Abstract
The ABO blood group is of great importance in blood transfusion and organ transplantation. However, the mechanisms regulating human ABO gene expression remain obscure. On the basis of DNase I–hypersensitive sites in and upstream of ABO in K562 cells, in the present study, we prepared reporter plasmid constructs including these sites. Subsequent luciferase assays indicated a novel positive regulatory element in intron 1. This element was shown to enhance ABO promoter activity in an erythroid cell–specific manner. Electrophoretic mobility–shift assays demonstrated that it bound to the tissue-restricted transcription factor GATA-1. Mutation of the GATA motifs to abrogate binding of this factor reduced the regulatory activity of the element. Therefore, GATA-1 appears to be involved in the cell-specific activity of the element. Furthermore, we found that a partial deletion in intron 1 involving the element was associated with Bm phenotypes. Therefore, it is plausible that deletion of the erythroid cell–specific regulatory element could down-regulate transcription in the Bm allele, leading to reduction of B-antigen expression in cells of erythroid lineage, but not in mucus-secreting cells. These results support the contention that the enhancer-like element in intron 1 of ABO has a significant function in erythroid cells.
Introduction
The ABO blood group system is of great importance in blood transfusion and organ transplantation. The system comprises complex carbohydrate structures that are biosynthesized by the A and B transferases encoded by the A and B genes, respectively.1 Since Yamamoto et al elucidated the molecular genetic basis of the ABO system, several weak phenotypes or subgroups have been found that are caused by single nucleotide polymorphisms, hybrid formation between the common alleles, and mutations outside of the catalytic domain of the enzyme.2–9 However, some weak phenotypes without any mutation in the coding region or splicing site are known to be present.10 For example, the molecular basis for reduced B-antigen expression in persons possessing the Bm phenotype is currently unclear. RBCs of persons with the Bm phenotype are agglutinated by anti-H, but not by anti-B or anti-A,B, whereas the saliva of Bm secretors contains approximately as much B substance as that of normal B secretors. However, the B antigens on RBCs can only be detected by sensitive techniques such as adsorption and elution of anti-B.1 Therefore, Bm could be characterized by rather selectively reduced expression of the B-transferase gene in erythropoietic tissue, but not in mucus-secreting cells.10 Conversely, the distribution of the A and B antigens is cell type specific; for example, they are absent from the CNS, muscle, and connective tissue. Moreover, ABH antigens are known to be expressed during the maturation of erythroid and epithelial cells.1 However, delineation of these phenomena has been speculative because of insufficient information about the regulatory mechanisms of ABO expression.
Gene expression is driven by promoters, enhancers, trans-acting factors, and other cis-regulatory elements upstream and downstream of the gene. Studies of the human ABO gene using cultured cells have demonstrated transcriptional regulatory elements.11–17 A proximal promoter and a negative regulatory region just upstream from it have been found within the ABO CpG island. The proximal promoter is rich in CpG and shows no apparent tissue specificity. However, a cell-type–specific promoter has been demonstrated at the 5′ boundary of the CpG island, although its role remains obscure. Furthermore, 4 tandem copies of a 43-bp repeat unit located 3.8 kb upstream of transcription starting exon 1 driven by the proximal promoter have been shown to have enhancer activity in human gastric cancer KATOIII cells, but not in human erythroleukemia HEL cells,11,14 suggesting that the enhancer potential of this element is dependent on cell type. Recently, genome-wide approaches for the discovery of enhancers have become available.18 Regulatory elements are often characterized by the presence of DNase I–hypersensitive sites (DHSs), which can mark positions where transcription factors bind to DNA. Other chromatin features found in distant regulatory elements are specific histone modification signatures such as increased levels of histone H3 monomethylation at lysine residue 4 and acetylation at lysine residue 9.19,20 In addition, these sequences are often conserved across species. All of these features can be used to identify putative functional elements, and these powerful strategies are now being applied widely.
In the present study, we identified an erythroid cell–specific positive regulatory element in intron 1 of ABO through investigation of the enhancer potential of DHSs in genomic DNA in and upstream of ABO using luciferase reporter assays with the human erythroleukemia cell line K562. Furthermore, we found that a partial deletion involving the element was linked to the Bm phenotypes, supporting the contention that ABO expression is controlled by the element in intron 1.
Methods
Cells
K562 cells (JCRB0019) and KATOIII cells (JCRB0611) were cultured as described previously.17 The human embryo fibroblast cell line OUMS-36T-1 (JCRB1006.1) was grown in DMEM containing 10% FBS (Invitrogen), 100 U/mL of penicillin, and 100 μg/mL of streptomycin.
ABO grouping
All persons included in this study were Japanese. ABO phenotypes, including the common types and subgroups, were serologically determined on the basis of their features.1 Phenotype Bm was determined using agglutination tests with anti-A, anti-B, and anti-A,B Abs, as well as Ulex europaeus, adsorption-elution tests with pooled antisera prepared from 20 persons with the A phenotype,21 agglutination inhibition tests for ABH substances in saliva, and plasma B-transferase assay.
PCR
Genomic DNA was prepared from K562 cells and peripheral blood samples by standard phenol-chloroform extraction. The DNA samples except for 1 Bm sample were from a library of donors available at the Japanese Red Cross Tokyo Blood Center. This study was approved by the Ethics Committees at Gunma University Graduate School of Medicine and the Japanese Red Cross Society.
For PCR amplification, genomic DNA (100 ng) was mixed in a 50-μL final volume containing 1 × PrimeSTAR GXL buffer with PrimeSTAR GXL DNA polymerase (TaKaRa), 0.2 mmol/L of dNTP, and 0.2 μmol/L of each primer.
Genomic DNA from K562 cells was used for the preparation of reporter constructs. Sequences of the primers used and PCR targets are shown in Figure 1 and Table 1. The PCR conditions used for amplification of regions −8.8, +5.8, and +11.5 are listed in Table 2. PCR products were cloned into cloning vector pUC118 using a Mighty Cloning Reagent Set (Blunt End; TaKaRa). The nucleotide sequences of the amplified fragments were determined with a BigDye Terminator Version 1.1 Cycle Sequencing Kit (Applied Biosystems) with both M13 forward and reverse primers, specific primers for the target, and the primers used for PCR. The sequencing run was performed on a Genetic Analyzer (model 310; Applied Biosystems).
A map of the 35-kb region of genomic DNA in and upstream of the human ABO gene. The top diagram indicates the ABO gene exons 1-7 as represented by vertical lines with coordinates in hg18. The second diagram from the top indicates DHSs that were constructed through Washington digital DNase genomic footprinting on the UCSC genome browser. DHSs are denoted by arrows and contiguous DHSs are integrated into one site and represented by a thick arrow. These DHSs are referred to as −8.8, −5.5, −3.8, −0.7, −0.1, +2.0, +5.8, and +11.5. The third diagram from the top indicates the locations of DNA fragments that were obtained by PCR amplification or restriction enzyme digestion of human genomic clone HG-1, followed by subcloning upstream of the ABO proximal promoter in reporter plasmids. The solid boxes represent PCR amplicons and the clear boxes indicate the genomic DNA fragments from the HG-1 clone. Transient transfection into K562 cells was performed using 2.5 μg of firefly luciferase reporter plasmid and 0.01 μg of pRL-SV40 Renilla luciferase reporter vector for each analysis. The diagram at the bottom shows the activities of individual reporter plasmids, in which the activity of reporter plasmid SN containing the promoter was assigned an arbitrary value of 1.0, indicated by a clear box. The results are expressed as an average of the relative activity observed. The mean values were calculated from more than 3 independent experiments. DNA regions −8.8, +5.8, and +11.5 were inserted upstream of the promoter sequence in both directions, and luciferase activity is expressed as the average activity of reporter plasmids in which the same DNA fragment was inserted in either direction.
