Abstract
miRNA deregulation has been implicated in the pathogenesis of mantle cell lymphoma (MCL). Using a high-throughput quantitative real-time PCR platform, we performed miRNA profiling on cyclin D1–positive MCL (n = 30) and cyclin D1–negative MCL (n = 7) and compared them with small lymphocytic leukemia/lymphoma (n = 12), aggressive B-cell lymphomas (n = 138), normal B-cell subsets, and stromal cells. We identified a 19-miRNA classifier that included 6 up-regulated miRNAs and 13 down regulated miRNA that was able to distinguish MCL from other aggressive lymphomas. Some of the up-regulated miRNAs are highly expressed in naive B cells. This miRNA classifier showed consistent results in formalin-fixed paraffin-embedded tissues and was able to distinguish cyclin D1–negative MCL from other lymphomas. A 26-miRNA classifier could distinguish MCL from small lymphocytic leukemia/lymphoma, dominated by 23 up-regulated miRNAs in MCL. Unsupervised hierarchical clustering of MCL patients demonstrated a cluster characterized by high expression of miRNAs from the polycistronic miR17-92 cluster and its paralogs, miR-106a-363 and miR-106b-25, and associated with high proliferation gene signature. The other clusters showed enrichment of stroma-associated miRNAs, and also had higher expression of stroma-associated genes. Our clinical outcome analysis in the present study suggested that miRNAs can serve as prognosticators.
Introduction
Mantle cell lymphoma (MCL) constitutes approximately 6% of all nonHodgkin lymphomas and occurs predominantly in men of advanced age.1,2 Several histologic variants of MCL, including the classic, small-cell, blastoid, and pleomorphic variants, have been reported1 and have various proliferation rates and genetic profiles.3,4 The putative cell-of-origin is considered to be a naive B cell in the mantle zones or primary follicles. However, 20%-30% of patients show mutated immunoglobulin variable-region heavy chain (IGVH) genes.2 The immunophenotype is characterized by expression of CD5 and the B cell–associated antigens CD20, CD22, CD79, and CD5, with strong expression of IgM and IgD, but by the lack of CD23, CD10, and BCL6.1,2 Historically, the majority of MCL patients exhibit an aggressive clinical course, but survival has improved with current management to a reported median survival time of 5-7 years.5 Recent studies have identified an indolent subtype of MCL that is associated with even longer survival times.6,7 The neoplastic cells in these patients exhibit hypermutated IGVH genes, a noncomplex karyotype, and lack SOX11 expression.
The genetic hallmark of MCL is the t (11;14)(q13;q32) mutation, resulting in the overexpression of cyclin D1. Nonetheless, small subsets of patients (< 5%) lack this genetic aberration but exhibit an almost indistinguishable gene-expression profile (GEP) and genomic profile compared with cyclin D1–positive patients.8,9 Several recurrent genetic abnormalities have been reported in MCL, including frequent losses of 9p21.3, 11q22-q23, and 22q11.22, and gains of 10p11.23 and 13q31.3.3,4,9 Specific mutations and deletions in p16 (CDKN2A), ATM, CHEK2, and TP53 have also been noted frequently in MCL.2 Partial uniparental disomy has also been reported in the regions that are frequently targeted by chromosomal deletions.10
Abnormal miRNA expression has been implicated in the pathogenesis of lymphoma, including the recurrent 13q31.3 gain9 harboring MIHG1, which encodes the miR17-92 cluster composed of 6 polycistronic miRNAs (miR-17, miR-18a, miR-19a, miR19b-1, miR-20a and miR-92a). Alteration in miRNA expression has been explored in B-cell lymphomas including MCL.11-13 In the present study, we performed a large-scale global analysis on multiple types of B-cell lymphoma to compare with MCL using an miRNA profiling platform based on high-throughput Taqman quantitative real-time RT-PCR. The study was aimed at identifying diagnostic and prognostic signatures in MCL, including cyclin D1–positive and cyclin D1-negative patients. The quantitative RT-PCR assay permits highly accurate quantitation of individual miRNAs over a wide dynamic range and distinguishes between closely related miRNA family members. We also explored the applicability of this platform to both cryopreserved and formalin-fixed paraffin-embedded (FFPE) tissues. We also compared the miRNA profiles with the corresponding GEP data to investigate the molecular mechanisms or pathways associated with deregulated miRNA expression.
Methods
Patient samples, cell lines, and normal primary cells
Frozen tumor specimens and fresh tonsils from routine tonsillectomy were obtained from patients under a protocol approved by the institutional review board of the University of Nebraska Medical Center (Omaha, NE). Tumor biopsies taken from a series of cyclin D1–positive mantle cell lymphoma (MCL) patients (n = 30) and small lymphocytic lymphoma/chronic lymphocytic leukemia (SLL/CLL) patients (n = 12) were studied for miRNA and gene-expression profiling (GEP). We compared these miRNA/GEP results with a series of diffuse large B-cell lymphoma (DLBCL) and Burkitt lymphoma (BL) patients (n = 138). We compared miRNA profiles obtained from cryopreserved tissues with corresponding FFPE tissues in 8 (of 30) MCL samples and 35 (of 138) DLBCL/BL samples. The other FFPE samples included cyclin D1–negative MCL (n = 7) and GEP for 6 patients have been reported previously.8,14 A panel of expert hematopathologists reviewed and confirmed the diagnosis of patients using the World Health Organization classification.1 The experimental details about cell lines and primary B cells are included in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
The detailed protocol on RNA isolation from fresh frozen and FFPE tissues for miRNA and/or GEP, miRNA profiling and GEP data analysis, immunologic and FISH analysis, and survival outcome analysis are available in supplemental Methods.
