27.8) |
| Intermediate | 594 (45.8) |
| High | 343 (26.4) |
| Donor type, n (%) | |
| Unrelated donor | 686 (52.8) |
| Matched related donor | 612 (47.2) |
| Stem cell source | |
| BM, n (%) | 921 (71.0) |
| Peripheral blood | 377 (29.0) |
| TBI dose | |
| None, n (%) | 531 (40.9) |
| ≤ 1200 Gy | 366 (28.2) |
| > 1200 Gy | 401 (30.9) |
| aGVHD IIb-IV, n (%) | 829 (58.5) |
| aGVHD III-IV, n (%) | 323 (25.0) |
| Variables . | Patient . |
|---|---|
| Number of patients | 1298 |
| Age, y | 38.2 ± 14 |
| Sex match (patient/donor), n (%) | |
| Male/male | 446 (34.3) |
| Male/female | 309 (23.8) |
| Female/male | 288 (22.2) |
| Female/female | 255 (19.7) |
| Race, n (%) | |
| White | 1116 (86.0) |
| Other | 182 (14.0) |
| Diagnosis, n (%) | |
| Acute leukemia | 465 (35.82) |
| CML | 486 (37.44) |
| MDS | 229 (17.64) |
| CLL, HD, and NHL | 79 (6.1) |
| Multiple myeloma | 39 (3.0) |
| Disease risk, n (%) | |
| Low | 361 (27.8) |
| Intermediate | 594 (45.8) |
| High | 343 (26.4) |
| Donor type, n (%) | |
| Unrelated donor | 686 (52.8) |
| Matched related donor | 612 (47.2) |
| Stem cell source | |
| BM, n (%) | 921 (71.0) |
| Peripheral blood | 377 (29.0) |
| TBI dose | |
| None, n (%) | 531 (40.9) |
| ≤ 1200 Gy | 366 (28.2) |
| > 1200 Gy | 401 (30.9) |
| aGVHD IIb-IV, n (%) | 829 (58.5) |
| aGVHD III-IV, n (%) | 323 (25.0) |
aGVHD indicates acute graft versus host disease; CML, chronic myelogenous leukemia; CLL, chronic lymphocytic leukemia; GWAS, genome-wide association study; HD, Hodgkin's disease; HLA, human leukocyte antigen; MDS, myelodysplastic syndrome; NHL, non-Hodgkin's lymphoma; and TBI, total body irradiation.
aGVHD was considered as 2 different phenotypes: grades 0-IIa versus grades IIb-IV, and grades 0-II versus grades III-IV. Grade IIa GVHD is a subset of grade II aGVHD that includes cases with upper gastrointestinal symptoms generally documented as aGVHD by biopsy, without diarrhea or with stool volume < 1000 mL/d, without rash or with rash involving < 50% of the body surface, and without liver involvement.3 Grade IIb aGVHD is a subset of grade II GVHD that includes cases with rash involving > 50% of the body surface (stage III skin disease) or with total serum bilirubin concentration between 2.0 and 2.9 mg/dL (stage I liver disease), with or without stage I gastrointestinal involvement. As previously reported, the overall incidence of grades II-IV GVHD is greater at our center than reported at other centers, primarily because of high sensitivity for the diagnosis of upper gastrointestinal GVHD.3 The incidence of grades IIb-IV GVHD at our center is similar to the overall incidence of grades II-IV GVHD at other centers.
Sample preparation, genotyping, and imputations
DNA specimens originated from blood mononuclear cells or EBV-transformed B-lymphocyte cell lines. Genomic DNA was extracted by a standard salting-out method with the use of the Puregene kit (QIAGEN). The quantity and purity of DNA in each sample was determined by UV absorption in a spectrophotometer (OD260) and by OD260/280 ratio, respectively. After DNA quantitation and initial screening for verification of sample identity, DNA samples were distributed into 96-well plates (92 samples per plate) at a targeted concentration of 50-100 ng/μL. Each plate of 92 samples contained 4 duplicate samples carried forward from a previously prepared plate as a quality control indicator. The plates were then sealed, frozen at −20°C, and shipped on dry ice to the Affymetrix Service Laboratory (ASL) in Santa Clara, CA, for amplification and hybridization.
All assays used the Affymetrix GeneChip Genome-Wide Human SNP Array 5.0, with hybridization and scanning at the ASL. After scanning of hybridized GeneChip arrays, the resulting raw probe intensities were evaluated with the Bayesian Robust Linear Model with Mahalanobis distance classifier (BRLMM) analysis tool adapted for the 5.0 Array (BRLMM-P). Data quality was assessed via 3 different methods: the Affymetrix “QC call rate,” the clustering call rate, and the sex call rate. Once the array data were cleared by internal ASL quality assessment, the genome-wide SNP data were transmitted to FHCRC, where they were subjected to further QC analysis. This analysis included assessment of overall sample quality using the DM and BRLMM-P call rate statistics generated by Affymetrix, adjustment of threshold standards to optimize genotyping accuracy, if necessary, and assessment and resolution of sample identity questions with the use of calculations for estimating genomic similarity between samples. Because sex and ABO type was known for all samples, PCR-based ABO and XY genotyping assays were implemented to determine sample quality and verify sample identity before submitting the DNA samples to the ASL for hybridization and scanning. A further evaluation of sample identity was performed via use of the Affymetrix-supplied genotypes obtained by clustering with the BRLMM-P algorithm for those samples passing the QC call rate threshold. The final SNP-based genotype data generated is referred to as the “GWAS-HCT” dataset.