A map of the 35-kb region of genomic DNA in and upstream of the human ABO gene. The top diagram indicates the ABO gene exons 1-7 as represented by vertical lines with coordinates in hg18. The second diagram from the top indicates DHSs that were constructed through Washington digital DNase genomic footprinting on the UCSC genome browser. DHSs are denoted by arrows and contiguous DHSs are integrated into one site and represented by a thick arrow. These DHSs are referred to as −8.8, −5.5, −3.8, −0.7, −0.1, +2.0, +5.8, and +11.5. The third diagram from the top indicates the locations of DNA fragments that were obtained by PCR amplification or restriction enzyme digestion of human genomic clone HG-1, followed by subcloning upstream of the ABO proximal promoter in reporter plasmids. The solid boxes represent PCR amplicons and the clear boxes indicate the genomic DNA fragments from the HG-1 clone. Transient transfection into K562 cells was performed using 2.5 μg of firefly luciferase reporter plasmid and 0.01 μg of pRL-SV40 Renilla luciferase reporter vector for each analysis. The diagram at the bottom shows the activities of individual reporter plasmids, in which the activity of reporter plasmid SN containing the promoter was assigned an arbitrary value of 1.0, indicated by a clear box. The results are expressed as an average of the relative activity observed. The mean values were calculated from more than 3 independent experiments. DNA regions −8.8, +5.8, and +11.5 were inserted upstream of the promoter sequence in both directions, and luciferase activity is expressed as the average activity of reporter plasmids in which the same DNA fragment was inserted in either direction.
Location of DNA regions and oligonucleotide primer pairs for PCR amplification of genomic DNA for preparation of reporter plasmid constructs
| DNA regions . | Position* . | 5′-primer . | 3′-primer . |
|---|---|---|---|
| −8.8 | −10536: −6495 | ATCTGGGAGAAGGAATGGCCAGCATT | TGGGCTCTCGTTAGCTCCTATGTTCTTTAGT |
| −5.5† | −6209: −4687 | (none) | |
| −3.8† | −3982: −3281 | (none) | |
| +2.0† | +1823: +2597 | (none) | |
| +5.8 | +4602: +7196 | ATTTCCCTTCTTCTCACTTTGGGTTTAATTTG | ATACTGGAGCAAGGAAAGGTCACCAGACAAT |
| +11.5 | +10803: +12797 | TGTTTACAGAAGCTTTGTTGCTAGTGGCCC | ATCAGCCCGACTGCTCCTGCAGACTCT |
| DNA regions . | Position* . | 5′-primer . | 3′-primer . |
|---|---|---|---|
| −8.8 | −10536: −6495 | ATCTGGGAGAAGGAATGGCCAGCATT | TGGGCTCTCGTTAGCTCCTATGTTCTTTAGT |
| −5.5† | −6209: −4687 | (none) | |
| −3.8† | −3982: −3281 | (none) | |
| +2.0† | +1823: +2597 | (none) | |
| +5.8 | +4602: +7196 | ATTTCCCTTCTTCTCACTTTGGGTTTAATTTG | ATACTGGAGCAAGGAAAGGTCACCAGACAAT |
| +11.5 | +10803: +12797 | TGTTTACAGAAGCTTTGTTGCTAGTGGCCC | ATCAGCCCGACTGCTCCTGCAGACTCT |
Position is presented by location relative to the ATG translation start site of exon 1 in ABO on genomic DNA of NCBI reference sequence NT_035014.4.
DNA regions −5.5, −3.8, and +2.0 were prepared from human genomic clone HG-1.
PCR conditions for amplification of genomic DNA
| PCR* . | Initial denaturation . | Cycles; denaturation . | Annealing . | Extension . | Incubation . |
|---|---|---|---|---|---|
| Region −8.8 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 60°C for 15 s | 68°C for 4 min | 68°C for 7 min |
| Region +5.8 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 60°C for 15 s | 68°C for 3 min | 68°C for 7 min |
| Region +11.5 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 60°C for 15 s | 68°C for 4 min | 68°C for 7 min |
| PCR1-PCR3 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 60°C for 15 s | 68°C for 16 min | 68°C for 7 min |
| PCR4 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 66°C for 15 s | 68°C for 8 min | 68°C for 7 min |
| PCR4† | 94°C for 3 min | 35 cycles; 98°C for 10 s | 66°C for 15 s | 68°C for 1 min | 68°C for 7 min |
| PCR5 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 66°C for 15 s | 68°C for 8 min | 68°C for 7 min |
| PCR6 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 66°C for 15 s | 68°C for 6 min | 68°C for 7 min |
| PCR7 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 66°C for 15 s | 68°C for 10 min | 68°C for 7 min |
| PCR8 and PCR9 | 94°C for 3 min | 30 cycles; 98°C for 1 min | 55°C for 48 s | 72°C for 3.4 min | 72°C for 10 min |
| PCR* . | Initial denaturation . | Cycles; denaturation . | Annealing . | Extension . | Incubation . |
|---|---|---|---|---|---|
| Region −8.8 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 60°C for 15 s | 68°C for 4 min | 68°C for 7 min |
| Region +5.8 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 60°C for 15 s | 68°C for 3 min | 68°C for 7 min |
| Region +11.5 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 60°C for 15 s | 68°C for 4 min | 68°C for 7 min |
| PCR1-PCR3 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 60°C for 15 s | 68°C for 16 min | 68°C for 7 min |
| PCR4 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 66°C for 15 s | 68°C for 8 min | 68°C for 7 min |
| PCR4† | 94°C for 3 min | 35 cycles; 98°C for 10 s | 66°C for 15 s | 68°C for 1 min | 68°C for 7 min |
| PCR5 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 66°C for 15 s | 68°C for 8 min | 68°C for 7 min |
| PCR6 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 66°C for 15 s | 68°C for 6 min | 68°C for 7 min |
| PCR7 | 94°C for 3 min | 35 cycles; 98°C for 10 s | 66°C for 15 s | 68°C for 10 min | 68°C for 7 min |
| PCR8 and PCR9 | 94°C for 3 min | 30 cycles; 98°C for 1 min | 55°C for 48 s | 72°C for 3.4 min | 72°C for 10 min |
Each reaction was performed by the 3-step PCR method. The PCR was started from the initial denaturation, followed by cycling reactions and incubation.
Alternative condition of PCR4 was used for direct sequencing of the shorter product derived from the Bm allele.