Results
Patient characteristics
The clinical characteristics of the MCL and SLL patients are summarized in Table 1. The median age of the MCL patients (n = 30) was 63 years (range, 37-88) at the time of diagnosis with a high ratio of male to female patients (5:1). These MCL patients exhibited an aggressive clinical course with a median overall survival (OS) of 2.98 years (supplemental Figure 1). These patients were also profiled for GEP and were classified as MCL with > 90% confidence. Most of the patients were CD5+ and/or CD43+ and expressed cyclin D1 or showed cyclin D1 translocation by FISH (supplemental Table 1A).
Characteristics of MCL (cyclin D1–positive) and SLL patients included in the study*
| Clinical feature . | MCL n = 30 . | SLL n = 12 . |
|---|---|---|
| Median age, y (range) | 63 (37-88) | 59 (40-90) |
| Sex, n (%) | ||
| Female | 5 (17) | 3 (27) |
| Male | 25 (83) | 8 (63) |
| Performance score, n (%) | ||
| < 70 | 3 (10) | 0 (0) |
| > 70 | 27 (90) | 11 (100) |
| Stage | ||
| I/II | 2 (6) | 2 (18) |
| III/IV | 28 (94) | 9 (82) |
| Serum lactate dehydrogenase, n (%) | ||
| Normal | 21 (70) | 9 (82) |
| Elevated | 9 (30) | 2 (11) |
| No. of extranodal sites, n (%) | ||
| < 2 | 26 (87) | 10 (91) |
| > 2 | 4 (13) | 1 (9) |
| Median survival, y | ||
| OS | 3.0 | |
| EFS | 1.5 | 6.2 |
| Clinical feature . | MCL n = 30 . | SLL n = 12 . |
|---|---|---|
| Median age, y (range) | 63 (37-88) | 59 (40-90) |
| Sex, n (%) | ||
| Female | 5 (17) | 3 (27) |
| Male | 25 (83) | 8 (63) |
| Performance score, n (%) | ||
| < 70 | 3 (10) | 0 (0) |
| > 70 | 27 (90) | 11 (100) |
| Stage | ||
| I/II | 2 (6) | 2 (18) |
| III/IV | 28 (94) | 9 (82) |
| Serum lactate dehydrogenase, n (%) | ||
| Normal | 21 (70) | 9 (82) |
| Elevated | 9 (30) | 2 (11) |
| No. of extranodal sites, n (%) | ||
| < 2 | 26 (87) | 10 (91) |
| > 2 | 4 (13) | 1 (9) |
| Median survival, y | ||
| OS | 3.0 | |
| EFS | 1.5 | 6.2 |
EFS indicates event-free survival.
The characteristics of 6 (of 7) cyclin D1–negative MCL patients have been described previously8 ; the seventh case, without GEP, showed MCL morphology and SOX11 expression consistent with other t(11;14)–negative MCL patients. One of the 12 SLL patients lacked complete clinical data.
Of the other MCL patients (n = 7) who were negative for t(11;14) and cyclin D1 expression, the GEP of 6 has been reported previously.8 The seventh patient, without GEP, showed MCL morphology and SOX11 expression consistent with other t(11;14)–negative MCL patients. Similar to cyclin D1–positive MCL patients, the median age at the time of diagnosis was 60 years (range, 51-65) with male predominance (5 of 7 patients), and also showed a similar immunophenotype, with expression of B-cell markers and CD5. The expression of SOX11 (7 of 7), cyclinD2 (3 of 5), or D3 (2 of 5) was noted in the cyclin D1–negative patients (supplemental Table 1B).
The median age of the SLL/CLL patients was 59 years (range, 40-90 years) at the time of diagnosis, with a ratio of male to female patients of 2:1. These patients had the characteristic morphology and immunophenotype, including a lack of cyclin D1 expression. The majority (70%) of SLL/CLL patients had not received any chemotherapy and the median follow-up time from diagnosis was 6.2 years.
Molecular classifier for MCL based on miRNA profile
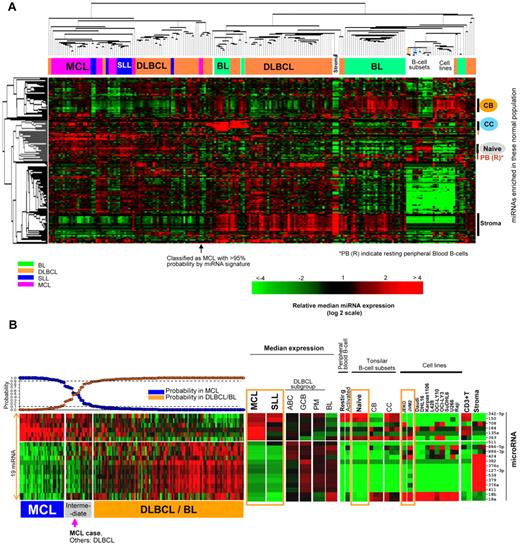
Unsupervised hierarchical clustering (HC) analysis revealed that the MCL and SLL patients formed a distinct cluster compared with other lymphoma entities (Figure 1A). Of the 30 MCL and 12 SLL/CLL patients, only one each clustered separately in the DLBCL cluster. However, both patients were classified molecularly as MCL or SLL with the miRNA classifiers after further analysis (see paragraph below), indicating that the tumors still maintained a substantial differentiation-associated miRNA profile.