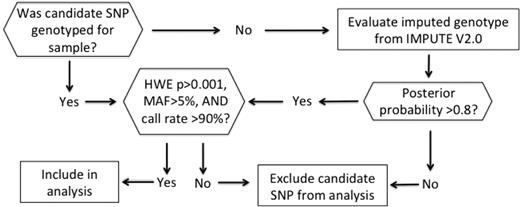
The candidate SNP genotype determination algorithm is summarized in Figure 1. If the original candidate SNP was not genotyped on our array, we evaluated the imputed genotype, which was obtained by imputing all the HapMap release 22 SNPs (combined panel of ethnic groups: CEU [Utah residents with Northern and Western European ancestry from the CEPH collection], YRI [Yoruba in Ibadan], CHB [Han Chinese in Beijing], and JPT [Japanese in Tokyo]) that were not genotyped on the Affymetrix 5.0 array using IMPUTE software (https://mathgen.stats.ox.ac.uk/impute/impute.html). At each unobserved HapMap SNP locus, 1 or more genotypes may be predicted with a positive posterior probability, which measures the probability of observing that genotype at the imputed locus. We assign the genotype with the maximum posterior probability as the imputed SNP genotype only if the maximum posterior probability exceeded 0.8. All genotyped and imputed SNPs that violated Hardy-Weinberg equilibrium with P < .001, had a minor allele frequency < 0.05, or had a < 90% call rate were excluded from the analyses.
Candidate SNP genotype determination algorithm. HWE indicates Hardy-Weinberg equilibrium; MAF, minor allele frequency; and SNP, single-nucleotide polymorphism.
Candidate SNP genotype determination algorithm. HWE indicates Hardy-Weinberg equilibrium; MAF, minor allele frequency; and SNP, single-nucleotide polymorphism.
Statistical analysis
All analyses were performed with Matlab v.2010b (MathWorks). All analyses were stratified by donor type (matched related donor [MRD] vs unrelated donor [URD]) and were examined for recipients and donors in separate univariate and multivariate Cox regression models, censored at the time of death or onset of recurrent malignancy. Multivariate Cox models adjusted for clinical factors recently found to be associated with the risk of aGVHD female donor and male recipient status and total body irradiation.4 For URDs, an additional in-model adjustment was made for HLA match. We also incorporated as covariates principal components derived from a principal component analysis of population stratification,5 where the first 4 components were included as covariates in the final adjusted analysis.
Each SNP was assessed for allelic and genotypic (recessive and dominant models) modes of transmission, stratified according to donor type (HLA-matched related versus unrelated). For a SNP with a major allele “a” and a minor allele “b,” the recessive model tests the hypothesis that the genotype “bb” is associated with an increased or decreased risk compared with the collective genotypes “ab and “aa.” The dominant model tests the hypothesis that the collective genotypes “bb” and “ab” are associated with an increased or decreased risk compared with the genotype “aa.” The allelic model tests the hypothesis that the minor allele b is associated with an increased or decreased risk compared with the major allele a, and the number of copies of the minor allele is modeled as an additive effect. Because the main goal of this analysis was to replicate published associations with candidate SNPs, a 2-sided P ≤ .05 was selected as the threshold of significance, despite the multiple comparisons made in this analysis.
Results
Candidate gene and SNP selection
A search of the PubMed database identified 41 publications that reported associations of donor or recipient genotypes for 40 candidate SNPs in 22 genes with the risk of aGVHD: CD31,6,7 CTLA4,8 ESR1,9 FAS,10 HMGB1,11 HPSE,12 HSPA1L,13,14 IL1α,9,15 IL1β,16 IL2,17 IL6,10,18,19 IL10,1,16,18,20,,,,–25 IL10RB,2,24 IL23R,26,–28 MADCAM1,29 MTHFR,30,,–33 NOD2,34,,–37 RFC1,38 TGFβ1,39,40 TNF,10,16,20,21,23,41,–43 TNFRII,23,42 and VEGFα.44 The median cohort size in these studies was 89 (range, 24-536). The median number of genetic variations examined in each study was 3.5 (range, 1-19); in none of the studies did authors adjust for multiple comparisons in the analyses.
Of the 40 published candidate SNP associations, usable genotype or imputed data were available for 16 (40%; Table 2). Six candidate SNPs were genotyped: CD31 (rs1131012), CTLA4 (rs3087243), HPSE (rs4364254), IL23R (rs11209026), IL6 (rs1800795), and TNF (rs1799964). Ten candidate SNPs were imputed and passed the criteria for analysis: CD31 (rs668), IL1β (rs16944), IL2 (rs2069762), IL10 (rs1800871, rs1800872), MTHFR (rs1801131), RFC1 (rs1057807), TNF (rs1800629, rs1800630), and TNFRII (rs1061622). Of the remaining 24 candidate SNPs, 10 candidate SNPs were imputed but did not pass the criteria for analysis: ESR1 (rs2234693), FAS (rs1800682), HPSE (rs4693608), IL10 (rs1800896), IL10RB (rs2834167), MTHFR (rs1801133), RFC1 (rs4975003, rs6833176), and VEGFα (rs699947, rs833061). Fourteen SNPs were not genotyped and could not be imputed: CD31 (rs12953), FAS (rs2234767), HMGB1 (rs41376448), HSPA1L (rs2075800), MADCAM1 (rs2302217), NOD2 (rs2066844, rs2066845, rs2066847), TGFβ1 (rs1800470), TNF (−488, rs1799724, rs361525), VEGFα (rs2010963, rs3025039).