Primers for PCR amplification of genomic DNA are listed in Table 3. PCR-amplified regions are indicated as PCR1-9 in Figure 6A. The PCR conditions used for PCR1-9 are listed in Table 2. PCR products 2 and 4 were cloned and sequenced as described in the previous paragraph, and the nucleotide sequences of PCR products 4-9 were determined by direct sequencing. The ABO genotype was also determined by restriction fragment–length polymorphism of PCR8 and PCR9.22
Oligonucleotide primers for PCR amplification of genomic DNA prepared from persons with Bm and ABm, as well as the common ABO types and other B subgroups
| Primer* . | Sequence (5′ → 3′) . | PCR† . |
|---|---|---|
| ABO−186S | 5′ −GCGCCGTCCCTTCCTAGCAG−3′ | 6 |
| ABO+3043S | 5′ −GGAACTCCTCTGCAGTCATTCCC−3′ | 1, 2 |
| ABO+10803S | 5′ −TGTTTACAGAAGCTTTGTTGCTAGTGGCCC−3′ | 3 |
| ABO+4419S | 5′ −TGGAATTGCTGTCTCCTCTTTTAGTCC−3′ | 4, 5, 7 |
| ABO+11044AS | 5′ −TCCCTGGCGTTTCCCATACAC−3′ | 6 |
| ABO+11078AS | 5′ −GGTCCCTCCTGACCCTGACAA−3′ | 4 |
| ABO+12797AS | 5′ −ATCAGCCCGACTGCTCCTGCAGACTCT−3′ | 2 |
| ABO+15374AS | 5′ −GAGACATGCTGCAGATGGTCAGG−3′ | 1, 3 |
| ABO+17909AS | 5′ −CGCCACCCCACTCTGCTTG−3′ | 5 |
| ABO+19915AS | 5′ −GGCGTATCTGCGATTGCGTGT−3′ | 7 |
| ABO+17621S | 5′ −GAATTCATGTGGGTGGCACCCTGCCA−3′ | 8 |
| ABO+17860AS | 5′ −GAATTCACTCGCCACTGCCTGGGTCTC−3′ | 8 |
| ABO+18812S | 5′ −GGATCCAGGGGTGCACGGCCGGCGGC−3′ | 9 |
| ABO+19109AS | 5′ −AGGTAATCCACCTCGCTGAGGAA−3′ | 9 |
| Primer* . | Sequence (5′ → 3′) . | PCR† . |
|---|---|---|
| ABO−186S | 5′ −GCGCCGTCCCTTCCTAGCAG−3′ | 6 |
| ABO+3043S | 5′ −GGAACTCCTCTGCAGTCATTCCC−3′ | 1, 2 |
| ABO+10803S | 5′ −TGTTTACAGAAGCTTTGTTGCTAGTGGCCC−3′ | 3 |
| ABO+4419S | 5′ −TGGAATTGCTGTCTCCTCTTTTAGTCC−3′ | 4, 5, 7 |
| ABO+11044AS | 5′ −TCCCTGGCGTTTCCCATACAC−3′ | 6 |
| ABO+11078AS | 5′ −GGTCCCTCCTGACCCTGACAA−3′ | 4 |
| ABO+12797AS | 5′ −ATCAGCCCGACTGCTCCTGCAGACTCT−3′ | 2 |
| ABO+15374AS | 5′ −GAGACATGCTGCAGATGGTCAGG−3′ | 1, 3 |
| ABO+17909AS | 5′ −CGCCACCCCACTCTGCTTG−3′ | 5 |
| ABO+19915AS | 5′ −GGCGTATCTGCGATTGCGTGT−3′ | 7 |
| ABO+17621S | 5′ −GAATTCATGTGGGTGGCACCCTGCCA−3′ | 8 |
| ABO+17860AS | 5′ −GAATTCACTCGCCACTGCCTGGGTCTC−3′ | 8 |
| ABO+18812S | 5′ −GGATCCAGGGGTGCACGGCCGGCGGC−3′ | 9 |
| ABO+19109AS | 5′ −AGGTAATCCACCTCGCTGAGGAA−3′ | 9 |
Numerals of primers represent locations of the 5′terminus relative to the ATG translation start site of exon 1 in ABO on genomic DNA of NCBI reference sequence NT_035014.4.
PCR amplification in which the primer was used is indicated.
Plasmids
Luciferase reporter plasmids PXm/SN, SN, and TK-luc have been described previously.11 In reporter plasmid SN, the ABO proximal promoter located between −150 and −2 relative to the translation start site was subcloned upstream of the luciferase gene. The GATA-1 expression vector was kindly provided by Prof K. Ohneda at Takasaki University of Health and Welfare.
DNA regions −8.8, +5.8, and +11.5 were PCR amplified and cloned into the pUC118 cloning vector as described in “PCR” (Table 1). Both the SacI and MluI sites just upstream of the ABO proximal promoter sequences in construct SN were used to insert these DNA fragments in either direction. Restriction enzyme–digested fragments of regions −8.8, +5.8, and +11.5 were also subcloned upstream of the promoter. Construct +5.8Δ/SN was prepared by removal of subregion C through ApaI and MscI-digestion of DNA fragment +5.8 and blunt-end ligation. Insertion of subregion C 5′ adjacent to DNA fragment +5.8 lacking subregion C led to preparation of construct C+5.8Δ/SN. Subregion C was inserted into the BamHI and SalI sites downstream of the luciferase gene to generate construct SN/C. Subregion C was subcloned at the SacI and MluI sites just upstream of the thymidine kinase (TK) promoter and SV40 promoter in constructs C/TK and C/SV, respectively. Mutations of GATA motifs in subregion C were generated using overlapping PCR mutagenesis in constructs CmG1/SN, CmG2/SN, and CmG3/SN.
DNA regions −5.5, −3.8, and +2.0 were prepared by restriction enzyme digestion of human genomic clone HG-111 and subcloned upstream of the ABO promoter in the same direction as that of the promoter (Table 1). Region −3.8 was subcloned in construct PXm/SN as described previously.11
Directions of the inserts of all of the constructs used in this study were verified by detailed restriction enzyme mapping and DNA sequence analysis as described above. For all constructs containing PCR-amplified fragments, sequencing was performed over the entire region of the amplified sequences as described in “PCR.” Plasmid DNA was purified using a HiSpeed Plasmid Maxi Kit (QIAGEN).
Transfection and luciferase assay
Transient transfections of K562 cells and KATOIII cells were performed as described previously.17 Transient transfection into OUMS-36T cells was performed using lipofectamine according to the protocol of the supplier; 1 μg of firefly luciferase reporter plasmid and 0.01 μg of pRL-SV40 Renilla luciferase reporter vector were used for each analysis. After collecting the cells, cell lysis and luciferase assays were performed using the Dual Luciferase Reporter Assay System (Promega) to measure the activities of firefly and Renilla luciferases. Variations in transfection efficiency were normalized to the activities of Renilla luciferase expressed from the cotransfected pRL-SV40 Renilla luciferase reporter.
Preparation of nuclear extracts and EMSA
Nuclear extracts were prepared from K562 cells using a nuclear extraction kit (Panomics) in accordance with the manufacturer's instructions. The electrophoretic mobility–shift assay (EMSA) was carried out according to the method described by Holmes et al,23 The double-stranded GATA consensus and mutant oligonucleotides were purchased from Santa Cruz Biotechology, and the double-stranded oligonucleotides G1, G2, mG1, and mG2 were obtained by annealing 2 chemically synthesized strands. The sequences of the various oligonucleotides are listed in Table 4. Probes were prepared as described previously.11 Two microliters of anti–GATA-1 IgG (N6; Santa Cruz Biotechnology) or anti–GATA-2 IgG (CG2-96 and H-116; Santa Cruz Biotechnology) was added to the nuclear extracts overnight at 4°C before addition of the radiolabeled probe. The DNA-protein complex was quantified with a BAS-1800 image analyzer (Fuji).
Sequences of oligonucleotides used for EMSA
| Oligonucleotide . | Sequences* . |
|---|---|
| G1 | 5′-GAAAGGGAGAGATAAGGCTCACTAG-3′ and 5′-GCTAGTGAGCCTTATCTCTCCCTTT-3′ |
| G2 | 5′-AGATTATCTGTTTAGTGACCTTGCAG-3′ and 5′-GCTGCAAGGTCACTAAACAGATAAT-3′ |
| mG1 | 5′-GAAAGGGAGACTTAAGGCTCACTAG-3′ and 5′-GCTAGTGAGCCTTAAGTCTCCCTTT-3′ |
| mG2 | 5′-AGATTAAGTGTTTAGTGACCTTGCAG-3′ and 5′-GCTGCAAGGTCACTAAACACTTAAT-3′ |
| GATA† | 5′-CACTTGATAACAGAAAGTGATAACTCT-3′ and 5′-AGAGTTATCACTTTCTGTTATCAAGTG-3′ |
| mGATA† | 5′-CACTTCTTAACAGAAAGTCTTAACTCT-3′ and 5′-AGAGTTAAGACTTTCTGTTAAGAAGTG-3′ |
| Oligonucleotide . | Sequences* . |
|---|---|
| G1 | 5′-GAAAGGGAGAGATAAGGCTCACTAG-3′ and 5′-GCTAGTGAGCCTTATCTCTCCCTTT-3′ |
| G2 | 5′-AGATTATCTGTTTAGTGACCTTGCAG-3′ and 5′-GCTGCAAGGTCACTAAACAGATAAT-3′ |
| mG1 | 5′-GAAAGGGAGACTTAAGGCTCACTAG-3′ and 5′-GCTAGTGAGCCTTAAGTCTCCCTTT-3′ |
| mG2 | 5′-AGATTAAGTGTTTAGTGACCTTGCAG-3′ and 5′-GCTGCAAGGTCACTAAACACTTAAT-3′ |
| GATA† | 5′-CACTTGATAACAGAAAGTGATAACTCT-3′ and 5′-AGAGTTATCACTTTCTGTTATCAAGTG-3′ |
| mGATA† | 5′-CACTTCTTAACAGAAAGTCTTAACTCT-3′ and 5′-AGAGTTAAGACTTTCTGTTAAGAAGTG-3′ |
The sequences of each oligonucleotide are shown. The double-stranded oligonucleotides were obtained by annealing 2 chemically synthesized strands. The underlining of oligonucleotides mG1 and mG2 indicates the sites of the specific mutations in the GATA motifs, similar to those in the mutant oligonucleotide mGATA.