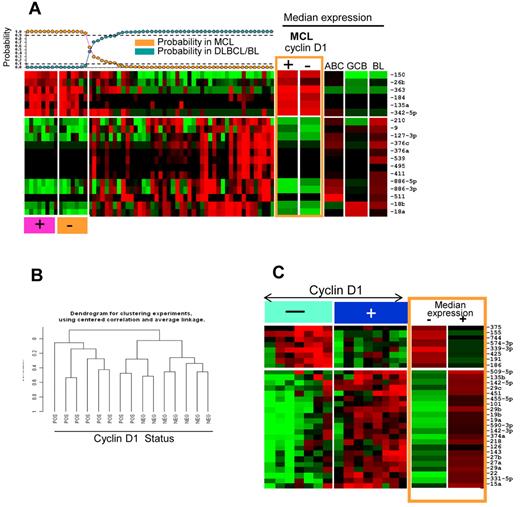
MicroRNA expression profile and classifier for MCL. (A) Unsupervised hierarchical clustering of lymphoma samples, normal cells, and cell lines. MCL, SLL, DLBCL, and BL formed largely distinct clusters. A stromal cell–associated miRNA signature was more highly expressed in DLBCL and BL patients. There were also significant differences in the expression of miRNA associated with naive B cells, resting B cells, CB cells, and CC cells, with miRNA associated with naive and resting B cells being more highly represented in MCL and SLL patients. (B) An miRNA classifier derived using a Bayesian algorithm resulted in a 19-miRNA classifier (6 up-regulated and 13 down-regulated miRNAs) was able to separate most MCL patients from DLBCL and BL patients. The expression of this miRNA classifier is illustrated in SLL, normal cells, and cell lines. Orange boxes highlight naive B-cell and MCL cell lines.
MicroRNA expression profile and classifier for MCL. (A) Unsupervised hierarchical clustering of lymphoma samples, normal cells, and cell lines. MCL, SLL, DLBCL, and BL formed largely distinct clusters. A stromal cell–associated miRNA signature was more highly expressed in DLBCL and BL patients. There were also significant differences in the expression of miRNA associated with naive B cells, resting B cells, CB cells, and CC cells, with miRNA associated with naive and resting B cells being more highly represented in MCL and SLL patients. (B) An miRNA classifier derived using a Bayesian algorithm resulted in a 19-miRNA classifier (6 up-regulated and 13 down-regulated miRNAs) was able to separate most MCL patients from DLBCL and BL patients. The expression of this miRNA classifier is illustrated in SLL, normal cells, and cell lines. Orange boxes highlight naive B-cell and MCL cell lines.
Further examination of the miRNA profile showed that 2 prominent miRNA signatures were differentially expressed among the major lymphoma entities: one signature reflected miRNAs highly expressed in stromal cells that were more highly expressed in DLBCL and BL. The other signature was associated with nondividing, quiescent cells (naive, resting peripheral blood cells) and was highly represented in MCL or SLL/CLL patients. These observations are consistent with the morphologic findings that MCL lacks a major stromal component compared with DLBCL and that most MCL cases are not as proliferative as other aggressive lymphomas.
We used a Bayesian algorithm to derive a miRNA classifier that differentiates MCL from DLBCL/BL and leave-one-out cross-validation for classification precision.15 This algorithm resulted in a 19-miRNA classifier, which included 6 up-regulated miRNAs (miR-135a, miR-708, miR-150, miR-363, miR-184, and miR-342-5p) and 13 down-regulated miRNAs (Figure 1B). When this signature was evaluated in normal B-cell subsets, the majority of the up-regulated miRNAs were also highly expressed in naive B cells, resting B cells, or CC cells, with the exception of miR-135a and miR-708, which were normally more highly expressed by CB/CC and stromal cells, respectively. However, they were also expressed by 2 MCL cell lines, JEKO and JVM2. The majority (8 of 13) of down-regulated miRNAs (miR-424, miR-382, miR-376c, miR-127-3p, miR-539, miR-379, miR-376a, and miR-411) were expressed by stromal cells at a high level, which is consistent with a low stromal content in MCL. The expression of the other down-regulated miRNAs was also low in naive and resting B cells.
Morphologic evaluation of the discrepant patients identified by miRNA classifier
We evaluated the precision of the miRNA classifier by leave-one-out cross-validation and observed that 2 patients defined by GEP as DLBCL (n = 89) were misclassified as MCL with a > 90% probability. These 2 patients were classified as ABC-DLBCL (n = 1) and unclassifiable-DLBCL (n = 1) by GEP analysis. However, after further review, the morphology of these patients was compatible with the paraimmunoblastic variant of SLL. In general, SLL/CLL patients had a high association (80%-90% probability) with the MCL classifier and therefore required a separate analysis (see “Comparison of miRNA expression profile between MCL and SLL”).
Only one MCL patient showed probability < 90% by the miRNA classifier (Figure 1B). This patient was different from the other one that clustered within DLBCL patients in unsupervised HC (Figure 1A), but by miRNA classifier showed > 95% probability as MCL. After review, this patient was shown to have blastoid-variant morphology.
Comparison of miRNA expression profile between MCL and SLL
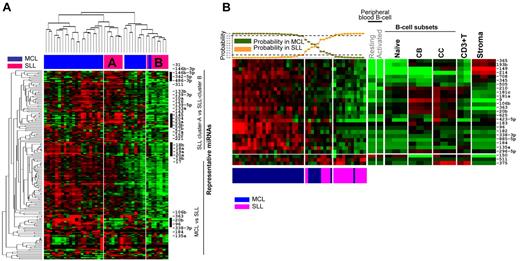
As expected from the close clustering of MCL and SLL patients (Figure 1A), the miRNAs in the MCL classifier (derived from comparison with DLBCL/BL) showed significant overlap with the SLL patients, with the exception of 4 miRNAs (miR-363, miR-184, miR-708, and miR-135a) that were highly expressed in MCL (Figure 1B). When only the MCL and SLL patients were analyzed by unsupervised HC, 2 separate clusters (A and B) of SLL (Figure 2A) were observed. The SLL cluster A showed mainly up-regulated miRNAs compared with patients in cluster B and was characterized by high expression of miRNAs shown to have a tumor-suppressive function (miR-1, miR-133a,16 miR-133b,16 miR-139-5p, miR-139-3p, miR-143,17 miR-10b,18 miR-145,16 and miR-23b) and stroma-related (miR-23a, miR-27a, miR-27b, miR-152, and miR-221). One each of 2 paraimmunoblastic SLL patients (identified above), clustered with SLL cluster A and cluster B, and showed no association with MCL clusters.
Differences in miRNA expression between MCL and SLL. (A) Unsupervised hierarchical clustering of MCL and SLL samples showed 2 separate clusters of SLL patients with each having unique miRNA profiles. (B) A 26-miRNA signature differentiates MCL from SLL, with 23 miRNAs including 2 polycistronic miRNA clusters, miR106b-25 (miR-106b and miR-25) and miR106a-363 (miR-363 and miR-20b) being significantly up-regulated in MCL compared with SLL.
Differences in miRNA expression between MCL and SLL. (A) Unsupervised hierarchical clustering of MCL and SLL samples showed 2 separate clusters of SLL patients with each having unique miRNA profiles. (B) A 26-miRNA signature differentiates MCL from SLL, with 23 miRNAs including 2 polycistronic miRNA clusters, miR106b-25 (miR-106b and miR-25) and miR106a-363 (miR-363 and miR-20b) being significantly up-regulated in MCL compared with SLL.
To separate MCL and SLL more definitively, we constructed a classifier consisting of 26 miRNAs that was dominated by 23 miRNAs up-regulated in MCL, including miR-184 and members of 2 polycistronic miRNA clusters, miR106b-25 (miR-106b and miR-25) and miR106a-363 (miR-363 and miR-20b), which are paralogs of the miR17-92 cluster. In the classifier, only miR-150, miR-511, and miR-375 were up-regulated significantly in SLL patients (Figure 2B). Five MCL patients showed a probability of < 90% (40%-70% for 4 patients and 20% for 1 patients) and 1 patient was classified as SLL, suggesting that a small subset of MCL patients may have a miRNA profile very similar to SLL patients. The latter 2 patients showed low expression of the proliferation gene signature (PS; log2 signal intensity 7.3, 7.6; range in 30 patients of PS is 6.9-9.1), and were part of the indolent MCL patient cluster (see “Identification of MCL subsets by miRNA profiling and correlation with GEP signatures”). Interestingly the 2 SLL paraimmunoblastic variants, despite their much higher proliferation, showed significant similarity with the SLL group (80% and 90% probability, respectively) in their miRNA expression profile (Figure 2B)
Identification of MCL subsets by miRNA profiling and correlation with GEP signatures
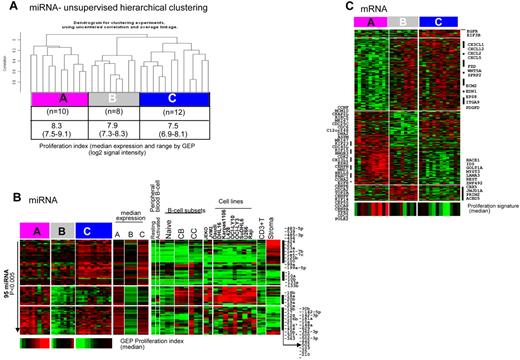
When MCL patients alone were analyzed by unsupervised HC, 3 distinct clusters were observed (Figure 3A) with a significant difference in expression of 95 miRNAs (P < .005) among these clusters. We applied previously defined MCL PS19 and observed a significant difference (P = .01 by Kruskal-Wallis Test) in the median expression of PS among the 3 miRNA-defined clusters, designated as cluster A (high PS), cluster B (medium PS), and cluster C (low PS; Figure 3A). The difference was more prominent between clusters A and C. The patients in cluster A showed up-regulation of miR17-92 cluster members and its paralogs miR-106a-363 and miR-106b-25, which is indicative of a proliferative miRNA profile and a subgroup of miRNAs closely associated with the CB, CC, and cell lines, but not with naive B cells (Figure 3B). The patients in cluster C showed up-regulation of miRNA with growth-inhibitory functions including miR-1, miR-133b,16 miR-10b,18 and stroma-associated markers (miR-23a, miR-23b, let-7c, let-7-b, and miR-125b). Of the differentially expressed miRNAs, those associated with stroma were significantly enriched in cluster C, illustrating the contribution of stroma in this subset of patients. The cluster-B patients showed low expression of majority of the miRNAs compared with clusters A and C, but high expression of miRNAs associated with stroma (miR-636, miR-539, and miR-485-3p) was noted specifically in this subset.
Unsupervised hierarchical clustering of MCL patients showed 3 distinct MCL clusters. (A) Significant differences in the PS among the 3 groups (P = .01 by Kruskal-Wallis test). PS included the same gene set identified in Rosenwald et al.19 (B) Differential miRNA expression (95 miRNAs, P < .005) among these clusters designated as cluster A (associated with high PS), cluster B (medium PS), and cluster C (low PS). (C) Differential gene expression (649 transcripts, P < .005) among the 3 clusters, with cluster A showing high expression of proliferation-associated genes and the other clusters showing higher expression of genes associated with stromal components.
Unsupervised hierarchical clustering of MCL patients showed 3 distinct MCL clusters. (A) Significant differences in the PS among the 3 groups (P = .01 by Kruskal-Wallis test). PS included the same gene set identified in Rosenwald et al.19 (B) Differential miRNA expression (95 miRNAs, P < .005) among these clusters designated as cluster A (associated with high PS), cluster B (medium PS), and cluster C (low PS). (C) Differential gene expression (649 transcripts, P < .005) among the 3 clusters, with cluster A showing high expression of proliferation-associated genes and the other clusters showing higher expression of genes associated with stromal components.
To further understand the biologic significance of these clusters, we also examined the GEP data. Interestingly, GEP-based unsupervised HC showed that patients placed in cluster A as defined by miRNA profile were almost identical to 1 of the 3 subclusters defined by GEP (all 8 patients in cluster A), supporting that patients in cluster A had distinctive molecular characteristics. When GEP data from the 3 miRNA-defined clusters were analyzed, 649 transcripts showed significant differential expression (P < .005). Functional analysis showed that transcripts encoding proteins with roles in cell-cycle progression and proliferation or in inhibition of apoptosis were highly up-regulated in cluster A, which is consistent with the association with a high PS (Figure 3C). The transcript level of the proliferation marker Ki67 was associated significantly with this group (P < .0001). In addition, we observed high expression of genes encoding proteins secreted by macrophages (eg, CHI3L1), including CD163 expressed in M2 macrophages, in this group. The patients in cluster C (the low-PS group) showed relatively high expression of genes encoding cytokines mainly associated with T cells (CX3CL1, CXCL12, CXCL2, and CXCL5) and genes associated with WNT signaling (FZD, WNT5A, and SFRP2) compared with cluster A (the high-PS group). The patients in this group also expressed transcripts encoding extracellular matrix–related proteins (ECM2, EDN1, EGFR, EPS8, ITGA9, and PDGFD). Interestingly, many up-regulated and down-regulated genes in cluster C showed similar expression pattern in cluster B, however, a unique gene signature was also noted in these groups. Information on the functional characteristics of these genes is limited in the literature (Figure 3C). Gene-set enrichment analysis complemented these results, with significant enrichment of the proliferation-related gene signatures in cluster A, whereas both of the other clusters showed enrichment of IL-6, TGF-β, hypoxia, VEFG, Hox10-induced, and quiescent/stem cell–like gene signatures. Compared with cluster C, cluster B had a higher TGF-β signature and showed more genotoxic stress with higher p21 and ATM signatures, whereas cluster C showed higher expression of WNT and IL-4 signaling pathway genes (supplemental Table 2). We performed immunohistochemistry with β-catenin on 3 representative patients from cluster C and 2 patients from cluster A. We observed strongly positive expression of β-catenin in stromal/endothelial cells in all 3 patients from cluster C. The majority of tumor cells were negative, whereas patients with a high proliferation signature (cluster A) showed only the occasional cell weakly positive for β-catenin (supplemental Figure 3), indicating that WNT activation may be attributed mostly to stromal components in cluster C patients.
Validation of GEP signatures.
To further validate the GEP findings of these 30 MCL patients, we performed validation of the gene signatures in another cohort of MCL patients (n = 82). In our initial analysis, we derived a specific gene-expression signature from the GEP data of the 30 MCL patients in this study (training data) that could distinguish cluster A and cluster C patients using Bayesian algorithm. This resulted in a 71-probe set with 2 gene signatures, one enriched in proliferation-related genes (signature 1) and the other enriched in stroma-related genes including WNT pathway genes (signature 2), as shown in supplemental Figure 4A. The mean expression levels of these 2 gene signatures were inversely correlated and the ratio of the mean expression of the 2 signatures was associated significantly with event-free survival (P < .01). We then analyzed the gene signatures in the independent MCL series (n = 82) using hierarchical clustering, and obtained 4 subsets of MCL patients, with the majority showing an inverse correlation between signature 1 and 2 expression. However, a small subset of patients expressed both signatures at similar levels (both high and both low). There was a significant association of OS with signature 1 that predicted a worse prognosis. When the ratio of expression of the 2 signatures was correlated with OS, higher ratios (signature 1 vs 2) were associated with poorer prognosis (supplemental Figure 4B).
Comparison of naive B cells and MCL miRNA profile and functional implications
We identified a miRNA signature associated significantly with naive B cells by comparison with other B-cell subsets, including CCs and CBs (supplemental Figure 2). The miRNAs associated significantly with naive B-cell subsets were miR-150, miR-223, miR-342-3p, miR-146-5p, miR-95, miR-342-5p, and miR-146b-3p, with at least 4 of the 7 miRNAs described in previous studies.20-22 These up-regulated miRNAs were also observed in resting B and T cells and showed marginal enrichment in MCL patients compared with those with other lymphoid entities (data not shown), but were largely absent from the lymphoid cell lines.
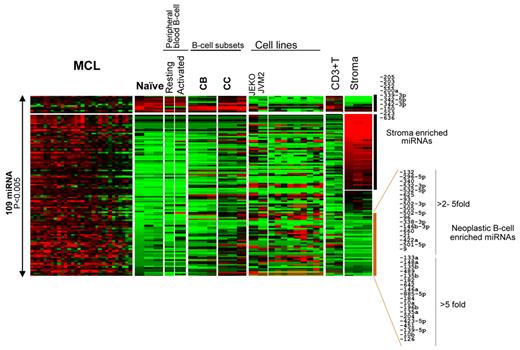
To identify miRNAs that may have pathogenetic significance, we compared the miRNA profile of normal naive B cells with that of MCL. Excluding differences that may be attributable to the stromal elements, we observed that the majority (> 80%) of differentially expressed miRNAs were up-regulated in MCL compared with naive B cells (Figure 4). The miRNAs that were up-regulated in MCL but not in naive B cells or stromal elements included miR-184, miR-21, miR-10b, and miR-135a, the oncogenic roles of which have been demonstrated in several malignancies,23-26 but the functional characteristics of many other up-regulated miRNAs is not known. Only a few miRNAs were down-regulated in MCL cells compared with normal naive B cells; however, these included some of the most abundant miRNAs (miR-150, miR-223, miR-222, and miR342-5p/3p) in naive B cells, suggesting that low expression of these miRNAs may be important for the pathogenesis of MCL (Figure 4).
Comparison of the MCL miRNA profile with other B-cell subsets, T-cells, and stromal elements. More than 80% of differentially expressed (P < .005) miRNAs were up-regulated in MCL cells compared with naive B cells.
Comparison of the MCL miRNA profile with other B-cell subsets, T-cells, and stromal elements. More than 80% of differentially expressed (P < .005) miRNAs were up-regulated in MCL cells compared with naive B cells.
Evaluation of cryopreserved and FFPE miRNA profile
The miRNA classifier obtained from cryopreserved tissues was evaluated in corresponding FFPE MCL (n = 8) and DLBCL/BL (n = 35) patients. Of the 19 miRNAs, 2 showed inconsistent expression in the FFPE samples, but the remaining 17 miRNAs were able to distinguish MCLs from DLBCLs and BLs with similar sensitivity and specificity. The expression pattern of this 17-miRNA signature was similar between cyclin D1–positive and cyclin D1–negative MCL patients (n = 7) and they readily distinguished from DLBCL/BL patients (Figure 5A).
miRNA expression in cyclin D1–positive and –negative MCL. (A) MCL classifier obtained from cryopreserved tissues showed similar predictive power in FFPE tissues and cyclin D1–negative MCL classified as MCL. (B) Unsupervised clustering based on miRNA profiles showed distinct clusters of cyclinD1-negative and cyclin D1–positive MCL patients. (C) Differential expression of miRNA between cyclin D1–negative and cyclin D–positive MCL patients.
miRNA expression in cyclin D1–positive and –negative MCL. (A) MCL classifier obtained from cryopreserved tissues showed similar predictive power in FFPE tissues and cyclin D1–negative MCL classified as MCL. (B) Unsupervised clustering based on miRNA profiles showed distinct clusters of cyclinD1-negative and cyclin D1–positive MCL patients. (C) Differential expression of miRNA between cyclin D1–negative and cyclin D–positive MCL patients.
Cyclin D1–negative versus cyclin D1–positive MCL.
Despite the substantial similarity in miRNA profiles, cyclin D1–positive and cyclin D1–negative MCL patients clustered separately in unsupervised HC, and showed 30 differentially expressed miRNAs (P < .05 and 4-fold differences; Figure 5B-C). The differences included down-regulation of miRNAs negatively regulated by MYC (miR-15a, miR-22, miR-29a, miR-29b, miR-29c, and miR-142-3p)27 and up-regulation of the oncomiR miR-15528 in cyclin D1–negative MCL patients. In contrast, cyclin D1–positive patients showed significant up-regulation of miR-27 and miR-19a, suggesting some distinct pathogenetic features in these 2 subgroups of MCL. In addition, we did not observe significant expression changes of miRNAs located on 11q13 (miR-1237, miR-192, miR-194-2, miR-612, miR-548, miR-139, and miR-326) encompassing the CCND1 locus between cyclin D1–positive and cyclin D1–negative MCL patients.
Association of miRNA profile with clinical outcome
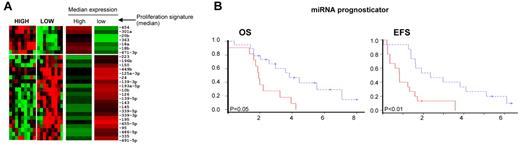
The correlation of miRNA expression with clinical outcome was evaluated by correlation with the mRNA-based proliferation signature and by the Bair et al29 unsupervised principle to identify miRNAs that predict OS or event-free survival with PS as a covariate in the analysis. When miRNAs were analyzed with respect to the mRNA-based PS, 28 miRNAs were significantly (P < .05) differentially expressed between the highest-tertile (n = 10) and lowest-tertile (n = 10) proliferative subgroups (Figure 6A). The high-proliferative group is characterized by high expression of miR-18a of the miR17-92 cluster and miR-18b, miR-20b, and miR-363 of the miR-106a-363 cluster, indicating a proliferative miRNA signature.30,31 The low-proliferative group included higher expression of miR-125-3p, miR-126, miR-10b, miR-143, and miR-145, many of which are highly expressed in stromal cells, suggesting that the low-proliferative group is associated with a higher microenvironment signature.
Correlation of miRNA expression with clinical outcome. (A) Differential miRNA expression between patients in the highest tertile (n = 10) and lowest tertile (n = 10) of the proliferation signature (P < .05). (B) Kaplan-Meier curves of miR-636 and miR-424 showing significant (P < .05) association with OS on univariate analysis. (C) Kaplan-Meier curves for risk groups using the 6-miRNA signature obtained for survival risk prediction using the method of Bair et al.29
Correlation of miRNA expression with clinical outcome. (A) Differential miRNA expression between patients in the highest tertile (n = 10) and lowest tertile (n = 10) of the proliferation signature (P < .05). (B) Kaplan-Meier curves of miR-636 and miR-424 showing significant (P < .05) association with OS on univariate analysis. (C) Kaplan-Meier curves for risk groups using the 6-miRNA signature obtained for survival risk prediction using the method of Bair et al.29
We also performed survival risk prediction and a multivariate proportional hazards model was developed using PS as one of the covariates. We identified a 6-miRNA signature (high expression of miR129-3p, miR-135a, miR-146a, miR-424, and miR-450-5p and low expression of miR-222), separating MCL into good (median OS, approximately 4 years) and poor (median OS, approximately 2 years) prognostic groups independently of miRNAs associated with PS (Figure 6C and supplemental Table 3B).
Discussion
In the present study, we profiled 187 patients with B-cell lymphoma and subsets of normal B cells with the goals of constructing a reliable miRNA classifier for MCL, identifying miRNA-based predictors of outcome, and determining possible roles of miRNA in the pathogenesis of MCL. Because the putative cell of origin for MCL is the naive, pre–germinal-center (pre-GC) B cell, we determined which miRNAs were associated with naive B cells and investigated their expression pattern in other quiescent B cells. These miRNAs included miR-150, miR-223, miR-29a, miR-29c, miR-101, miR-320, miR-331, let-7b, miR-26a, and miR-342. Some of these (miR-150, miR-29c, miR-101, miR-223, and miR-320) have been described previously as being enriched in naive B cells, using multiple platforms including deep sequencing.21,22 Some of these miRNAs may have a role in maintaining the quiescent state or uncommitted status of B cells in peripheral lymphoid organs. For example, the expression of miR-223 in naive B cells has been shown to block the differentiation of naive B cells into GC B cells by repressing LMO2 and MYBL1.21,22 Similarly, miR-150 controls B1-cell expansion and the humoral immune response in mice by targeting the key transcription factor MYB.32 However, the miRNA profile of MCL cells showed substantial differences from that of naive B cells. Because MCL cells contain stromal elements, we included the miRNA profiles of stromal cells and T cells isolated from the tonsils to facilitate interpretation of the data. Although some changes in miRNA profiles may occur after culturing the stromal cells, many miRNAs are associated with tissue of origin so we expected that this set of miRNAs would be maintained. Although the stromal cells we generated represent only a portion of the tumor microenvironment, the expression of this stromal miRNA signature is clearly correlated with the abundance of stroma in a tumor (Figure 1). After excluding stromal miRNAs, there were still many overexpressed miRNAs compared with naive B cells, suggesting that they may play a role in MCL. Of these, miR-135a has been demonstrated to have an oncogenic role by down-regulating adenomatous polyposis coli and activating the WNT pathway in colorectal cancer,25 but in classic Hodgkin lymphoma, it is associated with better prognosis and targets JAK2, resulting in down-regulation of Bcl-xl.33 miR-21 has been shown to be an oncomiR in a pre-B cell lymphoma mouse model,24 and its direct targets include several tumor-suppressor genes (PTEN, PDCD4, and ANP32A).34 Similarly, the other up-regulated miRNAs (miR-10a and miR-10b) are oncomiRs that induce cell motility and invasiveness by suppressing HOXD10.23 Interestingly, in the present study, miR-10a was down-regulated in cluster C (miRNA clustering Figure 3A-B) of MCL, and the HOX10-induced genes were correspondingly enriched in the same group when assessed by GEP. Consequently, low expression of miR-10a was associated significantly with better survival in MCL by univariate analysis (P = .03). Conversely, a few highly up-regulated miRNAs in naive B cells, such as miR-150, miR-223, and miR-342-5p, were also down-regulated in activated peripheral blood B cells as in MCL cells, suggesting that they regulate B-cell activation and that their down-regulation may be important in terminating quiescence.
Our analysis identified a robust MCL miRNA classifier, which included 6 up-regulated and 13 down-regulated miRNAs. The classifier was able to separate MCL from other aggressive lymphomas accurately in 29 of 30 patients with a > 90% probability. Interestingly, 2 DLBCL patients were misclassified, but after further morphologic examination, these 2 patients were diagnosed as having the paraimmunoblastic variant of CLL and showed similarity to MCL and SLL in their miRNA expression profiles. It is interesting that these 2 highly proliferative patients still retained sufficient similarity to MCL and SLL patients to be included in this category rather than in the DLBCL/BL category. The analysis of miRNA profiling performed on the corresponding FFPE tissues showed similar results, demonstrating that the classifier performed with similar accuracy in FFPE tissues.
Of the miRNAs in the classifier, we observed a 45-fold down-regulation of miR-150 after differentiation of naive B cells to CB cells. This observation suggests that miR-150 is a stage-specific marker of naive B cells and may block the transition of naive B cells to CB cells by down-regulating MYB.32 The association of miR-184 with MCL may be attributed in part to its presence in 15q25.1, which is frequently gained or amplified (> 20%) in MCL9 and has been associated functionally with cell proliferation and tumorigenesis.26 The other up-regulated miRNAs in the classifier with known roles include miR-135a33 and miR-363,31 which have been implicated in oncogenesis. However, the classifier contains highly selected miRNAs, and the study of miRNAs in tumor biology is more appropriately performed in the context of differentially expressed miRNAs compared with the normal counterpart and other B-cell lymphomas. Because the function of miRNAs may be context dependent, their reported functions may need to be validated in the cell type of interest.
In the present study, we have demonstrated that the miRNA classifier for cyclin D1–positive MCL was very similarly expressed in cyclin D1–negative patients in FFPE tissues, which were all classified as MCL. There were, however, sufficient differences that these 2 groups of patients tended to form their own clusters when analyzed as a group. Of the differentially expressed miRNAs, miR-155 was up-regulated whereas 6 miRNAs down-regulated by MYC, including tumor suppressor miR-15a, were down-regulated in cyclin D1–negative MCL patients. Cyclin D1–positive patients showed significant up-regulation of miR-27, miR-101, miR-142-5p, miR-19a, miR-19b, and 2 stroma-associated miRNAs, miR-126 and miR-143, compared with their cyclin D1–negative counterparts, suggesting a subtle distinction in their pathogenesis. However, different miRNAs may affect the same oncogenic pathways through different mechanisms, as illustrated by miR-155 (up-regulated in cyclin D1–negative patients) and miR-19 (up-regulated in cyclin D1–positive patients) that may both activate the PI3K pathway by suppressing SHIP1 and PTEN, respectively.
The SLL patients showed an miRNA expression pattern similar to that of MCL patients, probably due in part to their low proliferation, the non-GC B-cell origin of the neoplastic cells, and their low stromal content, although the majority of patients do form a separate cluster (Figure 1A). Most SLL patients can also be differentiated from MCL patients with an miRNA classifier, with miR-150 showing marked up-regulation in SLL, whereas 23 of the 26 other miRNAs showed higher expression in MCL, including miRNAs associated with the proliferation signature (miR-106b-25 and miR-20b), prosurvival signal (miR-181a) via Bim down-regulation,35 and 15q amplification (miR-184).9 Only one patient of each was misclassified into the other category; the misclassified SLL patient was an aggressive paraimmunoblastic variant, and the misclassified MCL patient belongs to cluster C, which included more indolent patients.
By unsupervised HC, the miRNA expression profile segregated the MCL patients into 3 clusters that appeared to have biologic and clinical differences. Cluster A showed high expression of proliferation-related miRNAs, including the miR17-92 cluster30 and its paralogs miR-106a-363 and miR-106b-25, which is also consistent with higher expression of proliferation-related genes. Contrary to cluster A, cluster C showed higher expression of stroma-associated miRNAs, including miR-23a, miR-23b, let-7c, let-7-b, and miR-125b and miRNAs with growth-inhibitory functions, including miR-1, miR-133b,16 and miR-10b.18 GEP analysis also showed low expression of proliferation-related genes and, interestingly, showed high expression of transcripts encoding extracellular matrix-related proteins (ECM2, EDN1, EGFR, EPS8, ITGA9, and PDGFD). Further examination of these gene signatures in another MCL cohort (n = 82) generally validated these findings. Most of the patients showed an inverse correlation between the signature enriched in proliferation-associated genes versus stromal-associated genes, and a high ratio of these 2 signatures was associated with poorer survival. This observation suggests that the stroma may have influence on tumor cell proliferation. The influence of the tumor microenvironment on the outcome of patients with follicular lymphoma has been demonstrated previously.36 Similarly, in DLBCL, the “stromal-1 signature,” which is related to extracellular-matrix deposition and mesenchymal and histiocytic cells, is associated with favorable outcome.37 We have also reported recently the contribution of the stromal signatures in the prognosis of patients with angioimmunoblastic T-cell lymphoma.38 The results of the present study suggest that a group of stroma-associated miRNAs may define a more indolent group of patients and warrant further investigation.
These MCL subgroups also showed differences in the expression of distinct signaling pathways when the associated GEP data were examined. Cluster C showed enrichment of pathways associated with “stemness/quiescence,” such as WNT and TGF-β signaling, whereas cluster B appeared to be associated with higher genotoxic stress (enriched ATM and p21 pathway genes). Because of the reported role of WNT signaling in CLL39 and of the nuclear staining of β-catenin in neoplastic cells in 52% of MCL patients,40 we further investigated the expression of β-catenin by immunohistochemistry. In our patients, β-catenin seemed to be mostly localized in stromal elements. Neither we nor our collaborators (E.C., personal communication) were able to demonstrate nuclear expression of β-catenin in neoplastic MCL cells, suggesting that WNT activation may be attributed mostly to stromal components in cluster C patients. The effects of WNT signaling on B cells are complex, with some studies indicating that WNT signaling is important in the proliferation of pro- and pre-B cells.41 WNT signaling has also been reported to be prosurvival in GC B cells and in CLL cells, but several other studies have reported that activation of canonical WNT signaling in stromal cells inhibits B-cell lymphopoiesis42 and WNT5a can signal through noncanonical WNT/Ca2+ pathways to negatively regulate B-cell proliferation.43 A more recent study has shown that activation of canonical WNT signaling in stromal cells blocks the proliferation and production of B and NK cells, as well as plasmacytoid dendritic cells,44 and induces the expression of extracellular matrix genes. Because WNT signaling is observed in the least proliferative subset of MCL and because β-catenin appears to be found in stromal cells, the possible role of microenvironmental miRNA in generating inhibitory signals to neoplastic MCL-cells is intriguing and warrants further experimental studies.
Among the miRNAs up-regulated in the high-PS group, miRNAs from the miR17-92 cluster and its 2 paralogs have been confirmed in promoting cell proliferation and inhibiting apoptosis.30 In our previous study, we have also shown that miR17-92 targets PHLPP2, an important negative regulator of the PI3K/AKT pathway, in addition to PTEN and BIM, and overexpression of miR17-92 leads to constitutive activation of PI3K/AKT pathway and also chemoresistance in MCL cell lines.30 In previous studies, high expression of miR17-5p and miR-20b in MCL was associated with short OS.12,13 In contrast, low expression of miR-29, the target genes of which includes CDK6, was associated with a poor prognosis.11 We observed that miR17-92 clusters and its paralogs, miRNA-363, and some of the other miRNAs were correlated with PS (Figure 6A). We generated a 6-miRNA prognosticator for MCL that worked independently of the PS. Of the 6 miRNAs, miR-222 and miR-146a showed a higher difference (> 3-fold) in expression between the 2 prognostic groups. miR-146a27 is repressed and miR-22245 is induced by MYC, and these miRNAs showed the expected correlation with MYC mRNA expression, suggesting that MYC may play a role in MCL prognosis, which is consistent with previous findings.46 The effect of MYC may be mediated in part though the control of miRNA expression. miR-222, which is coexpressed as a cluster with miR-221, targets the tumor-suppressor p27Kip147 and loss of miR-146a promotes tumorigenesis in mice.48 Aside from miRNAs that are correlated with the PS, our study suggests that there are other miRNAs that could serve as predictors of survival. However, because of the limited number of patients in the present study, this miRNA prognosticator will need to be validated and refined in future studies with more MCL patients, particularly in a clinical trial setting.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Martin Bast for coordinating the clinical data and the Lymphoma Research Foundation (MCL consortium) for providing the MCL cell lines. JEKO and JVM2 cells included in this study were provided by the Lymphoma Cell Bank Collection (ATCC).
This work was supported in part by the National Cancer Institute (grant 5U01/CA114778).
National Institutes of Health
Authorship
Contribution: J.I. and Y.S. designed and performed the experiments, analyzed the data, and wrote the manuscript; Y.L., C.L., Z.L., C.M.L., and K.D. designed and performed the experiments; K.F., E.S.J., T.C.G., S.B., L.M.S., L.R., A.R., J.D., E.C., G.O., R.M.B., R.R.T., J.R.C., R.D.G., D.D.W., and W.C.C. provided materials, reviewed the pathology results, or contributed to the GEP data; J.M.V. and J.O.A. provided the clinical data and other support; and T.W.M. and W.C.C. designed and supervised the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for Y.L. is Department of Internal Medicine, Institute of Lymphoma, Henan Cancer Hospital, Zhengzhou, China.
Correspondence: Wing C. Chan, MD, Codirector, Center for Research in Lymphoma and Leukemia, Department of Pathology and Microbiology, 983135 Nebraska Medical Center, Omaha, NE 68198-3135; e-mail: jchan@unmc.edu.
References
Author notes
J.I., Y.S., and Y.L. contributed equally to this work.