Summary of genotyped and imputed SNP data generated from the GWAS-HCT dataset using the Affymetrix 5.0 Array and the HapMap release 22 SNPs
| Gene . | Type . | rs number . | Chr . | Alleles* . | MAF . | Call rate . | p_HWE . | Average posterior probability . | Status . |
|---|---|---|---|---|---|---|---|---|---|
| CD31 | Genotyped | rs1131012 | 17 | C/T | 0.472 | 0.98 | 0.08 | 0.98 | Pass |
| CD31 | Imputed | rs668 | 17 | G/C | 0.494 | 0.966 | 0.7 | 0.978 | Pass |
| CTLA4 | Genotyped | rs3087243 | 2 | A/G | 0.446 | 1 | 0.814 | 1 | Pass |
| ESR1 | Imputed | rs2234693 | 6 | C/T | 0.35 | 0.642 | 0.121 | 0.831 | Fail |
| FAS | Imputed | rs1800682 | 10 | G/A | 0.464 | 0.899 | 0.218 | 0.948 | Fail |
| HPSE | Genotyped | rs4364254 | 4 | C/T | 0.296 | 0.999 | 0.349 | 0.999 | Pass |
| HPSE | Imputed | rs4693608 | 4 | G/A | 0.328 | 0.62 | 0.668 | 0.817 | Fail |
| IL10 | Imputed | rs1800871 | 1 | A/G | 0.238 | 0.951 | 0.352 | 0.972 | Pass |
| IL10 | Imputed | rs1800872 | 1 | T/G | 0.238 | 0.951 | 0.359 | 0.972 | Pass |
| IL10 | Imputed | rs1800896 | 1 | C/T | 0.472 | 0.799 | 0.245 | 0.898 | Fail |
| IL10RB | Imputed | rs2834167 | 21 | G/A | 0.162 | 0.353 | 0.027 | 0.722 | Fail |
| IL1B | Imputed | rs16944 | 2 | A/G | 0.359 | 0.944 | 0.44 | 0.962 | Pass |
| IL2 | Imputed | rs2069762 | 4 | C/A | 0.317 | 0.998 | 0.115 | 0.998 | Pass |
| IL23R | Genotyped | rs11209026 | 1 | A/G | 0.064 | 0.998 | 0.598 | 0.998 | Pass |
| IL6 | Genotyped | rs1800795 | 7 | C/G | 0.375 | 0.998 | 0.006 | 0.998 | Pass |
| MTHFR | Imputed | rs1801131 | 1 | G/T | 0.304 | 0.946 | 0.238 | 0.967 | Pass |
| MTHFR | Imputed | rs1801133 | 1 | A/G | 0.182 | 0.449 | 0.221 | 0.77 | Fail |
| RFC1 | Imputed | rs1057807 | 4 | G/A | 0.425 | 0.999 | 0.166 | 0.999 | Pass |
| RFC1 | Imputed | rs4975003 | 4 | C/G | 0.441 | 0.893 | 0.274 | 0.932 | Fail |
| RFC1 | Imputed | rs6844176 | 4 | C/T | 0.494 | 0.748 | 0.047 | 0.878 | Fail |
| TNF | Genotyped | rs1799964 | 6 | C/T | 0.205 | 1 | 0.063 | 1 | Pass |
| TNF | Imputed | rs1800629 | 6 | A/G | 0.135 | 0.926 | 0.726 | 0.965 | Pass |
| TNF | Imputed | rs1800630 | 6 | A/C | 0.152 | 0.996 | 0.153 | 0.997 | Pass |
| TNFRII | Imputed | rs1061622 | 1 | G/T | 0.227 | 0.964 | 0.12 | 0.971 | Pass |
| VEGFA | Imputed | rs699947 | 6 | A/C | 0.114 | 0.141 | 0.244 | 0.633 | Fail |
| VEGFA | Imputed | rs833061 | 6 | C/T | 0.183 | 0.094 | 0.283 | 0.607 | Fail |
| Gene . | Type . | rs number . | Chr . | Alleles* . | MAF . | Call rate . | p_HWE . | Average posterior probability . | Status . |
|---|---|---|---|---|---|---|---|---|---|
| CD31 | Genotyped | rs1131012 | 17 | C/T | 0.472 | 0.98 | 0.08 | 0.98 | Pass |
| CD31 | Imputed | rs668 | 17 | G/C | 0.494 | 0.966 | 0.7 | 0.978 | Pass |
| CTLA4 | Genotyped | rs3087243 | 2 | A/G | 0.446 | 1 | 0.814 | 1 | Pass |
| ESR1 | Imputed | rs2234693 | 6 | C/T | 0.35 | 0.642 | 0.121 | 0.831 | Fail |
| FAS | Imputed | rs1800682 | 10 | G/A | 0.464 | 0.899 | 0.218 | 0.948 | Fail |
| HPSE | Genotyped | rs4364254 | 4 | C/T | 0.296 | 0.999 | 0.349 | 0.999 | Pass |
| HPSE | Imputed | rs4693608 | 4 | G/A | 0.328 | 0.62 | 0.668 | 0.817 | Fail |
| IL10 | Imputed | rs1800871 | 1 | A/G | 0.238 | 0.951 | 0.352 | 0.972 | Pass |
| IL10 | Imputed | rs1800872 | 1 | T/G | 0.238 | 0.951 | 0.359 | 0.972 | Pass |
| IL10 | Imputed | rs1800896 | 1 | C/T | 0.472 | 0.799 | 0.245 | 0.898 | Fail |
| IL10RB | Imputed | rs2834167 | 21 | G/A | 0.162 | 0.353 | 0.027 | 0.722 | Fail |
| IL1B | Imputed | rs16944 | 2 | A/G | 0.359 | 0.944 | 0.44 | 0.962 | Pass |
| IL2 | Imputed | rs2069762 | 4 | C/A | 0.317 | 0.998 | 0.115 | 0.998 | Pass |
| IL23R | Genotyped | rs11209026 | 1 | A/G | 0.064 | 0.998 | 0.598 | 0.998 | Pass |
| IL6 | Genotyped | rs1800795 | 7 | C/G | 0.375 | 0.998 | 0.006 | 0.998 | Pass |
| MTHFR | Imputed | rs1801131 | 1 | G/T | 0.304 | 0.946 | 0.238 | 0.967 | Pass |