The double-stranded GATA consensus and mutant oligonucleotides GATA and mGATA were purchased from Santa Cruz Biotechology. The underlining of oligonucleotide mGATA indicates the sites of the specific mutations in the GATA motifs.
Results
Enhancement of ABO promoter activity by a region encompassing DNase I–hypersensitive sites in intron 1 of ABO
K562 cells have been used as a model of the common progenitor of erythroblasts and megakaryocytes,24 and were previously demonstrated to express ABO.13 Accordingly, this cell line is useful for investigations of the mechanisms underlying ABO expression. Publicly available expression data from the University of California-Santa Cruz (UCSC) Genome Bioinformatics site (http://genome.ucsc.edu) obtained from the March 2006 (NCBI36/hg18) assembly show genome-wide maps for DHSs in the cells. Among them, Washington digital DNase genomic footprinting indicated many DHSs in the genomic DNA from 15 kb upstream to the end of the human ABO gene.25,26 Because DNase I sensitivity was indicated by the absolute enrichment of in vivo cleavage sites across the genome, we focused on DHSs located at −8.9/−8.6 kb, −5.5 kb, −4.0/−3.6 kb, −0.7 kb, −0.1 kb, +2.0 kb, +5.5/+5.8 kb, and +11.5 kb from the ATG translation start site of exon 1. Hereafter, we refer to these DHSs as DHS −8.8, −5.5, −3.8, −0.7, −0.1, +2.0, +5.8, and +11.5, respectively (Figure 1). To verify the enhancer potential of these sites using luciferase reporter assays, PCR-amplified fragments or genomic DNAs from the HG-1 clone encompassing them were subcloned upstream of the ABO proximal promoter sequence in luciferase reporter plasmids. Transient transfections into K562 cells were carried out using these reporter vectors. DNA regions used in the reporter plasmids are denoted as regions −8.8, −5.5, −3.8, +2.0, +5.8, and +11.5 (Figure 1 and Table 1).
The ABO proximal promoter was chosen because its activity was constitutive and higher, whereas the activity of the distal promoter was shown to be twice as high as that of the pGL3-basic vector after transient transfection into K562 cells.
In the upstream region, the reporter plasmids containing regions −8.8, −5.5, and −3.8 did not elicit increased luciferase activity compared with the original vector SN (Figure 1). Five subregions ranging in size from 0.8-2.3 kb from region −8.8 also elicited no increase in luciferase activity (data not shown). DHS −3.8 corresponds to the location of tandem copies of a 43-bp repeat unit located 3.8 kb upstream of exon 1. Region −3.8, including the repetitive elements of reporter construct PXm/SN, did not exhibit enhancer potential in K562 cells (Figure 1), although extrapolation of the experimental results from gastric cancer cells to ABO expression in cells of erythroid lineage has created controversy regarding the extent to which the number of enhancer repeats influences transcription in erythroid tissue.7,27–32 DHS −0.7 is concurrent with the distal promoter at the 5′ boundary of the CpG island, whereas DHS −0.1 coincides with the proximal promoter within the island.13 Within intron 1, transfection of reporter constructs including region +5.8 with an orientation the same as or opposite to that of the promoter resulted in 3.2- or 2.5-fold higher luciferase activity, respectively, than did that of SN. In contrast, reporter activity was not elevated by other regions, including regions +2.0 and +11.5. Moreover, luciferase activity was also not increased by reporter plasmids containing 3 subregions ranging in size from 0.6-1.4 kb from region +11.5 (data not shown). Therefore, in the first intron of ABO, we found a region that was considered likely to be involved in the regulation of ABO transcription.
Identification of a positive regulatory element in intron 1
First, to choose the most appropriate direction for inserts, region +5.8 was introduced upstream of luciferase in either direction. The vectors used did not contain the promoter sequence. The resulting reporter containing the insert oriented in a direction opposite to that of luciferase showed lower luciferase activity than the pGL3-basic vector, whereas the other reporter displayed higher activity (Figure 2). Accordingly, we constructed a series of plasmids in which various parts of region +5.8 were subcloned upstream of the ABO promoter in a direction opposite to that of luciferase to better locate the sequence essential for enhancement. Transfection of a reporter construct including the entire region (+5.8/SN construct) resulted in a 2.5-fold increase of luciferase activity compared with that of SN. Removal of the sequence between positions +4602 and +6361 (subregion B-F) resulted in a significant reduction of luciferase activity compared with the entire sequence. Construct B-F/SN expressed a 3.5-fold increase of luciferase activity compared with SN. Deletion of one end from position +6361 to +6154, +5648, and +5056 resulted in a large decrease of luciferase activity, suggesting that subregions B and C were required for enhancement of the promoter activity. In addition, deletion of the other end from positions +4602 to +5056, +5272, +5653, and +6154 led to a large decrease in reporter activity, indicating that subregions C and F were associated with the increase in luciferase activity. When fragment C was inserted upstream from the promoter sequence (C/SN construct), the luciferase activity of this reporter plasmid was as high as that of the construct containing the entire region. Strikingly, deletion of subregion C from the entire sequence abrogated the increase of luciferase activity in construct +5.8Δ/SN. These results indicate that a cis-acting element critical for reporter expression resides within subregion C between +5653 and +6154 of intron 1. In contrast, a reporter plasmid containing fragment F upstream from the promoter sequence expressed higher activity than SN, but the reporter activity was not comparable to that directed by the entire region. In addition, introduction of fragment B upstream of the promoter sequence resulted in a loss of 50% of the luciferase activity. These data suggest that subregion C is indispensable for the regulatory activity of the entire region +5.8 and that subregions B and F might be involved in maximum enhancement of the promoter activity by the entire region.
Summary of the relative luciferase activities of reporter constructs containing various parts of DNA region +5.8 in the ABO gene. Transient transfection experiments were performed using reporter plasmids in which various parts of region +5.8 were subcloned upstream of the ABO promoter sequence. Below the map of restriction enzyme sites with positions relative to the translation start site of exon 1, region +5.8 is subdivided into regions A through F. Construct names are indicated to the left of the square, and the locations of the fragments that were inserted upstream of the ABO promoter sequence in a direction opposite to that of the promoter are shown. Arrows to the left represent the inserts oriented in a direction opposite to that of the promoter. The v-shaped segment represents deleted sequences. The +5653 to +6154 sequence (subregion C) is indicated by solid box with an arrow. Each construct as depicted on the left was transiently transfected into K562 cells and the luciferase activity obtained was normalized, as shown in the right panel. To facilitate comparison of the corresponding reporter activity of each construct, the activity of reporter plasmid SN was assigned an arbitrary value of 1.0. The results are expressed as an average of the relative activity observed. The mean values and SDs were calculated from more than 3 independent experiments.
Summary of the relative luciferase activities of reporter constructs containing various parts of DNA region +5.8 in the ABO gene. Transient transfection experiments were performed using reporter plasmids in which various parts of region +5.8 were subcloned upstream of the ABO promoter sequence. Below the map of restriction enzyme sites with positions relative to the translation start site of exon 1, region +5.8 is subdivided into regions A through F. Construct names are indicated to the left of the square, and the locations of the fragments that were inserted upstream of the ABO promoter sequence in a direction opposite to that of the promoter are shown. Arrows to the left represent the inserts oriented in a direction opposite to that of the promoter. The v-shaped segment represents deleted sequences. The +5653 to +6154 sequence (subregion C) is indicated by solid box with an arrow. Each construct as depicted on the left was transiently transfected into K562 cells and the luciferase activity obtained was normalized, as shown in the right panel. To facilitate comparison of the corresponding reporter activity of each construct, the activity of reporter plasmid SN was assigned an arbitrary value of 1.0. The results are expressed as an average of the relative activity observed. The mean values and SDs were calculated from more than 3 independent experiments.
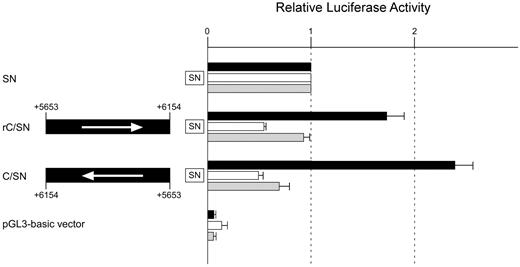
Characterization of subregion C
We further characterized the transcriptional activity of core subregion C. First, to examine whether the subregion activates promoter activity independently of orientation, a reporter assay was performed using construct rC/SN in which subregion C had been introduced in a direction opposite to that of the insert in plasmid C/SN. The luciferase activity was almost twice as high as that of construct SN (Figure 3), indicating that the subregion increases promoter activity irrespective of the insert direction. Next, to investigate whether subregion C activates the promoter independently of the distance from it, a reporter plasmid was prepared by addition of subregion C 5′ adjacent to region +5.8 lacking the subregion (C+5.8Δ/SN; Figure 2). Restoration of the sequence resulted in a 1.8-fold increase of luciferase activity compared with SN. Moreover, introduction of the sequence downstream of luciferase resulted in an approximately 1.5-fold increase of luciferase activity (SN/C; Table 5). These observations suggest that subregion C might exert positive regulatory activity independently of position. Third, to examine whether the function of the subregion is dependent on the promoter, luciferase assays were carried out using reporter plasmids in which the sequence had been subcloned upstream of the TK promoter or SV40 promoter. Approximately 2-fold higher luciferase activity was observed when the region was subcloned upstream of the TK promoter, whereas introduction of the sequence upstream of the SV40 promoter led to an approximately 1.3-fold increase of luciferase activity (Table 5). Therefore, these results indicate that the function of subregion C is dependent on the promoter. Finally, we examined whether the subregion exhibits cell type–specific activity. Transient transfection studies were performed using K562 cells and KATOIII cells expressing ABO and fibroblasts not expressing ABO (data not shown). Because the proximal ABO promoter shows activity independently of cell type,13 we calculated the relative ratios of luciferase activity between constructs SN and C/SN in those cells. A significant elevation of activity was demonstrated in K562 cells, whereas no elevation was observed in either KATOIII cells or fibroblasts (Figure 3). Therefore, subregion C appears to be a significant functional component specific to cells of erythroid lineage. On the basis of the data overall, we concluded that the +5653 to +6154 region in intron 1 has the potential to increase ABO promoter activity in an erythroid cell–specific manner.
Subregion C shows an enhancer potential specific to cells of erythroid lineage. Transient transfection experiments were carried out using K562 cells, KATOIII cells, and fibroblasts. Transfection into K562 cells was performed using 2.5 μg of firefly luciferase reporter plasmid and 0.01 μg of pRL-SV40 Renilla luciferase reporter vector for each analysis. Transfection into KATOIII cells was carried out using 6 μg of firefly reporter and 0.01 μg of pRL-SV40 Renilla reporter. Transfection into fibroblasts was performed using 1 μg of firefly reporter and 0.01 μg of pRL-SV40 Renilla reporter. DNA fragment C between positions +5653 and +6154 was inserted upstream of the ABO promoter sequence in the same direction as that of the promoter in construct rC/SN, and in the opposite direction in construct C/SN. Arrows to the left represent the inserts oriented in a direction opposite to that of the promoter. To facilitate comparison of the corresponding reporter activity of each construct among the cells, the activity of the SN vector was assigned an arbitrary value of 1.0 in each cell line. The results are expressed as an average of the relative activity observed. The mean values and SDs were calculated from more than 3 independent experiments. Solid box indicates K562 cells; clear box, KATOIII cells; and gray box, fibroblasts.
Subregion C shows an enhancer potential specific to cells of erythroid lineage. Transient transfection experiments were carried out using K562 cells, KATOIII cells, and fibroblasts. Transfection into K562 cells was performed using 2.5 μg of firefly luciferase reporter plasmid and 0.01 μg of pRL-SV40 Renilla luciferase reporter vector for each analysis. Transfection into KATOIII cells was carried out using 6 μg of firefly reporter and 0.01 μg of pRL-SV40 Renilla reporter. Transfection into fibroblasts was performed using 1 μg of firefly reporter and 0.01 μg of pRL-SV40 Renilla reporter. DNA fragment C between positions +5653 and +6154 was inserted upstream of the ABO promoter sequence in the same direction as that of the promoter in construct rC/SN, and in the opposite direction in construct C/SN. Arrows to the left represent the inserts oriented in a direction opposite to that of the promoter. To facilitate comparison of the corresponding reporter activity of each construct among the cells, the activity of the SN vector was assigned an arbitrary value of 1.0 in each cell line. The results are expressed as an average of the relative activity observed. The mean values and SDs were calculated from more than 3 independent experiments. Solid box indicates K562 cells; clear box, KATOIII cells; and gray box, fibroblasts.
Effects of ABO intron 1 on transcriptional activation when the +5653 to +6154 sequence was inserted upstream of various promoters
| Reporter construct . | Promoter . | Insert . | Relative activity† . | -fold activity over control . |
|---|---|---|---|---|
| Test plasmids | ||||
| C/SN | ABO proximal promoter | +5653: +6154‡ | 2.386 ± 0.170 | 2.4 |
| C/TK | TK promoter | +5653: +6154‡ | 8.041 ± 1.583 | 2.0 |
| C/SV | SV40 promoter | +5653: +6154‡ | 7.247 ± 0.170 | 1.3 |
| SN/C | ABO proximal promoter | +5653: +6154§ | 1.497 ± 0.140 | 1.5 |
| Controls | ||||
| SN | ABO proximal promoter | None | 1.000 | |
| TK-luc | TK promoter | None | 4.023 ± 0.718 | |
| pGL3-promoter | SV40 promoter | None | 5.555 ± 1.315 | |
| pGL3-basic | None | None | 0.060 ± 0.014 | |
| Reporter construct . | Promoter . | Insert . | Relative activity† . | -fold activity over control . |
|---|---|---|---|---|
| Test plasmids | ||||
| C/SN | ABO proximal promoter | +5653: +6154‡ | 2.386 ± 0.170 | 2.4 |
| C/TK | TK promoter | +5653: +6154‡ | 8.041 ± 1.583 | 2.0 |
| C/SV | SV40 promoter | +5653: +6154‡ | 7.247 ± 0.170 | 1.3 |
| SN/C | ABO proximal promoter | +5653: +6154§ | 1.497 ± 0.140 | 1.5 |
| Controls | ||||
| SN | ABO proximal promoter | None | 1.000 | |
| TK-luc | TK promoter | None | 4.023 ± 0.718 | |
| pGL3-promoter | SV40 promoter | None | 5.555 ± 1.315 | |
| pGL3-basic | None | None | 0.060 ± 0.014 | |
The constructs as depicted above were transiently transfected into K562 cells; 2.5 μg of luciferase reporter and 0.01 μg of Renilla luciferase control vector were used for each analysis.
The results are expressed as the average relative activity compared with that observed for reporter vector SN containing the ABO proximal promoter. SDs are indicated for a minimum of 3 repetitions.
The +5653 to +6154 sequence was subcloned upstream of the ABO proximal promoter, TK promoter, and SV40 promoter in an orientation opposite to the transcription direction in these promoters.
The +5653 to +6154 sequence was subcloned downstream of the luciferase gene in the same orientation as the transcription direction in the promoter.
Involvement of GATA-1 in the cell type–specific regulatory activity of subregion C
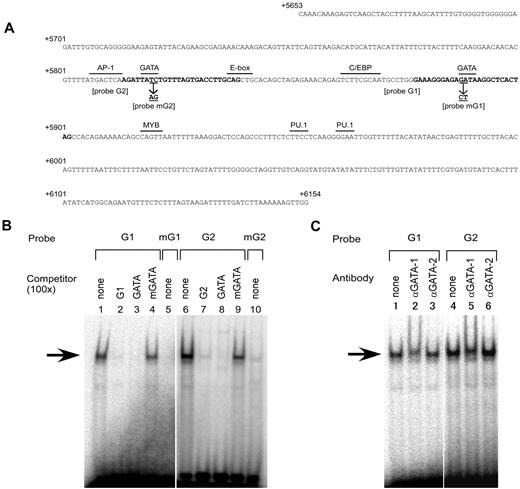
Inspection of the nucleotide sequence between +5653 and +6154 revealed 2 consensus binding sites for the hematopoietic transcription factors GATA-1, GATA-2, and GATA-3 (Figure 4A). One site was centered on position +5819 and the other on +5890. Therefore, the erythroid-specific potential of the region might be conferred through the binding of such tissue-specific factors. Because it was shown that GATA-1 and GATA-2 were expressed in K562 cells,33 we examined whether those factors bind to subregion C using EMSA with the labeled probes G1 or G2, corresponding to the putative GATA sites at position +5890 or +5819, respectively (Figure 4A). The oligonucleotide G1 or G2 probe produced a major up-shifted band when each probe was incubated with the nuclear extracts from K562 cells (Figure 4B lanes 1 and 6). Formation of the up-shifted complexes, indicated by arrows in Figure 4B, was decreased by addition of competing unlabeled self-oligonucleotides or oligonucleotide GATA containing the consensus motif for GATA transcription factors (Figure 4B lanes 2, 3, 7, and 8), but not by the addition of oligonucleotide mGATA containing the mutated version of the GATA motif to abrogate the binding of GATA factors34 (Figure 4B lanes 4 and 9). Consistently, formation of the DNA-protein complex was significantly reduced when an oligonucleotide mG1 or mG2 probe with the same mutation in the GATA motif was incubated with the nuclear extracts (Figure 4B lanes 5 and 10). To investigate whether GATA-1 or GATA-2 binds to the oligonucleotide G1 and G2 probes, anti–GATA-1 or anti–GATA-2 Ab was added to the nuclear extracts. For both probes, addition of the anti–GATA-1 Ab resulted in loss of the DNA-protein complex and the appearance of another complex that migrated slightly more slowly (Figure 4C lanes 2 and 5), whereas no change was observed after addition of the anti–GATA-2 Ab (Figure 4C lanes 3 and 6). In addition, another Ab against GATA-2 failed to supershift or neutralized the formation of the DNA-protein complex (data not shown). These results indicate that GATA-1 likely binds to both GATA sites in subregion C in vitro.
GATA-1 specifically binds to 2 GATA motifs in subregion C. (A) Nucleotide sequence of subregion C. The sequence from position +5653 to +6154 is shown relative to the ATG translation start site of ABO. The sequence was derived from the genomic DNA of K562 cells (JN863720). The motifs for several relevant transcription factors and the E-box are indicated by over bars. The sequences typed in bold indicate the oligonucleotides G1 and G2 used in EMSA. The positions and identities of the mutations in the GATA motifs used in the EMSA and transfection experiments are represented by underlining and arrows. (B) Oligonucleotides derived from subregion C bind a nuclear protein. EMSAs were performed using the nuclear extracts from K562 cells. DNA-protein interaction was investigated using radiolabeled probes G1 and G2 in the presence or absence of a 100-fold molar excess of competing unlabeled oligonucleotides (lanes 1-4 and 6-9). Oligonucleotides GATA and mGATA represent a GATA consensus oligonucleotide and a mutant oligonucleotide containing “GA” to “CT” substitutions within the GATA motif, respectively. Mutated versions of oligonucleotides G1 and G2 and mG1 and mG2, respectively, were used as radiolabeled probes (lanes 5 and 10). The major shifted complex is indicated by an arrow. (C) Anti–GATA-1 Ab diminishes the DNA-protein complex. The nuclear extracts prepared from K562 cells were preincubated with anti–GATA-1 Ab (N6) or anti–GATA-2 Ab (CG2-96) before the addition of the radiolabeled probes.
GATA-1 specifically binds to 2 GATA motifs in subregion C. (A) Nucleotide sequence of subregion C. The sequence from position +5653 to +6154 is shown relative to the ATG translation start site of ABO. The sequence was derived from the genomic DNA of K562 cells (JN863720). The motifs for several relevant transcription factors and the E-box are indicated by over bars. The sequences typed in bold indicate the oligonucleotides G1 and G2 used in EMSA. The positions and identities of the mutations in the GATA motifs used in the EMSA and transfection experiments are represented by underlining and arrows. (B) Oligonucleotides derived from subregion C bind a nuclear protein. EMSAs were performed using the nuclear extracts from K562 cells. DNA-protein interaction was investigated using radiolabeled probes G1 and G2 in the presence or absence of a 100-fold molar excess of competing unlabeled oligonucleotides (lanes 1-4 and 6-9). Oligonucleotides GATA and mGATA represent a GATA consensus oligonucleotide and a mutant oligonucleotide containing “GA” to “CT” substitutions within the GATA motif, respectively. Mutated versions of oligonucleotides G1 and G2 and mG1 and mG2, respectively, were used as radiolabeled probes (lanes 5 and 10). The major shifted complex is indicated by an arrow. (C) Anti–GATA-1 Ab diminishes the DNA-protein complex. The nuclear extracts prepared from K562 cells were preincubated with anti–GATA-1 Ab (N6) or anti–GATA-2 Ab (CG2-96) before the addition of the radiolabeled probes.
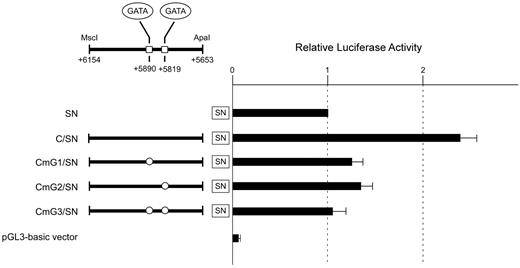
To evaluate whether these GATA sites are crucial for the regulatory activity of subregion C, the same mutations in the GATA motifs used in the EMSA were incorporated into 3 constructs, followed by transient transfection into K562 cells. The mutation of the GATA site centered on +5819 or +5890 resulted in a loss of 75% or 82%, respectively, relative to the wild-type sequence (Figure 5). Furthermore, mutation of both sites abrogated the increase of luciferase activity by the subregion. These results indicated that either GATA site is indispensable for the regulatory activity of subregion C. Therefore, the erythroid cell–specific activity of subregion C seems to be dependent on these GATA sites.
Mutations of the GATA motifs reduce the regulatory potential of subregion C. The wild-type subregion C construct C/SN or mutant constructs CmG1/SN, CmG2/SN, and CmG3/SN carrying “GA” to “CT” substitutions in the GATA motif were transiently transfected into K562 cells. Boxes indicate the locations of putative GATA-binding sites. The clear circles represent mutated sequences of the GATA sites. To facilitate comparison of the corresponding reporter activity of each construct, the activity of reporter plasmid SN was assigned an arbitrary value of 1.0. The results are expressed as an average of the relative activity observed. The mean values and SDs were calculated from more than 3 independent experiments.
Mutations of the GATA motifs reduce the regulatory potential of subregion C. The wild-type subregion C construct C/SN or mutant constructs CmG1/SN, CmG2/SN, and CmG3/SN carrying “GA” to “CT” substitutions in the GATA motif were transiently transfected into K562 cells. Boxes indicate the locations of putative GATA-binding sites. The clear circles represent mutated sequences of the GATA sites. To facilitate comparison of the corresponding reporter activity of each construct, the activity of reporter plasmid SN was assigned an arbitrary value of 1.0. The results are expressed as an average of the relative activity observed. The mean values and SDs were calculated from more than 3 independent experiments.
Conversely, the luciferase activity of reporter C/SN was not increased when GATA-1 protein was overexpressed in KATOIII cells and fibroblasts in which the factor was not expressed (data not shown). These findings suggest that one or more factors other than GATA-1 might be required for the regulatory function of the subregion.
Association of the Bm phenotype with a partial deletion of intron 1 involving the erythroid cell–specific regulatory element
It has been suggested that the Bm phenotype results from reduction of B gene expression in BM cells, but not in mucus-secreting cells.10 Because this discrepancy can be explained by deletion of the erythroid cell–specific regulatory element, intron 1 including the element was investigated by PCR analysis using various combinations of primers with genomic DNA prepared from a person with the Bm phenotype (Figure 6A). Compared with a single main product from a control person with the B phenotype, an additional product of smaller size was observed in PCR1 and PCR2 from a person with the Bm phenotype (Figure 6B). Conversely, a single main band of similar length was observed in PCR3 from both persons. Nucleotide determination of the shortened PCR fragment in PCR2 revealed deletion of nucleotides located between positions +5137 and +10914, a stretch that includes the element (Figure 6A). Subsequently, we prepared a set of primers flanking the deletion to display the defect definitively, followed by PCR4. Electrophoresis of the PCR-amplified products demonstrated 2 bands of 0.9 and 6.7 kb in Bm, and one band of 6.7 kb in B (Figure 6B). Both the O and B alleles were demonstrated in the person with the Bm phenotype by restriction fragment-length polymorphism and direct sequencing of PCR8 and PCR9 comprising exons 6 and 7, respectively (Figure 6A). These results indicated a partial deletion within intron 1 of the B or O allele in this person. PCR5-PCR7 were carried out to examine the allele in which the deletion was located, B or O (Figure 6A). Because the extension times in these PCR reactions were not appropriate for amplification of the target in the allele lacking the deletion, a single major band appeared in each PCR (data not shown). Direct sequencing of these PCR products demonstrated that the nucleotide sequences of exons 1-7 matched entirely those of the B101 allele and showed no mutation in either the exon-intron boundaries or the proximal promoter. These results suggest that a partial deletion of intron 1 involving the regulatory element is present in the B allele, which we refer to as the Bm allele hereafter.
A partial deletion of intron 1 involving the regulatory element is found in genomic DNAs derived from persons with Bm. (A) Schematic representation of the genomic organization of the human ABO gene, locations of the erythroid cell–specific regulatory element, PCR-amplified fragments, and a partial deletion of intron 1 in the Bm allele. The top diagram indicates ABO exons 1-7 as solid boxes. It is noteworthy that the direction of ABO is opposite of that in Figure 1. The clear box indicates the location of the erythroid cell–specific regulatory element. Positions are indicated relative to the ATG translation start site in exon 1. PCR-amplified fragments in PCR1-PCR9 are indicated by broken lines, and the primers used are represented as thick lines at both ends of individual broken lines. The relationship between the PCR amplifications and primers used is shown in Table 3. Deleted nucleotides in intron 1 of the Bm allele are indicated by a V-shaped segment. (B) A partial deletion of intron 1 was found in genomic DNA derived from a person with Bm. PCR1-PCR4 were carried out with genomic DNAs derived from persons with B and Bm using various sets of primers, followed by electrophoresis through 0.5% agarose gel and staining with ethidium bromide. Gene Ladder Wide 2 (Nippon Gene) was used as a molecular size marker. (C) Partial deletion found in genomic DNAs obtained from 10 persons with Bm or ABm. PCR4 amplifications were carried out using genomic DNAs derived from persons with A, B, O, AB, Bm, and ABm, followed by electrophoresis through 0.5% agarose gel and staining with ethidium bromide. Representative PCR products from 10 persons with Bm or ABm are displayed. The shorter PCR4 products obtained from those persons were sequenced, confirming that the deletion was located between +5137 and +10914.
A partial deletion of intron 1 involving the regulatory element is found in genomic DNAs derived from persons with Bm. (A) Schematic representation of the genomic organization of the human ABO gene, locations of the erythroid cell–specific regulatory element, PCR-amplified fragments, and a partial deletion of intron 1 in the Bm allele. The top diagram indicates ABO exons 1-7 as solid boxes. It is noteworthy that the direction of ABO is opposite of that in Figure 1. The clear box indicates the location of the erythroid cell–specific regulatory element. Positions are indicated relative to the ATG translation start site in exon 1. PCR-amplified fragments in PCR1-PCR9 are indicated by broken lines, and the primers used are represented as thick lines at both ends of individual broken lines. The relationship between the PCR amplifications and primers used is shown in Table 3. Deleted nucleotides in intron 1 of the Bm allele are indicated by a V-shaped segment. (B) A partial deletion of intron 1 was found in genomic DNA derived from a person with Bm. PCR1-PCR4 were carried out with genomic DNAs derived from persons with B and Bm using various sets of primers, followed by electrophoresis through 0.5% agarose gel and staining with ethidium bromide. Gene Ladder Wide 2 (Nippon Gene) was used as a molecular size marker. (C) Partial deletion found in genomic DNAs obtained from 10 persons with Bm or ABm. PCR4 amplifications were carried out using genomic DNAs derived from persons with A, B, O, AB, Bm, and ABm, followed by electrophoresis through 0.5% agarose gel and staining with ethidium bromide. Representative PCR products from 10 persons with Bm or ABm are displayed. The shorter PCR4 products obtained from those persons were sequenced, confirming that the deletion was located between +5137 and +10914.
To determine whether the deletion is related to Bm phenotypes, PCR4 was carried out using genomic DNAs obtained from 111 persons with Bm and ABm as well as 1010 persons with the common ABO phenotypes and other B subgroups including B3, A1B3, and Bx. Figure 6C shows representative electrophoretograms of the PCR products for A, B, O, AB, Bm, and ABm. Two products were detectable in Bm and ABm, whereas the shortened fragment was not observed in the common phenotypes. After addition of the person with Bm described in Figure 6B, the deletion was observed in 111 persons with Bm and ABm, except for 1 person with the Bm phenotype (Table 6). In sharp contrast, the deletion was never observed in persons with the common phenotypes and the above B subgroups. A Fisher exact test confirmed that the partial deletion of intron 1 was associated significantly with the Bm phenotypes (Table 6). It is plausible that deletion of the erythroid cell–specific enhancer-like element could down-regulate transcription in the Bm allele, leading to reduction of B-antigen expression in cells of erythroid lineage, but not in mucus-secreting cells.
Summary of the partial deletion of intron 1 found in persons with phenotypes Bm and ABm
| Phenotypes . | Presence of deletion* . | Absence of deletion . |
|---|---|---|
| Bm | 72† | 1 |
| ABm | 39 | 0 |
| B3 | 0 | 1‡ |
| A1B3 | 0 | 2§ |
| Bx | 0 | 2¶ |
| A | 0 | 341 |
| B | 0 | 227 |
| O | 0 | 304 |
| AB | 0 | 133 |
| Phenotypes . | Presence of deletion* . | Absence of deletion . |
|---|---|---|
| Bm | 72† | 1 |
| ABm | 39 | 0 |
| B3 | 0 | 1‡ |
| A1B3 | 0 | 2§ |
| Bx | 0 | 2¶ |
| A | 0 | 341 |
| B | 0 | 227 |
| O | 0 | 304 |
| AB | 0 | 133 |
A partial deletion of intron1 was detectable by the appearance of a 0.9-kb band in PCR4. Discrete variables were compared between 2 phenotypes, Bm including Bm and ABm, and B including B and AB, using the Fisher exact test (P < .00001).
Seventy-one people were heterozygous for the deletion; 1 person was assumed to be homozygous on the basis of PCR4 and direct sequencing of exons 6 and 7.
Nucleotide substitution of C with T at position 278 of the cDNA was demonstrated on the B allele by sequencing, based on the method reported previously.5
Nucleotide substitution of C with T at position 278 was demonstrated on the B allele in one person, while C was substituted with T at 1054 in the other.
Nucleotide substitution of C with T at position 721 was demonstrated on the B allele.
Discussion
In the present study, we identified a novel positive regulatory element in intron 1 of ABO using transient transfection experiments with luciferase reporter plasmids that were prepared on the basis of DHSs in the genomic DNA from 15 kb upstream to the end of ABO in K562 cells. The element appeared to enhance the activity of the ABO promoter in an erythroid cell–specific manner. EMSA and supershift assays showed that the element binds to the tissue-restricted transcription factor GATA-1. Mutation of the GATA motifs to abrogate binding of this factor reduced the regulatory activity of the element in K562 cells. Therefore, it seems likely that ABO expression is regulated by the element, the erythroid cell–specific regulatory activity of which is dependent on GATA-1 binding. Furthermore, we found that a partial deletion in intron 1 involving this element was associated with the Bm phenotype. Therefore, reduction of B-antigen expression on erythrocytes of persons with the Bm phenotype could be caused by deletion of the element in intron 1 of ABO. Therefore, these results suggest that this enhancer-like element is essential for ABO expression in erythroid cells.
In Bm phenotypes, B-antigen expression is reduced on RBCs, whereas a large amount of B substance is present in the saliva of secretors.1 However, Bm erythrocytes contain abundant H sites, which can be converted into B sites by in vitro treatment with α-1,3-galactosyltransferase or B-transferase derived from healthy persons with the B phenotype. B-transferase activity was detected in serum of persons with the Bm phenotype, although the activity was distinctly reduced in all cases. The Bm trait is inherited as a rare allele at the ABO locus, although a few nonhereditary cases have also been reported.35,36 Consistent with speculation by Hansen et al,10 deletion of the erythroid cell–specific regulatory element on the Bm allele was significantly associated with the Bm phenotypes, thus explaining the discrepancy of B-antigen expression between RBCs and secretions. To strengthen our argument, it will be necessary in a future study to compare the amount of the transcript between the Bm and B alleles using RNA obtained from the erythroid-lineage progenitor cells expressing a large amount of the ABO transcripts.
Specific histone modification signatures are found in distant regulatory elements, as described in the “Introduction.” Comparison of the present data with publicly available genome-wide data on histone modifications in K562 cells on ENCODE Histone Modifications by Broad Institute ChIP-seq from the UCSC genome browser (http://genome.ucsc.edu) in the March 2006 (NCBI36/hg18) assembly25,37 revealed that subregion C was correlated with H3 monomethylation at lysine residue 4 as well as H3 acetylation at lysine residue 9. This concurrence suggested the enhancer potential of the region. Moreover, sequence conservation is an effective predictor of the locations of enhancers in the genome. Recently, Chain and Net Alignments from the UCSC Genome Browser have become publicly available: for the chimpanzee, on March 2006 (CGSC 2.1/panTro2); orangutan, July 2007 (WUGSC 2.0.2/ponAbe2); Rhesus monkey, January 2006 (MGSC Merged 1.0/rheMac2); and marmoset, June 2007 (WUGSC 2.0.2/calJac1). Comparison of the genomic DNAs in the ABO gene between humans and these primate species demonstrated high conservation between the ATG translation start codon and the stop codon of the ABO gene, except for a few regions. DHS +5.8 is conserved among humans, chimpanzees, and orangutans, showing similar expression of the A and B antigens on RBCs. However, it is intriguing that the site is not conserved in Rhesus monkeys and marmosets, in which the A and B antigens are expressed only slightly on RBCs.38 Therefore, a comparative approach could indicate involvement of the site in ABO expression in erythroid lineage cells. In addition, GATA sites are conserved in the monkeys in which the A and B antigens are expressed on erythrocytes, suggesting that these sites play an important role in ABO regulation.
Hematopoiesis is controlled by numerous transcription and signal factors with tightly integrated functions.39 Foundational studies defined the involvement of members of a family of developmental regulators, including GATA transcription factors. GATA-1, GATA-2, and GATA-3 are termed the hematopoietic GATA factors based on their important activities in controlling distinct and overlapping aspects of hematopoiesis.40 GATA-1 is expressed in erythroid precursors, megakaryocytes, eosinophils, mast cells, and testis tissue. GATA-2 is expressed in hematopoietic stem cells, multipotent hematopoietic progenitors, erythroid precursors, megakaryocytes, eosinophils, mast cells, differentiated endothelial cells, and specific neurons. GATA-3 is expressed in hematopoietic stem cells, T lymphocytes, neurons, kidney, and mammary gland. After ABO antigen expression during hematopoiesis, it has been shown that A-antigen expression gradually increases during erythroid maturation in 2-phase liquid culture,41,42 and FACS has demonstrated expression of A antigens on colony cells derived from erythroid blast-forming units and CFUs.43 Therefore, it is likely that GATA-1 plays a role in the regulation of ABO expression at an earlier stage during erythropoiesis. Although the addition of anti–GATA-2 Ab did not result in any change in the DNA-protein complex (Figure 4C), the possibility that GATA-2 has the capacity to bind to subregion C could not be excluded, because the amount of GATA-2 protein was lower than that of GATA-1 protein in K562 cells (data not shown), so the DNA-GATA-2 complex might have been difficult to detect. Further investigation of GATA-2 is required, because GATA-1 and GATA-2 share many chromatin sites in erythroid precursor cells and function redundantly to promote the development of primitive erythroblasts, although they also exist at specific sites and exert distinct actions in certain contexts.44,45
Antigens of the ABO blood groups are carbohydrates, and carbohydrate chains are built up by sequential addition of various monosaccharides, each extension of the chain being catalyzed by a specific glycosyltransferase. ABH antigens are present at the terminals of repeated N-acetylglucosamine units. It has been shown that transcription factor C/EBPα is involved in regulation of the gene encoding I β-1,6-N-acetylglucosaminyltransferase (Iβ6GlcNAcT; IGnT), which biosynthesizes branched lactosaminoglycan structures.46,47 Information on the transcription factors associated with regulation of ABO or IGnT expression might shed light on how the genes encoding these glycosyltransferases associated with synthesis of the ABH antigen and its precursors are regulated through densely interconnected transcriptional circuits during hematopoiesis.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
Authorship
Contribution: R.S., T.N., and Y.K. conceived, designed, coordinated, and performed the research, analyzed the data, and wrote the manuscript; K.T. and R.K. performed the research; J.T. helped to design the experiments; H.T. and T.Y. analyzed the data; K. Ito, T.M., and A.Y. determined the ABO blood group phenotypes serologically and provided one blood sample; and K. Isa, K.O., and M.U. determined the phenotypes serologically and performed the PCR analysis.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The nucleotide sequences reported herein have been submitted to GenBank as accession number JN863720.
Correspondence: Yoshihiko Kominato, Department of Legal Medicine, Gunma University Graduate School of Medicine, Maebashi, 371-8511, Japan; e-mail: kominato@med.gunma-u.ac.jp.
References
Author notes
R.S. and T.N. contributed equally to this work.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal