Abstract
BM stromal cells (BMSCs) are key players in the microenvironmental support of multiple myeloma (MM) cell growth and bone destruction. A spliced form of the X-box–binding protein-1 (XBP1s), a major proximal effector of unfolded protein response signaling, is highly expressed in MM cells and plays an indispensable role in MM pathogenesis. In the present study, we found that XBP1s is induced in the BMSCs of the MM microenvironment. XBP1s overexpression in healthy human BMSCs enhanced gene and/or protein expression of VCAM-1, IL-6, and receptor activator of NF-κB ligand (RANKL), enhancing BMSC support of MM cell growth and osteoclast formation in vitro and in vivo. Conversely, deficiency of XBP1 in healthy donor BMSCs displayed a range of effects on BMSCs that were opposite to those cells with overexpression of XBP1s. Knock-down of XBP1 in MM patient BMSCs greatly compromised their increased VCAM-1 protein expression and IL-6 and RANKL secretion in response to TNFα and reversed their enhanced support of MM-cell growth and osteoclast formation. Our results demonstrate that XBP1s is a pathogenic factor underlying BMSC support of MM cell growth and osteoclast formation and therefore represents a therapeutic target for MM bone disease.
Introduction
Multiple myeloma (MM) is a severely debilitating, incurable, and essentially uniformly fatal neoplastic disease of B-cell origin.1 It is the most frequent cancer affecting the skeleton, with 90% of patients developing osteolytic lesions.2 Complex cell-cell interactions among MM tumor cells and their microenvironment are essential for tumor growth, survival, and MM-induced osteolytic lesions.3,4 BM stromal cells (BMSCs) are considered a key player in the bone microenvironmental support of MM cell growth and bone destruction.4 BMSCs are the major cell type that produces IL-6, a key inflammatory cytokine required for the growth and survival of MM cells.3 In addition, BMSCs highly express VCAM-1, a cell-surface sialoglycoprotein that is required for MM cell adhesion to BMSCs and chemoresistance of MM cells.5 Further, BMSCs also produce many osteoclastogenic cytokines, including TNFα and receptor activator of NF-κB ligand (RANKL), that promote osteoclast (OCL) differentiation and activity.2 In MM, BMSCs become hyperresponsive to TNFα stimulation and increase the expression of VCAM-1 and the secretion of inflammatory cytokines, including IL-6, TNFα, and RANKL.6 However, it remains unclear how the inflammatory signature of BMSCs is induced and/or maintained. We reasoned that BMSCs in MM must develop enhanced protein folding, trafficking, and secretory capacities to accommodate the increased protein synthesis of the cytokines and growth factors. Unfolded protein response (UPR) signaling is a central regulator for these events and for intracellular protein homeostasis.7,8
Inositol-requiring enzyme-1α (IRE1α)/X-box-binding protein 1 (XBP1) signaling is the most ancient UPR signaling branch that is conserved across species from Caenorhabditis elegans to humans.9,10 IRE1 is an endoplasmic reticulum (ER) transmembrane kinase/endoribonuclease, which upon ER stress, executes unconventional splicing of Xbp1 mRNA to generate spliced Xbp1 (Xbp1s).11 Xbp1s encodes an active transcription factor, XBP1s, which drives the expression of a wide range of gene targets that are involved in ER biogenesis, protein folding, and trafficking, as well as clearance of unfolded/misfolded proteins.11,12
XBP1s is implicated in a wide variety of human physiologic and pathologic processes such as lipogenesis13 and adipogenesis.12,14 Most relevant to the current study, XBP1s is highly expressed in plasma cells.15 XBP1s overexpression in B-lineage cells stimulates plasma cell differentiation16 and enhances mRNA expression and protein secretion of IL-6,17,18 In contrast, XBP1-deficient B cells fail to colonize the BM and do not sustain Ab production.19 Moreover, recent studies have demonstrated that Eμ-directed XBP1s overexpression drives MM pathogenesis.20 An increased ratio of Xbp1s versus unspliced Xbp1 (Xbp1u) predicts poor survival of MM patients.21 Further, both myeloma-specific XBP1 peptides and an IRE1α inhibitor have cytotoxic effects on human MM cells.22,23 These studies demonstrated that IRE1/XBP1s signaling is essential for the growth and survival of MM cells and their prototype plasma cells. However, it remains completely unknown whether XBP1s signaling regulates the capacity of the cells in the MM microenvironment to support MM cell growth and induce osteolytic lesions. In the present study, we used both loss- and gain-of-function strategies and a variety of cell culture models, including human BMSCs (hBMSCs) from both healthy donors and MM patients, and established that XBP1s signaling is a pathogenic factor that regulates the protective effects of the BMSCs of the MM microenvironment on MM tumor growth and OCL formation. Further, overexpression of XBP1s in the BM microenvironment enhances MM cell growth in vivo. These results support targeting XBP1s signaling in the MM microenvironment to disrupt its support of MM tumor growth and bone resorption to treat MM and its bone complications.
Methods
Cell culture and related reagents
Human kidney 297T cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen), 100 units/mL of penicillin, and 100 μg/mL of streptomycin (P/S).24 The human BM stromal cell lines KM101 cells were cultured in α-MEM (Invitrogen) supplemented with 10% FBS and P/S. IRE1α+/+, IRE1α−/−, XBP1+/+, and XBP1−/− mouse embryonic fibroblast (MEF) cells were kindly provided by Dr Randy Kaufman (University of Michigan, Ann Arbor, MI) and maintained in DMEM supplemented with 10% FBS, l-glutamine, P/S, and additional essential and nonessential amino acids (Invitrogen).25 The mouse MM cell line 5TGM1 was grown in RPMI 1640 medium6 supplemented with 10% FBS and P/S. The IL-6–dependent human MM cell line ANBL-6 was cultured in RPMI 1640 medium supplemented with 10% FBS and 1 ng/mL of recombinant human IL-6 (R&D Systems). All cells were cultured at 37°C in a 5% CO2 incubator.
Isolation of primary hBMSCs was performed with informed consent in accordance with the Declaration of Helsinki. hBMSCs and murine BMSCs (mBMSCs) were prepared via a long-term Dexter culture method, as described previously.6 CD 138+ cells were undetectable by immunofluorescence in hBMSCs (supplemental Figure 1B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), indicating that these cells were free of human primary MM cell contamination. These studies were approved by the University of Pittsburgh institutional review board and by the VA Pittsburgh Healthcare System institutional animal care and use committee.
For the MM cell growth assay, mBMSCs (4 × 103/well in 96-well plates) were cocultured with 5TGM1 cells. KM101 cells (4 × 103/well in 96-well plates) or primary hBMSCs (2 × 103/well in 96-well plates) were cocultured with 5TGM1 (2 × 104/well in 96-well plates) or ANBL-6 cells (2 × 104/well in 96-well plates) in RPMI for 3 days as described previously.6 IRE1α+/+, IRE1α−/−, XBP1+/+, or XBP1−/− MEF cells were cocultured with 5TGM1 in RPMI for 3 days. MM cells then were harvested by pipetting and counted using a hemocytometer.
For cell adhesion assays, healthy donor BMSCs with or without Flag-hXBP1s overexpression (8 × 103/well in 96-well plates) were cocultured with MM cells (ie, MM1.S-GFP or ANBL6 at 8 × 104/well) for 60 minutes. Floating MM cells were then harvested by washing 3 times and counted using a hemocytometer. The percentages of the adhered MM cells versus total MM cells used were then calculated.
For the OCL formation assay, primary hBMSCs (2 × 103/well in 96-well plates) or mBMSCs (106/well in 6-well plates) were cocultured with healthy donor nonadherent BM monocytes (hBMMs; 105/well) or murine BMMs (mBMMs; 107/well) for 21 or 12 days, respectively, as described previously.26 OCL formation was assessed by counting the number of tartrate-resistant acid phosphatase (TRAP)–positive multinucleated cells.
Constructs
The Flag-CMV10-hXBP1s plasmid was constructed by inserting the full-length hXBP1s cDNA into Flag-CMV10 (a gift from Dr Shunqian Jing, University of Pittsburg) at HindIII and BamH I sites. Lentivirus constructs (FUGW, pLL3.7, VSVG, RSV-REV, and PRRE) were kindly provided by Dr Tao Chen (University of Pittsburgh, Pittsburg, PA). For XBP1s overexpression, the Flag-tagged hXBP1s cDNA and the pCMV-GFP-WRE sequences were inserted into the FUGW construct. The lentiviral plasmids that carry red fluorescent protein (RFP) were constructed by replacing green fluorescent protein (GFP) with RFP in the FUGW and FUGW-Flag-hXBP1s constructs. For XBP1 knock-down, pLL3.7 was used as described previously (http://web.mit.edu/jacks-lab/protocols/pll37cloning.htm). The sequences of the control shRNA and mouse Xbp1 (mXBP1) shRNA used were published previously.23,27 The hXBP1 shRNA target sequences include clones 1 (5′-GGAACAGCAAGTGGTAGAT-3′), 2 (5′-CATGGATTCTGGCGGTATT-3′), and 3 (5′-GAAGTAGACATGGAATTTA-3′).
Transfection and infection studies
Lipofectamine reagent (Invitrogen) was used for transfection according to the manufacturer's instructions.
For hXBP1s overexpression or XBP1 knockdown experiments, we modified the lentiviral systems FUGW and pLL3.7, which were described previously.28,29 Briefly, 293T cells (6.0 × 106/per 10-cm culture dish) were transfected with the FUGW or FUGW-Flag-hXBP1s construct (10 μg) or the pLL3.7 control shRNA or XBP1 shRNA construct (10 μg) and VSVG (2.5 μg), RSV-REV (3.5 μg), and PRRE (6.5 μg) constructs in antibiotic-free DMEM using lipofectamine 2000 reagent. Forty-eight hours later, the supernatant was harvested. The BMSCs were seeded into 6-well plates and, 24 hours later, infected with lentivirus-containing medium, including the 293T cell supernatant (1 mL) and fresh α-MEM (1 mL) with 4 μg/mL of polybrene. After a 60-minute centrifugation, the cells were incubated in the infection medium for 12-14 hours and then cultured in fresh α-MEM at 37°C and 5% CO2 for 48-72 hours.
Real-time PCR
Real-time PCR was performed as described previously.30 RNA (18S) was used as an internal control. The PCR primers used in this study are listed in Table 1.
PCR primers
| Gene . | Sequence* . | Size (bp) . |
|---|---|---|
| hXBP1s | 198 | |
| Sense | 5′-GAGTCCGCAGCAGGTG-3′ | |
| Antisense | 5′-TCCTTCTGGGTA GACCTCTGGGAG -3′ | |
| hXBP1t | 164 | |
| Sense | 5′-CCCATGGATTCTGGCGGTATTGAC -3′ | |
| Antisense | 5′-TCCTTCTGGGTAGACCTCTGGGAG -3′ | |
| mXBP1s | 150 | |
| Sense | 5′-GAGTCCGCAGCAGGTG-3′ | |
| Antisense | 5′-GTGTCAGAGTCCATGGGA-3′ | |
| mXBP1t | 67 | |
| Sense | 5′-AAGAACACGCTTGGGAATGG-3′ | |
| Antisense | 5′-ACTCCCCTTGGCCTCCAC-3′ | |
| hOPG | 225 | |
| Sense | 5′-GGAACCCCAGAGCGAAATACA-3′ | |
| Antisense | 5′-CCTGAAGAATGCCTCCTCACA-3′ | |
| hRANKL | 203 | |
| Sense | 5′-CAGAAGATGGCACTCACTGCA-3′ | |
| Antisense | 5′-CACCATCGCTTTCTCTGCTCT -3′ | |
| h18S rRNA | 154 | |
| Sense | 5′-ATCCCTGAAAAGTTCCAGCA-3′ | |
| Antisense | 5′-CCCTCTTGGTGAGGTCAATG-3′ | |
| m18S rRNA | 137 | |
| Sense | 5′-CGCTTCCTTACCTGGTTGAT-3′ | |
| Antisense | 5′-GAGCGACCAAAGGAACCATA-3′ |
| Gene . | Sequence* . | Size (bp) . |
|---|---|---|
| hXBP1s | 198 | |
| Sense | 5′-GAGTCCGCAGCAGGTG-3′ | |
| Antisense | 5′-TCCTTCTGGGTA GACCTCTGGGAG -3′ | |
| hXBP1t | 164 | |
| Sense | 5′-CCCATGGATTCTGGCGGTATTGAC -3′ | |
| Antisense | 5′-TCCTTCTGGGTAGACCTCTGGGAG -3′ | |
| mXBP1s | 150 | |
| Sense | 5′-GAGTCCGCAGCAGGTG-3′ | |
| Antisense | 5′-GTGTCAGAGTCCATGGGA-3′ | |
| mXBP1t | 67 | |
| Sense | 5′-AAGAACACGCTTGGGAATGG-3′ | |
| Antisense | 5′-ACTCCCCTTGGCCTCCAC-3′ | |
| hOPG | 225 | |
| Sense | 5′-GGAACCCCAGAGCGAAATACA-3′ | |
| Antisense | 5′-CCTGAAGAATGCCTCCTCACA-3′ | |
| hRANKL | 203 | |
| Sense | 5′-CAGAAGATGGCACTCACTGCA-3′ | |
| Antisense | 5′-CACCATCGCTTTCTCTGCTCT -3′ | |
| h18S rRNA | 154 | |
| Sense | 5′-ATCCCTGAAAAGTTCCAGCA-3′ | |
| Antisense | 5′-CCCTCTTGGTGAGGTCAATG-3′ | |
| m18S rRNA | 137 | |
| Sense | 5′-CGCTTCCTTACCTGGTTGAT-3′ | |
| Antisense | 5′-GAGCGACCAAAGGAACCATA-3′ |
Western blot and ELISA
To determine the secreted RANKL (sRANKL) protein levels, conditioned medium (2 mL) was collected and concentrated to 50 μL by centrifuging with a Centricon Centrifugal Filter Device 10 000 NMWL (Millipore). Total cell lysates were prepared with RIPA buffer containing protease inhibitor cocktail and phosphatase inhibitor cocktails I/II (Sigma-Aldrich). The centrifuged, conditioned medium (CM) and total cell lysates then were separated by 10% sodium dodecylsulfate polyacrylamide gel electrophoresis and analyzed by Western blot analysis as described previously.6
The Abs used include the following: anti–VCAM-1 (sc-1504; 1:1000), anti-XBP1 (sc-7160; 1:1000), and anti-sRANKL (sc-9073; 1:1000; Santa Cruz Biotechnology); anti–β-actin (A5316; 1:1000) HRP-conjugated anti-Flag (A8592; 1:2000; Sigma-Aldrich); and HRP-conjugated IgG secondary Abs (1:2000; GE Healthcare Life Sciences).
To determine IL-6 secretion by BMSCs, supernatants from cell cultures were collected and analyzed with IL-6 ELISA kits (eBioscience) as described previously.6 The serum concentration of IgG2b was measured with the IgG2b ELISA kit (Quidel).
In vivo MM cell growth and bone destruction analyses
SCID NIHIII female mice (4 weeks of age) were injected in the tibias with 20 μL of 5 × 104 KM101 cells (with or without Flag-hXBP1s overexpression) alone or with 105 5TGM1-GFP-TK cells in saline. All research protocols were approved by the Pittsburgh VA Healthcare System institutional animal care and use committee. At 4 weeks after injection, the animals were killed and the tibias were dissected out. Images of dissected tibias were acquired on a vivaCT 40 scanner (Scanco Medical) at a resolution of 21-μm isotropic, reconstructed, and segmented for 3-dimensional display using the instrument's analysis algorithm software. The in vivo growth of 5TGM1-GFP-TK cells was detected using the LT-9MACIMSYSPLUS fluorescence imaging system (Lightools Research).
Statistical analysis
Each experiment was performed at least 3 times and results are presented as means ± SD. Statistical significance was determined by Mann-Whitney test. A P value of less than .05 was considered statistically significant.
Results
XBP1s is induced in BMSCs of the MM microenvironment
We observed that the primary BMSCs from MM patients displayed elevated protein levels of endogenous XBP1s compared with the BMSCs from healthy donors and from the human BMS cell line KM101 cells (Figure 1A and supplemental Figure 1A). Further, MM patient BMSCs exhibited significantly increased mRNA levels of Xbp1s (4.6-fold) compared with healthy donor BMSCs (Figure 1B left). In contrast, the difference in total Xbp1 mRNA levels between healthy donor and MM patient BMSCs remained insignificant (Figure 1B middle). Therefore, there was a 3.4-fold up-regulation of Xbp1 mRNA splicing, as reflected by the ratios of Xbp1s to total Xbp1, in the MM patient BMSCs compared with the healthy donor hBMSCs (Figure 1B right). These results demonstrate that XBP1s signaling is activated in MM patient BMSCs. In addition, we observed that the CM of the MM cell line MM1.S cells induced XBP1s protein expression to a same extent as direct contact of BMSCs and MM1.S (Figure 1C), suggesting that MM cell CM is sufficient to induce XBP1s in BMSCs and direct cell-cell contact is not required.
XBP1s is induced in MM patient BMSCs and TNF-α induces XBP1s up-regulation in healthy hBMSCs. (A) Western blot demonstrating XBP1s protein levels in primary BMSCs from MM patients and healthy donors. (B) Real-time PCR analysis demonstrating mRNA levels of Xbp1s (left), total Xbp1 (middle), and mRNA splicing of total Xbp1 (right) in the BMSCs of MM patients and healthy donors. The latter was determined as the ratio of Xbp1s to total Xbp1 mRNA levels (n = 3; mean ± SD; *P < .05; NS indicates not statistically significant). (C) Western blot demonstrating XBP1s protein levels in healthy donor BMSCstreated with MM1.S CM or cocultured with MM1.S cells (MM1.S). (D) Western blot demonstrating the effects of TNFα (10 ng/mL for 24 hours) on XBP1s protein levels in human BMS cell line KM101. (E) Real-time PCR results demonstrating the effects of TNFα (10 ng/mL for 24 hours) on mRNA levels of Xbp1s and total Xbp1 and mRNA splicing of total Xbp1 in KM101 cells (n = 3; mean ± SD; *P < .05). (F) Western blot demonstrating the effect of TNFα (10 ng/mL for 24 hours) on XBP1s protein levels in healthy donor BMSCs. (G) Real-time RT-PCR results demonstrating the effects of TNFα (10 ng/mL for 24 hours) on mRNA levels of Xbp1s and total Xbp1 and splicing of total Xbp1 mRNA in healthy donor BMSCs (n = 3; mean ± SD; *P < .05).
XBP1s is induced in MM patient BMSCs and TNF-α induces XBP1s up-regulation in healthy hBMSCs. (A) Western blot demonstrating XBP1s protein levels in primary BMSCs from MM patients and healthy donors. (B) Real-time PCR analysis demonstrating mRNA levels of Xbp1s (left), total Xbp1 (middle), and mRNA splicing of total Xbp1 (right) in the BMSCs of MM patients and healthy donors. The latter was determined as the ratio of Xbp1s to total Xbp1 mRNA levels (n = 3; mean ± SD; *P < .05; NS indicates not statistically significant). (C) Western blot demonstrating XBP1s protein levels in healthy donor BMSCstreated with MM1.S CM or cocultured with MM1.S cells (MM1.S). (D) Western blot demonstrating the effects of TNFα (10 ng/mL for 24 hours) on XBP1s protein levels in human BMS cell line KM101. (E) Real-time PCR results demonstrating the effects of TNFα (10 ng/mL for 24 hours) on mRNA levels of Xbp1s and total Xbp1 and mRNA splicing of total Xbp1 in KM101 cells (n = 3; mean ± SD; *P < .05). (F) Western blot demonstrating the effect of TNFα (10 ng/mL for 24 hours) on XBP1s protein levels in healthy donor BMSCs. (G) Real-time RT-PCR results demonstrating the effects of TNFα (10 ng/mL for 24 hours) on mRNA levels of Xbp1s and total Xbp1 and splicing of total Xbp1 mRNA in healthy donor BMSCs (n = 3; mean ± SD; *P < .05).
Because we have shown that TNFα, a major component of MM cell CM, can cause MM cells to induce an inflammatory signature in BMSCs,6 we then invesetigated whether TNFα could also induce XBP1s in BMSCs. TNFα (10 ng/mL for 24 hours) strongly induced XBP1s protein levels in human BM stromal cell line KM101 cells (Figure 1D). At the same time, TNFα stimulated Xbp1s and total Xbp1 mRNA by 8.2-fold (Figure 1E left) and 1.3-fold (Figure 1E middle), respectively, indicating that TNFα induced an approximately 6.2-fold up-regulation of Xbp1 mRNA splicing in KM101 cells (Figure 1E right). Consistently, TNFα also induced XBP1s protein in BMSCs from healthy donors (Figure 1F). This was accompanied by a 4.3- and 1.8-fold induction of Xbp1s (Figure 1G left) and total Xbp1 (Figure 1G middle) mRNA levels, respectively, indicating a 2.3-fold increase in Xbp1 mRNA splicing (Figure 1G right). These data demonstrate that TNFα can case MM cell CM to induce Xbp1 mRNA splicing and, consequently, XBP1s protein levels in hBMSCs.
Elevation of XBP1s in BMSCs induces the BMSC inflammatory signature and promotes BMSC support of MM cell growth
We also investigated the pathophysiologic significance of elevated XBP1s in BMSCs in MM disease. For this purpose, we established healthy donor BMSCs and KM101 cells that overexpressed either the Flag-tagged human XBP1s (Flag-hXBP1s) or an empty vector (EV) construct (control) via lentiviral infection (supplemental Figure 1C left and right, respectively). Lentivirus infection did not induce ER stress or endogenous XBP1s protein expression in these cells (supplemental Figure 1C). ELISA assays demonstrated that overexpression of XBP1s stimulated baseline IL-6 secretion by approximately 2.4-fold in healthy donor BMSCs (Figure 2A). Moreover, it was observed that TNFα induced 1.8-, 2.0-, and 2.6-fold increases in IL-6 protein secretion in the EV-expressing healthy donor BMSCs at 24, 48, and 72 hours, respectively, whereas it stimulated 1.5-, 2.5-, and 4.2-fold increases in IL-6 secretion in Flag-hXBP1s–overexpressing healthy donor BMSCs at the corresponding time points (Figure 2A).
XBP1s overexpression in hBMSCs enhances their support of MM cell growth. (A) Overexpression of XBP1s enhanced the baseline and TNFα (10 ng/mL)–induced IL-6 secretion in healthy donor BMSCs, as determined by ELISA (n = 3; mean ± SD; *P < .05). (B) Overexpression of Flag-hXBP1s increased the baseline and TNFα (10 ng/mL for 24 hours)–induced VCAM-1 protein expression in healthy donor BMSCs. The schematic quantitative representation of Western blot results was performed by normalizing VCAM-1 protein levels to those of β-actin. The relative protein levels of VCAM-1 in the control BMSCs without TNFα is arbitrarily defined as 1 (right; n = 3; mean ± SD; *P < .05). (C) Overexpression of Flag-hXBP1s enhanced the baseline and TNFα-induced VCAM-1 protein expression in KM101 cells. The experiments were performed and analyzed as described in panel B. (D) Overexpression of hXBP1s in healthy donor BMSCs significantly enhanced the adhesion of MM1.S and ANBL6 cells to the hBMSCs (n = 3; mean ± SD; *P < .05). (E-F) Overexpression of Flag-hXBP1s enhanced the capacity of healthy donor BMSCs (E) and KM101 cells (F) to support MM cell growth. The cells with or without Flag-hXBP1s overexpression were cocultured with either 5TGM1 or the ANBL-6 MM cell line for 3 days. MM cells growth was determined by counting MM cells (n = 3; mean ± SD; *P < .05).
XBP1s overexpression in hBMSCs enhances their support of MM cell growth. (A) Overexpression of XBP1s enhanced the baseline and TNFα (10 ng/mL)–induced IL-6 secretion in healthy donor BMSCs, as determined by ELISA (n = 3; mean ± SD; *P < .05). (B) Overexpression of Flag-hXBP1s increased the baseline and TNFα (10 ng/mL for 24 hours)–induced VCAM-1 protein expression in healthy donor BMSCs. The schematic quantitative representation of Western blot results was performed by normalizing VCAM-1 protein levels to those of β-actin. The relative protein levels of VCAM-1 in the control BMSCs without TNFα is arbitrarily defined as 1 (right; n = 3; mean ± SD; *P < .05). (C) Overexpression of Flag-hXBP1s enhanced the baseline and TNFα-induced VCAM-1 protein expression in KM101 cells. The experiments were performed and analyzed as described in panel B. (D) Overexpression of hXBP1s in healthy donor BMSCs significantly enhanced the adhesion of MM1.S and ANBL6 cells to the hBMSCs (n = 3; mean ± SD; *P < .05). (E-F) Overexpression of Flag-hXBP1s enhanced the capacity of healthy donor BMSCs (E) and KM101 cells (F) to support MM cell growth. The cells with or without Flag-hXBP1s overexpression were cocultured with either 5TGM1 or the ANBL-6 MM cell line for 3 days. MM cells growth was determined by counting MM cells (n = 3; mean ± SD; *P < .05).
VCAM-1 is highly expressed by BMSCs and is required for MM cell adhesion to BMSCs and for MM cell growth.5 MM patient BMSCs displayed enhanced TNFα-induced VCAM-1 protein expression.3,6 We found that overexpression of XBP1s led to a 1.4-fold up-regulation of the base level of VCAM-1 in healthy donor BMSCs (Figure 2B). Further, in EV-expressing healthy donor BMSCs and KM101 cells, 24 hours of TNFα treatment stimulated 1.7- and 5.1-fold increases in VCAM-1 protein levels, respectively. In contrast, in their corresponding counterparts that express Flag-hXBP1s, TNFα induced approximately 3.2- and 9.5-fold increases in VCAM-1 protein expression, respectively (Figure 2B-C). Further, overexpression of XBP1s in healthy donor BMSCs promoted the adhesion of human MM cells (MM1.S and ANBL6) to the cocultured healthy donor BMSCs by approximately 60% compared with EV overexpression in healthy donor BMSCs (Figure 2D).
Finally, consistent with their increased VCAM-1 and IL-6 protein levels and enhanced capacity to promote MM tumor cell adhesion, the Flag-hXBP1s–overexpressing healthy donor BMSCs displayed approximately 35% and 23% greater support of the growth of murine and human MM cells (ie, 5TGM1 and ANBL-6 cells, respectively) compared with the EV-expressing healthy donor BMSCs (Figure 2E). Similarly, the Flag-hXBP1s–overexpressing KM101 cells also lent significantly greater support to the growth of 5TGM1 and ANBL-6 cells by 44% and 26% compared with their EV-expressing counterparts (Figure 2F).
These results demonstrate that elevated XBP1s is sufficient to cause MM cells to induce an inflammatory signature in BMSCs and to enhance the capacity of hBMSCs to support MM cell growth in vitro.
XBP1s overexpression in BMSCs promotes BMSC support of osteoclast formation
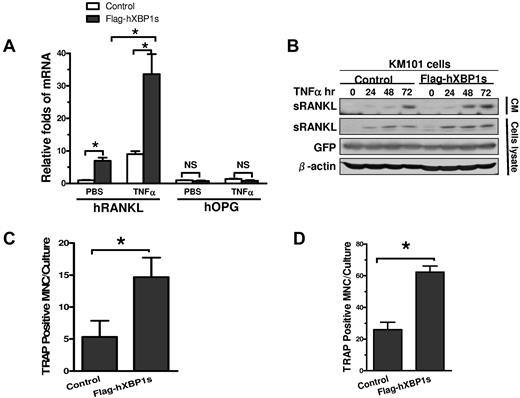
BMSCs promote OCL formation in MM bone disease by producing osteoclastogenic cytokines such as RANKL.3 Real-time PCR analysis demonstrated that overexpression of Flag-hXBP1s in KM101 cells enhanced baseline and TNFα-induced gene expression of RANKL in these cells by approximately 7.0- and 3.7-fold compared with EV overexpression, respectively (Figure 3A). In contrast, it did not affect the mRNA levels of osteoprotegerin (OPG), an important inhibitor of osteoclastogenesis,31 resulting in dramatic increases in RANKL/OPG ratios, an important indicator for osteoclastogenic potential of BMSCs, under both experimental conditions (Figure 3A).
XBP1s overexpression in BMSCs induces RANKL secretion and OCL formation. (A) Real-time PCR demonstrating the effects of overexpression of Flag-hXBP1s on gene expression of RANKL and OPG in KM101 cells with or without TNFα treatment (n = 3; mean ± SD; *P < .05). (B) Effects of overexpression of Flag-hXBP1s on the baseline and TNFα-induced sRANKL protein expression in both the conditioned medium and total cell lysates of KM101 cells as determined by anti-RANKL Western blot analysis. (C) Effects of overexpression of Flag-hXBP1s on the capacity of healthy donor BMSCs to support OCL formation. The healthy donor BMSCs with or without Flag-hXBP1s overexpression were cocultured with hBMMs for 21 days. The number of TRAP-positive multinucleated cells was calculated (n = 3; mean ± SD; *P < .05). (D) Effects of overexpression of Flag-hXBP1s on the capacity of mBMSCs to support OCL formation. The mBMSCs with or without Flag-hXBP1s overexpression were cocultured with mBMMs for 12 days (n = 3; mean ± SD; *P < .05).
XBP1s overexpression in BMSCs induces RANKL secretion and OCL formation. (A) Real-time PCR demonstrating the effects of overexpression of Flag-hXBP1s on gene expression of RANKL and OPG in KM101 cells with or without TNFα treatment (n = 3; mean ± SD; *P < .05). (B) Effects of overexpression of Flag-hXBP1s on the baseline and TNFα-induced sRANKL protein expression in both the conditioned medium and total cell lysates of KM101 cells as determined by anti-RANKL Western blot analysis. (C) Effects of overexpression of Flag-hXBP1s on the capacity of healthy donor BMSCs to support OCL formation. The healthy donor BMSCs with or without Flag-hXBP1s overexpression were cocultured with hBMMs for 21 days. The number of TRAP-positive multinucleated cells was calculated (n = 3; mean ± SD; *P < .05). (D) Effects of overexpression of Flag-hXBP1s on the capacity of mBMSCs to support OCL formation. The mBMSCs with or without Flag-hXBP1s overexpression were cocultured with mBMMs for 12 days (n = 3; mean ± SD; *P < .05).
We also determined the effect of XBP1s on RANKL protein secretion in BMSCs in both the presence and absence of TNFα treatment. Flag-hXBP1s– or EV-overexpressing KM101 cells were treated with TNFα for 0, 24, 48, or 72 hours. The CM and cell lysates were subjected to anti-RANKL Western blot analysis. The Flag-hXBP1s-KM101 cells displayed increased TNFα-induced sRANKL protein levels at all time points examined in both the CM and cell lysates compared with the EV-KM101 cells (Figure 3B).
Finally, we determined whether BMSCs with elevated XBP1s protein levels induce greater OCL formation. We established both healthy donor hBMSCs (supplemental Figure 1C) and mBMSCs (supplemental Figure 1D) that expressed Flag-hXBP1s or EV via lentiviral infection. The resultant cells were cocultured with hBMMs or mBMMs in osteoclastogenic medium for 21 or 12 days, respectively. Quantification of TRAP-positive, multinucleated OCLs revealed that there was approximately 2.7-fold more OCL in the cocultures with Flag-hXBP1s–overexpressing healthy donor BMSCs compared with those containing EV-expressing BMSCs (Figure 3C). Similar results were found with the mBMSCs and mBMMs coculture systems (Figure 3D). These results demonstrate that overexpression of XBP1s is sufficient to give BMSCs a greater capacity to induce OCL formation.
Deficiency of IRE1α or XBP1 compromises BMSC support of MM cell growth
To further establish the role of XBP1s in regulating BMSCs in MM disease, we determined whether deficiency of XBP1 or IRE1α compromised BMSC support of MM cell growth. First, we used MEFs null for either XBP1 or IRE1α (XBP1−/− or IRE1α−/−) and their corresponding wild-type (WT) MEF cells (XBP1+/+or IRE1α+/+). As shown by Western blot, unlike WT cells, XBP1−/− cells failed to elevate XBP1s protein levels in response to thapsigargin, a stress activator that disturbs Ca++ homeostasis in the ER (supplemental Figure 2A). This result confirmed the deficiency of XBP1 in MEF cells. XBP1−/− MEF cells exhibited significantly less TNFα-induced VCAM-1 protein expression (Figure 4A) and dramatically reduced baseline IL-6 protein secretion (Figure 4B) compared with XBP1+/+ MEF cells. Consistently, the XBP1−/− MEF cells displayed a significantly lower capacity (17%) to support the growth of murine MM cells (5TGM1) compared with WT cells (Figure 4C).
Deficiency of XBP1 or IRE1α compromises MEF and murine BMSC support of MM cell growth. (A) Western blot demonstrating the effect of XBP1 deficiency on baseline and TNFα (10 ng/mL for 24 hours)–induced VCAM-1 protein expression in MEF cells. The basal level VCAM-1 protein expression in XBP1+/+ MEF cells was normalized to β-actin and defined as 1. (B) ELISA assay shows that IL-6 secretion was decreased significantly in XBP1−/− MEF cells compared with XBP1+/+ MEF cells (n = 3; mean ± SD; *P < .05). (C) Effect of XBP1 deficiency on the capacity of MEF cells to support MM cell growth (n = 3, mean ± SD, *P < .05). (D) The effects of TNFα (10 ng/mL for 24 hours) on baseline and TNFα-induced VCAM-1 protein expression in IRE1α+/+ and IRE1α−/− MEF cells, determined by Western blotting. The basal level VCAM-1 protein expression in IRE1α+/+ MEF cells was normalized to β-actin and defined as 1. (E) ELISA assay shows that IL-6 secretion was significantly decreased in IRE1α−/− MEF cells compared with that in IRE1α+/+ MEF cells (n = 3; mean ± SD; *P < .05). (F) Effect of IRE1α deficiency on the capacity of MEF cells to support 5TGM1 cell growth (n = 3, mean ± SD, *P < .05). (G) Effects of murine XBP1 knock-down on the baseline and TNFα (10 ng/mL for 24 hours)–induced VCAM-1 protein expression in mBMSCs. The basal level VCAM-1 protein expression in mBMSCs with control shRNA was normalized to β-actin and defined as 1. (H) Effects of murine XBP1 knockdown on IL-6 secretion in mBMSCs (n = 3; mean ± SD; *P < .05). (I) XBP1 knockdown reduced mBMSC support of 5TGM1 cell growth (n = 3, mean ± SD, *P < .05).
Deficiency of XBP1 or IRE1α compromises MEF and murine BMSC support of MM cell growth. (A) Western blot demonstrating the effect of XBP1 deficiency on baseline and TNFα (10 ng/mL for 24 hours)–induced VCAM-1 protein expression in MEF cells. The basal level VCAM-1 protein expression in XBP1+/+ MEF cells was normalized to β-actin and defined as 1. (B) ELISA assay shows that IL-6 secretion was decreased significantly in XBP1−/− MEF cells compared with XBP1+/+ MEF cells (n = 3; mean ± SD; *P < .05). (C) Effect of XBP1 deficiency on the capacity of MEF cells to support MM cell growth (n = 3, mean ± SD, *P < .05). (D) The effects of TNFα (10 ng/mL for 24 hours) on baseline and TNFα-induced VCAM-1 protein expression in IRE1α+/+ and IRE1α−/− MEF cells, determined by Western blotting. The basal level VCAM-1 protein expression in IRE1α+/+ MEF cells was normalized to β-actin and defined as 1. (E) ELISA assay shows that IL-6 secretion was significantly decreased in IRE1α−/− MEF cells compared with that in IRE1α+/+ MEF cells (n = 3; mean ± SD; *P < .05). (F) Effect of IRE1α deficiency on the capacity of MEF cells to support 5TGM1 cell growth (n = 3, mean ± SD, *P < .05). (G) Effects of murine XBP1 knock-down on the baseline and TNFα (10 ng/mL for 24 hours)–induced VCAM-1 protein expression in mBMSCs. The basal level VCAM-1 protein expression in mBMSCs with control shRNA was normalized to β-actin and defined as 1. (H) Effects of murine XBP1 knockdown on IL-6 secretion in mBMSCs (n = 3; mean ± SD; *P < .05). (I) XBP1 knockdown reduced mBMSC support of 5TGM1 cell growth (n = 3, mean ± SD, *P < .05).
Moreover, in agreement with the role of IRE1α in generating Xbp1s, IRE1α-deficient MEFs also failed to elevate Xbp1 protein levels in response to thapsigargin (supplemental Figure 2B). Like XBP1−/− MEF cells, IREα−/− cells exhibited significantly reduced TNFα-induced VCAM-1 protein expression (Figure 4D) and baseline IL-6 protein secretion (Figure 4E) compared with IREα+/+ cells. Further, these cells recapitulated the XBP1−/− MEFs in their reduced support of MM cell growth by 24% compared with the IRE1α+/+ MEFs (Figure 4F). The striking phenotypic similarity between the XBP1−/− and IREα−/− MEFs demonstrates that IRE1α/XBP1s signaling is critical for BMSC support of MM cell growth.
To determine the impact of XBP1 deficiency on BMSCs, we constructed GFP-tagged lentiviral shRNA for mXbp1. This construct, when injected into the mBMSCs, reduced the endogenous Xbp1s mRNA levels to less than 50% of its original levels (supplemental Figure 2C) and also decreased the Thg-induced XBP1s protein level (supplemental Figure 2D). The mBMSCs that carried mXbp1 shRNA exhibited reduced IL-6 secretion (Figure 4H), and VCAM-1 protein expression both in the absence and presence of TNFα treatment compared with the cells expressing control shRNA (Figure 4G). Consistently, the mXBP1 shRNA–expressing cells also displayed significantly reduced (24%) support of the growth of cocultured 5TGM1 cells compared with the control shRNA-expressing mBMSCs (Figure 4I).
Xbp1 knockdown in MM patient BMSCs reverses their heightened inflammatory signatures and enhanced support of MM cell growth
Having demonstrated that XBP1s protein expression is induced in MM patient BMSCs (Figure 1A), we investigated whether elevated XBP1s protein is responsible for the pathologic behavior of MM patient BMSCs. For this purpose, we constructed 3 lentiviral constructs that carry GFP-tagged human Xbp1 shRNAs. Western blot analysis demonstrated that all 3 constructs efficiently knocked down endogenous XBP1s in a model cell culture system of HEK293 cells (supplemental Figure 3A). Using constructs 1 and 3, we successfully reduced endogenous total Xbp1 mRNA levels and, consequently, XBP1s protein levels in healthy donor BMSCs (supplemental Figure 3B). Western blot analysis demonstrated that hXbp1 shRNAs reduced the baseline and TNFα-induced VCAM-1 protein levels at 24, 48, and 72 hours in healthy donor BMSCs by 19%, 23%, 44%, and 51%, respectively. In contrast, hXbp1 shRNAs reduced the baseline and TNFα-induced VCAM-1 protein levels in MM patient BMSCs at the corresponding time points by 73%, 36%, 63%, and 60%, respectively, which led to a reduction in the patient BMSC VCAM-1 protein expression to levels comparable with those of healthy donor BMSCs at 48 and 72 hours of TNFα treatment (Figure 5A-B).
Knock-down of XBP1 in hBMSCs compromises their support of MM cell growth. (A) Western blot demonstrating the effects of XBP1 knockdown on the baseline and TNFα-induced VCAM-1 protein expression in BMSCs from 3 independent healthy donors and 4 independent MM patients. (B) A schematic quantitative representation of Western blot results shown in panel A. The VCAM-1 protein levels were normalized to those of β-actin. The baseline relative VCAM-1 protein levels of the healthy donor BMSCs in the absence of hXbp1 shRNA are arbitrarily defined as 1. Error bars represent means ± SD. (C) Effects of XBP1 knockdown on the baseline and TNFα-induced IL-6 secretion in the condition medium of both healthy donor and MM patient BMSCs as determined by ELISA (n = 3, mean ± SD, *P < .05). (D-E) Effects of XBP1 knockdown on the capacity of healthy donor and MM patient BMSCs to support the growth of human ANBL6 cells (D) or primary human MM cells (E; n = 3, mean ± SD, *P < .05).
Knock-down of XBP1 in hBMSCs compromises their support of MM cell growth. (A) Western blot demonstrating the effects of XBP1 knockdown on the baseline and TNFα-induced VCAM-1 protein expression in BMSCs from 3 independent healthy donors and 4 independent MM patients. (B) A schematic quantitative representation of Western blot results shown in panel A. The VCAM-1 protein levels were normalized to those of β-actin. The baseline relative VCAM-1 protein levels of the healthy donor BMSCs in the absence of hXbp1 shRNA are arbitrarily defined as 1. Error bars represent means ± SD. (C) Effects of XBP1 knockdown on the baseline and TNFα-induced IL-6 secretion in the condition medium of both healthy donor and MM patient BMSCs as determined by ELISA (n = 3, mean ± SD, *P < .05). (D-E) Effects of XBP1 knockdown on the capacity of healthy donor and MM patient BMSCs to support the growth of human ANBL6 cells (D) or primary human MM cells (E; n = 3, mean ± SD, *P < .05).
We also observed that hXbp1 shRNAs reduced baseline and TNFα-induced IL-6 secretion in healthy and patient BMSCs significantly (Figure 5C). Further, as shown previously6 and confirmed here, MM patient BMSCs displayed enhanced TNFα-induced IL-6 secretion (10 451 pg/mL) compared with healthy donor BMSCs (7046 pg/mL; Figure 5C). MM patient BMSCs expressing the hXbp1 shRNAs secreted IL-6 at 6322 pg/mL, a level that is even lower than that of the healthy donor BMSCs. The latter results indicate that the hXbp1 shRNAs abolished the increased IL-6 production in response to TNFα in MM patient BMSCs.
Finally, we determined whether knockdown of Xbp1 reversed the enhanced capacity of MM patient BMSCs to support MM cell growth. As described previously6 and confirmed here, the MM patient BMSCs exhibited greater support of the growth of 5TGM1 and ANBL-6 cells: by 22% (supplemental Figure 3D) and 27% (Figure 5D), respectively, compared with the healthy donor BMSCs. However, in the presence of hXbp1 shRNAs, the MM patient BMSCs displayed a level of support of MM cell growth comparable with (supplemental Figure 3D) or even less than (Figure 5D) that of the healthy donor BMSCs. Further, we confirmed these observations using primary human MM cells. MM patient BMSCs provided greater support of primary human MM cell growth (2.2-fold) compared with the healthy donor BMSCs (Figure 5E). However, the enhanced support of MM patient BMSCs was abolished by hXbp1 shRNAs (Figure 5E). These results show that XBP1s is a pathologic factor that affects the BMSC support of MM cell growth.
Xbp1 knockdown reduces BMSC secretion of RANKL and support of OCL formation
The heightened production of RANKL and enhanced capacity to induce osteoclastogenesis are pathologic features of MM BMSCs. We therefore investigated whether elevated XBP1s protein levels are responsible for these pathologic effects. Western blot results demonstrated that hXbp1 shRNAs inhibited the baseline and TNFα-induced sRANKL protein levels at 24, 48, and 72 hours by 46%, 21%, 13%, and 43%, respectively, in healthy donor BMSCs. In contrast, hXbp1 shRNAs inhibited the baseline and TNFα-induced sRANKL protein levels at the corresponding time points by 54%, 61%, 59%, and 62%, respectively, in MM patient BMSCs, which reduced the sRANKL protein expression to levels comparable with those of healthy donor BMSCs (Figure 6A-B).
Knockdown of XBP1 in BMSCs compromises BMSC secretion of RANKL and support of OCL formation. (A) Western blot demonstrating the effects of XBP1 knockdown on the baseline and TNFα-induced sRANKL protein levels in the total lysates of the BMSCs of both healthy donors (n = 3; left) and MM patients (n = 4; right). β-actin served as the loading control. (B) Schematic quantitative representation of results shown in panel A. The sRANKL protein levels were normalized to those of β-actin. The baseline relative sRANKL protein levels of the healthy donor BMSCs in the absence of hXbp1 shRNAs are arbitrarily defined as 1. Error bars represent means ± SD. (C) Effect of XBP1 knockdown on the capacity of MM patient BMSCs to support OCL formation (n = 3; mean ± SD; *P < .05).
Knockdown of XBP1 in BMSCs compromises BMSC secretion of RANKL and support of OCL formation. (A) Western blot demonstrating the effects of XBP1 knockdown on the baseline and TNFα-induced sRANKL protein levels in the total lysates of the BMSCs of both healthy donors (n = 3; left) and MM patients (n = 4; right). β-actin served as the loading control. (B) Schematic quantitative representation of results shown in panel A. The sRANKL protein levels were normalized to those of β-actin. The baseline relative sRANKL protein levels of the healthy donor BMSCs in the absence of hXbp1 shRNAs are arbitrarily defined as 1. Error bars represent means ± SD. (C) Effect of XBP1 knockdown on the capacity of MM patient BMSCs to support OCL formation (n = 3; mean ± SD; *P < .05).
Further, we cocultured human OCL precursors with healthy and MM BMSCs expressing either control shRNA or hXbp1 shRNAs in osteoclastogenic medium for 21 days. Quantification of the TRAP-positive, multinucleated OCLs revealed that MM patient BMSCs exhibited an approximately 4.7-fold greater support of OCL formation compared with healthy donor BMSCs, and this enhancement was reduced significantly by hXbp1 shRNAs (Figure 6C). Similarly, we found that mXbp1 shRNA reduced the capacity of mBMSCs to support OCL formation by murine OCL precursors to 35% of its original level (supplemental Figure 2F). These results reveal that XBP1s is a pathologic factor that affects the BMSC support of OCL formation.
Elevation of XBP1s in BMSCs promotes BMSC support of MM cell growth and bone absorption in vivo
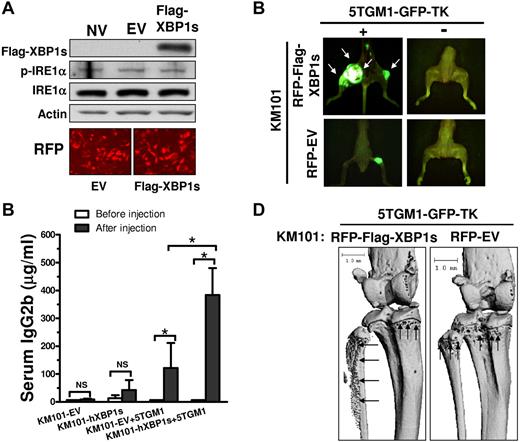
Finally, we investigated whether elevated XBP1s protein in BMSCs promotes MM cell growth and bone destruction in vivo. We established KM101 cells that overexpressed either RFP-tagged human XBP1s (RFP-Flag-hXBP1s) or RFP-tagged EV construct (RFP-EV Control) via lentiviral infection (Figure 7A and supplemental Figure 4). We mixed the infected KM101 cells with 5TGM1-GFP-TK cells and injected the cell mixtures into the tibias of SCID mice. As negative controls, the infected KM101 cells in the absence of 5TGM1-GFP-TK cells were also injected into the control SCID mice. At 4 weeks after injection, we examined MM tumor growth and bone destruction by fluorescence imaging of GFP and μQCT analysis, respectively. Many of the SCID mice developed MM tumors in the hXBP1s-KM101 plus 5TGM1-GFP-TK animal group (4 of 4 mice) compared with the EV-KM101 plus 5TGM1-GFP-TK animal group (2 of 4 mice); in addition, tumor masses were increased in both number and size in the former compared with the latter (Figure 7B). Consistently, the hXBP1s-KM101 plus 5TGM1-GFP-TK animal group displayed a significantly increased serum concentration of IgG2b, a monoclonal protein marker secreted by the MM cells,32 as measured via ELISA (Figure 7C). Further, the SCID mice with RFP-hXBP1s-KM101 plus 5TGM1-GFP-TK cells also exhibited greater bone destruction in vivo compared with those with RFP-EV-KM101 plus 5TGM1-GFP-TK cells by μQCT analysis (Figure 7D). The injection of infected KM101 cells alone failed to induce osteolytic lesions (data not shown).
hXBP1s overexpression in BMSCs enhances their support of MM cell growth and bone resorption in vivo. (A) Western blot demonstrating that lentiviral infection of Flag-hXBP1s led to enhanced expression of hXBP1s in KM101 cells. EV-expressing lentiviruses did not induce endogenous XBP1s protein level and phosphorylation of IRE1α in the KM101 cells (top). RFP fluorescence images indicate high infection efficiency (bottom). (B) Overexpression of XBP1s in KM101 cells enhanced MM cell growth in SCID mice. The tumor growth at 4 weeks after injection was reflected by fluorescence imaging of GFP. White arrows indicate MM tumor masses formed by the injected 5TGM1-GFP-TK cells. One of 4 representative mice is shown for each group. (C) ELISA analyses demonstrating the serum concentrations of IgG2b in the injected SCID mice before and after cell injections at 4 weeks after injection (n = 3; mean ± SD; *P < .05. NS indicates not statistically significant. (D) μQCT analysis demonstrating more severe and extensive osteolytic lesions in the SCID mice with XBP1s-KM101 cells plus GFP-5TGM1 cells compared with those with EV-KM101 plus GFP-5TGM1 cells. Black arrows indicate bone destruction.
hXBP1s overexpression in BMSCs enhances their support of MM cell growth and bone resorption in vivo. (A) Western blot demonstrating that lentiviral infection of Flag-hXBP1s led to enhanced expression of hXBP1s in KM101 cells. EV-expressing lentiviruses did not induce endogenous XBP1s protein level and phosphorylation of IRE1α in the KM101 cells (top). RFP fluorescence images indicate high infection efficiency (bottom). (B) Overexpression of XBP1s in KM101 cells enhanced MM cell growth in SCID mice. The tumor growth at 4 weeks after injection was reflected by fluorescence imaging of GFP. White arrows indicate MM tumor masses formed by the injected 5TGM1-GFP-TK cells. One of 4 representative mice is shown for each group. (C) ELISA analyses demonstrating the serum concentrations of IgG2b in the injected SCID mice before and after cell injections at 4 weeks after injection (n = 3; mean ± SD; *P < .05. NS indicates not statistically significant. (D) μQCT analysis demonstrating more severe and extensive osteolytic lesions in the SCID mice with XBP1s-KM101 cells plus GFP-5TGM1 cells compared with those with EV-KM101 plus GFP-5TGM1 cells. Black arrows indicate bone destruction.
Discussion
A thorough understanding of the molecular changes in the MM microenvironment is essential for disrupting the protective effects of the MM microenvironment on tumor cell growth and bone destruction. In the present study, we have shown that the UPR signaling molecule XBP1s is induced in MM patient BMSCs compared with the healthy donor BMSCs (Figure 1A). We also showed that MM-conditioned medium elevated the XBP1s protein levels in hBMSCs (Figure 1C). TNFα, a prominent inflammatory cytokine in MM-conditioned medium, mimicked MM-conditioned medium and triggered splicing of Xbp1 (Figure 1D-E). In contrast, direct cell-cell contact was not required for XBP1s up-regulation in BMSCs (Figure 1C). These observations demonstrate for the first time that XBP1 signaling is activated in the BMSCs of the MM microenvironment.
We determined the pathophysiologic consequences of elevated XBP1s protein levels in BMSCs from the MM microenvironment. Using loss- and gain-of-function strategies both in vitro and in vivo, we found that elevation of XBP1s in BMSCs is sufficient to cause MM cells to induce inflammatory signature of BMSCs and to promote BMSC support of MM cell adhesion, growth, and OCL formation and thus bone destruction. Further, induction of XBP1s protein levels is an important pathogenic factor underlying the pathologic behaviors of MM patient BMSCs, such as their heightened inflammatory signature and enhanced support of MM cell growth and OCL formation, compared with the healthy donor BMSCs. It will be important in future studies to determine whether other cellular components of the MM microenvironment, such as macrophages, also display enhanced XBP1s protein expression and if XBP1s also plays a role in their support of MM cell growth and osteoclastogenesis.
In determining the molecular mechanisms underlying XBP1s regulation of BMSC support of MM cell growth and OCL formation, we found that modulating XBP1s in BMSCs affects gene expression, protein expression, and/or protein production of multiple molecules that are essential for MM cell growth and OCL formation (ie, VCAM-1, IL-6, and RANKL). Given the pivotal role of XBP1s in controlling protein folding, trafficking, and secretion, it is conceivable that XBP1s also can regulate other BMSC-derived factors in addition to VCAM-1, IL-6, and RANKL. MM cell growth and OCL formation depend on a multitude of BMSCs-derived inflammatory cytokines and growth factors. The results of the present study suggest that XBP1 signaling could modulate multiple downstream targets that are implicated in MM cell adhesion, growth, and OCL formation. These pleiotropic effects of XBP1s on BMSCs explain the effectiveness of Xbp1 knockdown in reversing the pathologic behaviors of MM patient BMSCs to those of healthy donor BMSCs, and make XBP1 signaling an attractive target for disrupting the protective effects of the MM microenvironment on MM cell growth and OCL formation.
There may be distinct molecular mechanisms underlying XBP1s regulation of varied inflammatory molecules in BMSCs. For example, in the case of VCAM-1, modulating XBP1s strongly affected VCAM-1 protein expression in BMSCs, especially under stimulation by TNFα. However, we did not detect a change in VCAM-1 gene expression in BMSCs in response to altered XBP1s protein levels (data not shown). These results suggest that XBP1s modulation of VCAM-1 protein expression in BMSCs occurs most likely in a posttranscriptional mechanism yet to be determined. In contrast, XBP1s regulation of IL-6 and RANKL may involve transcriptional regulation. Overexpression of XBP1s in primary BMSCs of healthy donors elevated the baseline protein secretion of IL-6 (Figure 2B). This observation is consistent with previous findings that XBP1 regulates IL-6 production during B-cell differentiation.17,33 We also observed elevated IL-6 mRNA levels in hBMSCs (data not shown), which is consistent with previous studies showing that XBP1s can bind IL-6 promoter at the NF-κB site to regulate its gene expression.34 In addition, XBP1s overexpression stimulated RANKL gene expression by 7.0-fold, whereas it failed to affect gene expression of OPG, an osteoclastogenic inhibitor (Figure 3A), suggesting the selective regulation of RANKL gene expression by XBP1s. This result reveals for the first time a functional linkage between XBP1s and RANKL gene expression. Similar to its transcriptional control of IL-6, XBP1s may bind the RANKL promoter directly to regulate its gene expression. Further, although modulating XBP1s affected baseline protein levels of VCAM-1, IL-6, and RANKL in BMSCs, the impact of modulating XBP1s on these molecules became more prominent with stimulation by TNFα. These results suggest that XBP1s may act in synergy with TNFα signaling to regulate these molecules. Our studies are consistent with previous findings that XBP1s synergizes with inflammatory cytokine signaling (eg, TLRs), to enhance gene expression and protein secretion of IL-6 in macrophages.34 Future studies are warranted to investigate the detailed molecular mechanisms by which XBP1s acts on its own or in synergy with TNFα to regulate gene expression, protein expression, and/or protein secretion of VCAM-1, IL-6, and RANKL in BMSCs.
In summary, in the present study, we have established a novel role of XBP1s in regulating MM microenvironment support of MM cell growth and OCL formation. Our results provide the impetus for future studies to investigate therapeutic agents that target XBP1s in the MM microenvironment as a means to treat MM and its skeleton complications.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Randy Kaufman, Kezhong Zhang, Noriyoshi Kurihara, Jing Wang, Tao Chen, and Shunqian Jin for providing plasmids, cells, Abs and technical consultation, and Ms Donna Gaspich for assistance in manuscript preparation.
This work was supported by the Multiple Myeloma Research Foundation (to H.O.), a National Institutes of Health/National Institute of Dental and Craniofacial Research grant (DE017439 to H.O.), and by a Multiple Myeloma Research Foundation and VA Merit Review Grant (to G.D.R).
National Institutes of Health
Authorship
Contribution: G.X. designed and performed the experiments and wrote the manuscript; K.L. performed the experiments; J.A. and K.P. prepared the clinical samples and performed the in vivo experiments; S.L. and G.D.R. collected the clinical samples, provided critical suggestions, and edited the manuscript; and H.O. conceived and supervised the study, designed the experiments, analyzed the data, and wrote and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hongjiao Ouyang, VA Pittsburgh Healthcare System, R&D 151-U, 2W-143, University Dr C, Pittsburgh, PA 15240; e-mail: hoo1@pitt.edu.