| MTHFR | Imputed | rs1801133 | 1 | A/G | 0.182 | 0.449 | 0.221 | 0.77 | Fail |
| RFC1 | Imputed | rs1057807 | 4 | G/A | 0.425 | 0.999 | 0.166 | 0.999 | Pass |
| RFC1 | Imputed | rs4975003 | 4 | C/G | 0.441 | 0.893 | 0.274 | 0.932 | Fail |
| RFC1 | Imputed | rs6844176 | 4 | C/T | 0.494 | 0.748 | 0.047 | 0.878 | Fail |
| TNF | Genotyped | rs1799964 | 6 | C/T | 0.205 | 1 | 0.063 | 1 | Pass |
| TNF | Imputed | rs1800629 | 6 | A/G | 0.135 | 0.926 | 0.726 | 0.965 | Pass |
| TNF | Imputed | rs1800630 | 6 | A/C | 0.152 | 0.996 | 0.153 | 0.997 | Pass |
| TNFRII | Imputed | rs1061622 | 1 | G/T | 0.227 | 0.964 | 0.12 | 0.971 | Pass |
| VEGFA | Imputed | rs699947 | 6 | A/C | 0.114 | 0.141 | 0.244 | 0.633 | Fail |
| VEGFA | Imputed | rs833061 | 6 | C/T | 0.183 | 0.094 | 0.283 | 0.607 | Fail |
Chr indicates chromosome; HWE, Hardy-Weinberg equilibrium MAF, minor allele frequency; and SNP, single-nucleotide polymorphism.
The first allele is designated as the minor allele, and the second allele is designated as the major allele.
Validation of previous published IL10 SNP associations with aGVHD
We previously published associations between aGVHD risk and SNPs on IL10 (rs1800896, rs1800871, and rs1800872)1 and IL10RB (rs2834167)2 in a cohort that overlapped significantly with the current cohort. Because these SNPs were not directly genotyped on the array, we evaluated whether the original association could be replicated with the use of imputed genotypes. Although the genotypes of all 4 SNPs were imputable, only the IL10 SNPs rs1800871 and rs1800872 passed the selection criteria for analysis (Table 2). These 2 successfully imputed IL10 SNPs are in significant linkage disequilibrium (D′ > 0.9). Among MRD transplantations, the patient's genotypes for both SNPs were significantly associated with a 30% decrease of the risk for grade III-IV aGVHD in the allelic model (Table 3; hazard ratio [HR] 0.72, P = .048). These findings are consistent with the findings of our original publication (Table 4).1
Summary of significant associations (P < .05) between aGVHD and candidate SNPs
| Gene . | Phenotype . | Genome . | Donor type . | SNP . | Alleles . | MAF* . | Model . | HR . | Unadjusted CI . | P . | HR . | Adjusted CI† . | P . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL2 | III-IV | DNR | URD | rs2069762 | C/A | 0.34 | Allelic | 1.16 | (0.96-1.40) | .1293 | 1.15 | (0.94-1.40) | .1811 |
| Recessive | 1.04 | (0.70-1.55) | .8466 | 0.98 | (0.64-1.50) | .9310 | |||||||
| Dominant | 1.32 | (1.00-1.74) | .0487 | 1.33 | (0.99-1.78) | .0557 | |||||||
| IL6 | IIb-IV | DNR | URD | rs1800795 | C/G | 0.39 | Allelic | 1.21 | (1.07-1.36) | .0028 | 1.19 | (1.04-1.36) | .0106 |
| Recessive | 1.48 | (1.19-1.84) | .0004 | 1.44 | (1.15-1.81) | .0013 | |||||||
| Dominant | 1.17 | (0.97-1.40) | .0966 | 1.13 | (0.93-1.38) | .2231 | |||||||
| IL6 | IIb-IV | PT | MRD | rs1800795 | C/G | 0.34 | Allelic | 0.87 | (0.73-1.04) | .1362 | 0.8 | (0.66-0.97) | .0240 |
| Recessive | 0.84 | (0.57-1.25) | .3983 | 0.81 | (0.54-1.20) | .2923 | |||||||
| Dominant | 0.84 | (0.66-1.06) | .1444 | 0.74 | (0.57-0.95) | .0195 | |||||||
| IL6 | III-IV | DNR | MRD | rs1800795 | C/G | 0.36 | Allelic | 1.26 | (0.98-1.63) | .0719 | 1.27 | (0.97-1.67) | .0864 |
| Recessive | 1.09 | (0.65-1.82) | .7360 | 1.06 | (0.63-1.79) | .8177 | |||||||
| Dominant | 1.54 | (1.05-2.26) | .0257 | 1.59 | (1.05-2.41) | .0279 | |||||||
| IL10 | III-IV | PT | MRD | rs1800872 | T/G | 0.24 | Allelic | 0.72 | (0.52-1.00) | .0481 | 0.73 | (0.52-1.04) | .0796 |
| Recessive | 0 | (0.00,Inf) | .9817 | 0 | (0.00,Inf) | .9815 | |||||||
| Dominant | 0.82 | (0.56-1.19) | .2933 | 0.85 | (0.58-1.25) | .4035 | |||||||
| IL10 | III-IV | PT | MRD | rs1800871 | A/G | 0.24 | Allelic | 0.72 | (0.52-1.00) | .0481 | 0.73 | (0.52-1.04) | .0796 |
| Recessive | 0 | (0.00,Inf) | .9817 | 0 | (0.00,Inf) | .9815 | |||||||
| Dominant | 0.82 | (0.56-1.19) | .2933 | 0.85 | (0.58-1.25) | .4035 | |||||||
| CTLA4 | III-IV | PT | URD | rs3087243 | A/G | 0.45 | Allelic | 1.17 | (0.97-1.42) | .1042 | 1.23 | (1.01-1.50) | .0374 |
| Recessive | 1 | (0.72-1.41) | .9782 | 1.05 | (0.74-1.49) | .7870 | |||||||
| Dominant | 1.5 | (1.09-2.07) | .0138 | 1.65 | (1.18-2.30) | .0035 | |||||||
| HPSE | IIb-IV | DNR | URD | rs4364254 | C/T | 0.29 | Allelic | 1.07 | (0.93-1.22) | .3377 | 1.11 | (0.97-1.27) | .1453 |
| Recessive | 0.91 | (0.66-1.26) | .5736 | 0.95 | (0.68-1.33) | .7767 | |||||||
| Dominant | 1.15 | (0.97-1.37) | .1158 | 1.21 | (1.01-1.45) | .0415 | |||||||
| MTHFR | III-IV | PT | MRD | rs1801131 | G/T | 0.29 | Allelic | 0.88 | (0.67-1.17) | .3870 | 0.84 | (0.63-1.11) | .2157 |
| Recessive | 1.29 | (0.76-2.19) | .3408 | 1.22 | (0.71-2.08) | .4679 | |||||||
| Dominant | 0.71 | (0.49-1.04) | .0764 | 0.66 | (0.45-0.96) | .0309 |
| Gene . | Phenotype . | Genome . | Donor type . | SNP . | Alleles . | MAF* . | Model . | HR . | Unadjusted CI . | P . | HR . | Adjusted CI† . | P . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL2 | III-IV | DNR | URD | rs2069762 | C/A | 0.34 | Allelic | 1.16 | (0.96-1.40) | .1293 | 1.15 | (0.94-1.40) | .1811 |
| Recessive | 1.04 | (0.70-1.55) | .8466 | 0.98 | (0.64-1.50) | .9310 | |||||||
| Dominant | 1.32 | (1.00-1.74) | .0487 | 1.33 | (0.99-1.78) | .0557 | |||||||
| IL6 | IIb-IV | DNR | URD | rs1800795 | C/G | 0.39 | Allelic | 1.21 | (1.07-1.36) | .0028 | 1.19 | (1.04-1.36) | .0106 |
| Recessive | 1.48 | (1.19-1.84) | .0004 | 1.44 | (1.15-1.81) | .0013 | |||||||
| Dominant | 1.17 | (0.97-1.40) | .0966 | 1.13 | (0.93-1.38) | .2231 | |||||||
| IL6 | IIb-IV | PT | MRD | rs1800795 | C/G | 0.34 | Allelic | 0.87 | (0.73-1.04) | .1362 | 0.8 | (0.66-0.97) | .0240 |
| Recessive | 0.84 | (0.57-1.25) | .3983 | 0.81 | (0.54-1.20) | .2923 | |||||||
| Dominant | 0.84 | (0.66-1.06) | .1444 | 0.74 | (0.57-0.95) | .0195 | |||||||
| IL6 | III-IV | DNR | MRD | rs1800795 | C/G | 0.36 | Allelic | 1.26 | (0.98-1.63) | .0719 | 1.27 | (0.97-1.67) | .0864 |
| Recessive | 1.09 | (0.65-1.82) | .7360 | 1.06 | (0.63-1.79) | .8177 | |||||||
| Dominant | 1.54 | (1.05-2.26) | .0257 | 1.59 | (1.05-2.41) | .0279 | |||||||
| IL10 | III-IV | PT | MRD | rs1800872 | T/G | 0.24 | Allelic | 0.72 | (0.52-1.00) | .0481 | 0.73 | (0.52-1.04) | .0796 |
| Recessive | 0 | (0.00,Inf) | .9817 | 0 | (0.00,Inf) | .9815 | |||||||
| Dominant | 0.82 | (0.56-1.19) | .2933 | 0.85 | (0.58-1.25) | .4035 | |||||||
| IL10 | III-IV | PT | MRD | rs1800871 | A/G | 0.24 | Allelic | 0.72 | (0.52-1.00) | .0481 | 0.73 | (0.52-1.04) | .0796 |
| Recessive | 0 | (0.00,Inf) | .9817 | 0 | (0.00,Inf) | .9815 | |||||||
| Dominant | 0.82 | (0.56-1.19) | .2933 | 0.85 | (0.58-1.25) | .4035 | |||||||
| CTLA4 | III-IV | PT | URD | rs3087243 | A/G | 0.45 | Allelic | 1.17 | (0.97-1.42) | .1042 | 1.23 | (1.01-1.50) | .0374 |
| Recessive | 1 | (0.72-1.41) | .9782 | 1.05 | (0.74-1.49) | .7870 | |||||||
| Dominant | 1.5 | (1.09-2.07) | .0138 | 1.65 | (1.18-2.30) | .0035 | |||||||
| HPSE | IIb-IV | DNR | URD | rs4364254 | C/T | 0.29 | Allelic | 1.07 | (0.93-1.22) | .3377 | 1.11 | (0.97-1.27) | .1453 |
| Recessive | 0.91 | (0.66-1.26) | .5736 | 0.95 | (0.68-1.33) | .7767 | |||||||
| Dominant | 1.15 | (0.97-1.37) | .1158 | 1.21 | (1.01-1.45) | .0415 | |||||||
| MTHFR | III-IV | PT | MRD | rs1801131 | G/T | 0.29 | Allelic | 0.88 | (0.67-1.17) | .3870 | 0.84 | (0.63-1.11) | .2157 |
| Recessive | 1.29 | (0.76-2.19) | .3408 | 1.22 | (0.71-2.08) | .4679 | |||||||
| Dominant | 0.71 | (0.49-1.04) | .0764 | 0.66 | (0.45-0.96) | .0309 |
Allo indicates allogeneic; CI, confidence interval; DNR, donor, HR, hazard ratio MAF, minor allele frequency; MRD, matched related donor; NR, not reported; PT, patient; SNP, single-nucleotide polymorphism; and URD, unrelated donor.
The first allele is designated as the minor allele, and the second allele is designated as the major allele.
Models adjusted for total body irradiation (yes vs no), sex mismatch, and 4 principal components for population stratification. Matched related donor analyses are also adjusted for HLA match.
Summary of previous publications reporting associations with candidate genes that meet the P < .05 threshold in the current validation study
| Gene . | Reference . | Phenotype . | Genome . | Donor type . | SNP . | Alleles* . | MAF . | HR . | Unadjusted CI . | P . |
|---|---|---|---|---|---|---|---|---|---|---|
| IL2 | MacMillan et al17 | II-IV | PT | URD | rs2069762 | G/T | 0.3 | 2.1 | (1.0-4.5) | .05 |
| IL6 | Mullighan et al10 | I-IV | DNR | MRD | rs1800795 | G/C | 0.4 | 4.4 | (1.6-11.7) | .001 |
| IL6 | Karabon e tal18 | II-IV | DNR | NR | rs1800795 | G/C | 0.5 | 3.4 | (1.1-13.0) | .03 |
| IL6 | Ambruzova et al19 | II-IV | PT | Allo | rs1800795 | G/C | 0.5 | 2.2 | (1.1-4.4) | .03 |
| IL10 | Lin et al1 | III-IV | PT | MRD | rs1800871† | A/C | 0.3 | |||
| IL10 | Lin et al1 | III-IV | PT | MRD | rs1800872 | A/C | 0.3 | 0.4 | (0.2-0.9) | .02 |
| CTLA4 | Perez-Garcia et al8 | II-IV | DNR | MRD | rs3087243 | A/G | 0.5 | 1.5 | (1.0-2.3) | .03 |
| HPSE | Ostrovsky et al12 | II-IV and III-IV | PT | Allo | rs4364254 | T/C | 0.4 | NR | NR | < .01 |
| MTHFR | Robien et al30 | I-IV | PT | Allo | rs1801131 | A/C | 0.4 | 3.6 | (1.0-12.7) | < .01 |
| Gene . | Reference . | Phenotype . | Genome . | Donor type . | SNP . | Alleles* . | MAF . | HR . | Unadjusted CI . | P . |
|---|---|---|---|---|---|---|---|---|---|---|
| IL2 | MacMillan et al17 | II-IV | PT | URD | rs2069762 | G/T | 0.3 | 2.1 | (1.0-4.5) | .05 |
| IL6 | Mullighan et al10 | I-IV | DNR | MRD | rs1800795 | G/C | 0.4 | 4.4 | (1.6-11.7) | .001 |
| IL6 | Karabon e tal18 | II-IV | DNR | NR | rs1800795 | G/C | 0.5 | 3.4 | (1.1-13.0) | .03 |
| IL6 | Ambruzova et al19 | II-IV | PT | Allo | rs1800795 | G/C | 0.5 | 2.2 | (1.1-4.4) | .03 |
| IL10 | Lin et al1 | III-IV | PT | MRD | rs1800871† | A/C | 0.3 | |||
| IL10 | Lin et al1 | III-IV | PT | MRD | rs1800872 | A/C | 0.3 | 0.4 | (0.2-0.9) | .02 |
| CTLA4 | Perez-Garcia et al8 | II-IV | DNR | MRD | rs3087243 | A/G | 0.5 | 1.5 | (1.0-2.3) | .03 |
| HPSE | Ostrovsky et al12 | II-IV and III-IV | PT | Allo | rs4364254 | T/C | 0.4 | NR | NR | < .01 |
| MTHFR | Robien et al30 | I-IV | PT | Allo | rs1801131 | A/C | 0.4 | 3.6 | (1.0-12.7) | < .01 |
Allo indicates allogeneic; CI, confidence interval; DNR, donor; HR, hazard ratio; MAF, minor allele frequency; MRD, matched related donor; NR, not reported; PT, patient; SNP, single-nucleotide polymorphism; and URD, unrelated donor.
The first allele is designated as the minor allele, and the second allele is designated as the major allele.
Assessed as component of a haplotype.
Associations between IL6, IL2, CTLA4, HPSE, and MTHFR SNPs and aGVHD
Results for the remaining 14 evaluable candidate SNPs are summarized in Figure 2A-D. These analyses revealed that rs1800795 in IL6, rs2069762 in IL2, rs3087243 in CTLA4, rs4364254 in HPSE, and rs1801131 in MTHFR had unadjusted or adjusted models that met the P < .05 threshold (Table 3). The previously published associations for these SNPs are summarized in Table 4 for comparison. We report in the next 3 paragraphs the details of these associations in our cohort.
Forest plots for unadjusted and adjusted analyses of aGVHD and candidate SNPs. Forest plots for unadjusted (squares) and adjusted (circles) analyses of aGVHD and candidate SNPs among MRD (A) and URD (B) HCT. Hazard ratios are represented on the x-axis in logarithmic scale. Positive values indicate an increased risk for aGVHD, and negative values indicate a decreased risk for aGVHD. Gene/rs numbers in bold and closed squares and circles indicate the association exceeded the P = .05 threshold.
Forest plots for unadjusted and adjusted analyses of aGVHD and candidate SNPs. Forest plots for unadjusted (squares) and adjusted (circles) analyses of aGVHD and candidate SNPs among MRD (A) and URD (B) HCT. Hazard ratios are represented on the x-axis in logarithmic scale. Positive values indicate an increased risk for aGVHD, and negative values indicate a decreased risk for aGVHD. Gene/rs numbers in bold and closed squares and circles indicate the association exceeded the P = .05 threshold.
Of all the SNPs that met the P < .05 threshold, the associations with rs1800795 in IL6 were most consistently observed throughout the genetic models. In the unadjusted and adjusted URD analyses, the donor genotype for rs1800795 was associated with a 20% to 50% increase in the risk for grade IIb-IV aGVHD in the allelic (unadjusted P = .003; adjusted P = .011) and recessive (unadjusted P < .001; adjusted P = .001) models (Table 3). For MRD HCT, the donor genotype for rs1800795 was associated with a 60% increase in risk for grade III-IV aGVHD in both dominant unadjusted (P = .026) and adjusted (P = .028) models. We also found the patient C allele for rs1800795 in IL6 was associated with a 20% to 26% decrease in risk of grade IIb-IV aGVHD but only in the adjusted MRD allelic (P = .024) and dominant (P = .020) models (Figure 2A).
We found that the IL2 polymorphism rs2069762 in the donor genotype was associated in the dominant genetic mode with a 1.3-fold increase in risk of grade III-IV aGVHD after URD HCT (P = .049; Table 3, Figure 2B). The P value shifted slightly to .056 in multivariate analysis, but the point estimate remained unchanged at 1.3. In both unadjusted and adjusted analyses, the patient genotypes in our cohort indicated a consistent shift toward slightly greater risks of grade IIb-IV and III-IV aGVHD after URD HCT in the allelic and recessive models, but the P values did not exceed the .05 threshold. For CTLA4, the donor rs3087243 genotype was associated with an increased risk for grade III-IV aGVHD after URD HCT in both the unadjusted (HR 1.5, P = .014) and adjusted (HR 1.65, P = .004) analyses (Table 3, Figure 2B). However, unlike the published association, the point estimates in our analysis of the donor rs3087243 genotype in MRD HCT suggested a reduced risk for grade IIb-IV aGVHD in all models, with no appreciable association with risk for grade III-IV aGVHD (Figure 2A).
In the recently published HSPE analysis, the patient-donor disparity for SNP rs4693608 and rs4364254 genotypes was examined.12 Because the imputed rs4693608 genotype did not meet our quality control threshold, we were not able to perform a disparity analysis, and therefore we restricted our analysis to rs4364254, which was directly genotyped. The donor genotype for rs4364254 was significantly associated with a 1.2-fold increase (P = .042) in risk for grade IIb-IV aGVHD after URD HCT in the dominant multivariate model (Table 3, Figure 2B). For the MTHFR gene, we identified an association between rs1801131 and grade III-IV aGVHD. But unlike the original publication, the patient genotype was associated with a 35% decrease in risk (P = .031) in the dominant model after MRD HCT (Table 3, Figure 2A).
Discussion
We conducted an independent replication analysis of 14 candidate SNP genotypes previously documented in the published literature to be associated with the risk of aGVHD. To our knowledge, this is the first attempt to replicate such a large number of published genetic associations with the use of genotyped and imputed genome wide array data generated from a relatively large cohort of HCT patients and donors. Our analytical approach and the results of these analyses raise several key questions regarding candidate gene genetic association studies, some of which are of particular importance to the field of HCT.
The idea of an agnostic genome-wide approach toward discovery of risk variants is particularly challenging for HCT because of the cost of comprehensively screening both the patient and donor genomes and the need to accumulate a sample size of at least 5000 transplantations (10 000 total samples from patients and donors) to attain > 80% power for detecting significant associations with phenotype of > 30% frequency and odds ratios ranging from 1.5 to 1.8 for SNPs with minor allele frequencies > 20%.45 As we await the assembly of such a cohort, a bridging approach might be to use genome wide data from an intermediate sized cohort to conduct candidate gene studies, using both genotyped and imputed data.
The imputation approach, which has been widely adopted in genetic association studies,46 facilitates the use of actual genotypes for a limited set of SNPs to access information for a much larger number of SNPs across the entire genome. Our validation analysis of the imputed genotypes for rs1800871 and rs1800872 in IL10 demonstrate that imputed genotype data can be robust and correlates well with actual genotype data. This approach opens the possibility of evaluating candidate genes in pathways of high biologic relevance. Although this approach is limited in scope, it is financially and statistically more realistic for the limited sample size of most HCT cohorts. However, this approach is restricted by the quantity of usable imputed data, which depends on the extent and quality of the genome-wide SNP data generated by the commercial array. In our study, the SNP data generated by the Affymetrix 5.0 array only had a success rate of 40% for useable high-quality actual and imputed genotypes for the specified candidate SNPs, thus highlighting the limitation of this approach, at least with this array.
Our IL6 results suggest that the association with the IL6 SNP is likely worthy of more in-depth analysis. We found significant associations with the IL6 SNP genotype in both the patient and donor genomes, as well as with both aGVHD phenotypes. Associations with the donor and patient IL6 genotypes have been previously reported, all with risks that are similar in magnitude in comparison with our findings.10,18,19 In particular, the association of patient genotype has been validated under our multivariate analysis, in terms of all factors such as genome, donor type, and association model. However, the association of donor genotype is only arguably validated because the original findings were discrepant with regards to the assignment of the minor allele. Although both studies identified the association to be with the donor's G allele, Mullighan et al10 assigned the G allele as the major allele and Karabon et al18 assigned the G allele as the minor allele. The complementary nature of the C and G alleles, in conjunction with their similar allele frequencies, may have resulted in these differences in minor allele assignment in the genotyping process. However, because the minor allele's frequency is close to 50% in both study populations and this appears to vary among different ethnic groups in the HapMap database, it is possible that because of regional ethnic/racial differences in previous and current study populations, the minor allele frequencies may vary to the point where the minor allele assignment can differ between studies.
To judge whether these IL6 findings are significant, the totality of the IL6 association data should be considered. An association with polymorphisms in the promoter region of IL6 is biologically plausible. SNP rs1800795 is located at position −174 in the 5′ flanking region of the IL6 gene.47 This SNP has been documented to result in up-regulation of IL6 gene transcription and higher circulating IL6 levels.47 This SNP has also been reported to influence the risk for many other phenotypes, including juvenile arthritis,47 type 2 diabetes,48 bronchiolitis obliterans syndrome after lung transplantation,49 and kidney allograft survival.50 Multiple studies have documented increased plasma or serum concentrations of IL6 in patients with aGVHD.18 IL6 is a pleiotropic interleukin that in the context of an allogeneic HCT functions as both an acute phase mediator of inflammation and also as a critical factor in the differentiation of regulatory and Th17 effector cells. These effects may be mediated through variable IL6 expression encoded by both the host and donor genomes. It is unclear why donor cells genetically predisposed to lower circulating IL6 levels will increase the risk for aGVHD. It is feasible that a patient predisposed to chronically elevated IL6 levels (the phenotype associated with the major allele) may experience down regulation of the IL6 receptor and thereby reduce their responsiveness to IL6 and risk for aGCHD. This hypothesis requires further investigation.
The results for IL2, CTLA4, HPSE, and MTHFR indicate that the definition of “replication” is worthy of additional discussion in the context of genetic association studies in HCT. For each of these genes, we were able to identify an association with the candidate SNP that met the P < .05 threshold. However, certain aspects for each association were inconsistent with the original published findings. In the strictest sense, these associations did not represent replications of the original results. For IL2, our association was with the donor genome, which was inconsistent with the patient genome association in the original publication. For CTLA4, our association was observed with URD HCT and not with MRD HCT, which was the transplantation type for which the original association was documented.
For HSPE, we identified a SNP association with aGVHD, but this association was not reported as statistically significant in the original study. In fact, Ostrovsky et al reported that rs4364254 trended toward a lower cumulative risk for grades II-IV and III-IV aGVHD in the recessive model.12 Unfortunately, we were not able to impute the genotypes for rs4693608, the key HSPE SNP that was associated with aGVHD risk. For MTHFR, we observed an association with the same genome and donor type, but the observed effect was opposite to that of the original publication. At best, the associations we observed can be considered as new discoveries; at worst, they represent false-positive associations. Thus, our results cannot be used alone to judge whether a previously published association is valid. Instead, our results should be combined with previously published association studies to judge the likelihood that an association remains worthy of additional investigation.
If our IL6 findings are regarded as the only true replication of previously reported findings, then the replication rate in our analysis is only 7%. This low rate of replication is consistent with the observed replication rate of 3.6% in a comprehensive review of 166 candidate SNPs with putative associations that had been evaluated 3 or more times in the published literature.51 Guidelines for replicating genotype-phenotype associations are now well defined. These guidelines indicate that the likelihood of replicating a genetic association depends on many variables that can be related to the study population, genetic approach, phenotype definition and statistical power. We believe our results are valid because we took every measure to remain faithful to these guidelines.
Although we are unable to ensure that the ethnic background of our cohort is comparable with those of the published studies, we incorporated the principal component approach to account for the potential confounding effects of population stratification. Because there is little that we can do to control for center-to-center differences in clinical practice, we attempted to minimize imbalance in clinical risk factors by stratifying our analyses according to donor type, which provides some degree of standardization regarding treatment regimens. We also attempted to minimize center-specific differences in phenotype ascertainment by analyzing each SNP with 2 aGVHD phenotypes, increasing the likelihood that our aGVHD phenotypes approximate those described at other centers. Finally, our cohort was larger than all of the previously studied cohorts, and hence was at least equally powered to detect the original associations.
In summary, our study demonstrates the advantages and disadvantages of a novel approach via the use of imputed SNP data in genetic analyses and sets high standards for the conduct and reporting conventions of genetic association studies in the HCT population. If HCT investigators apply these standards uniformly, their findings may be more easily interpreted and replicated by other investigators in this field, thereby significantly improving the credibility of future genetic association reports. This has the potential to increase the likelihood that non-HLA genetic studies can contribute meaningfully to advancing the current understanding of the biology of aGVHD and improve our management of this disease.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grants AI33484, AI149213, CA015704, CA18029, HL087690, HL088201, HL094260, HL105914, and K23HL69860.
National Institutes of Health
Authorship
Contribution: J.W.C., L.P.Z., P.J.M., B.E.S., M.B., E.H.W., and J.A.H. were responsible for the study concept and design; X.C.Z., W.H.F., H.W., L.P.Z., and B.E.S. were responsible for data acquisition, statistical analysis, imputation, and informatics analyses; and J.W.C., X.C.Z., L.P.Z., P.J.M., B.E.S., M.B., E.H.W., and J.A.H. were responsible for data interpretation and writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jason W. Chien, MD, MSc, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, Suite D5-280, Seattle, WA 98109-1024; e-mail: jchien@fhcrc.org.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal