Abstract
In response to antigens and cytokines, mouse B cells undergo class-switch recombination (CSR) and differentiate into Ig-secreting cells. T-bet, a T-box transcription factor that is up-regulated in lymphocytes by IFN-γ or IL-27, was shown to regulate CSR to IgG2a after T cell–independent B-cell stimulations. However, the molecular mechanisms controlling this process remain unclear. In the present study, we show that inactivation of the Ets-1 transcription factor results in a severe decrease in IgG2a secretion in vivo and in vitro. No T-bet expression was observed in Ets-1–deficient (Ets-1−/−) B cells stimulated with IFN-γ and lipopolysaccharide, and forced expression of T-bet in these cells rescued IgG2a secretion. Furthermore, we identified a transcriptional enhancer in the T-bet locus with an activity in B cells that relies on ETS-binding sites. After IFN-γ stimulation of Ets-1−/− B cells, activated Stat1, which forms a complex with Ets-1 in wild-type cells, no longer binds to the T-bet enhancer or promotes histone modifications at this site. These results demonstrate that Ets-1 is critical for IgG2a CSR and acts as an essential cofactor for Stat1 in the regulation of T-bet expression in B cells.
Introduction
In response to antigen-driven stimulation, naive B cells undergo class-switch recombination (CSR), a process that joins a functional IgH V(D)J exon, initially associated with the μ constant (C) region, to one of the downstream Cγ, Cϵ, or Cα genes.1 CSR is necessary for the secretion of Abs with the original antigen specificity but different immune effector functions. It requires the expression of activation-induced cytidine deaminase that activates recombination between repetitive DNA sequences called switch (S) regions located upstream of each Cγ, Cϵ, or Cα gene.2 CSR needs transcription through the S and C regions that targets C genes for recombination by the selective activation of isotype-specific intronic promoters (I) in response to antigens, cytokines, and costimulatory signals.3
In mice, IgG2a is the predominant isotype elicited during virus infection to mediate Ab-dependent cellular cytotoxicity.4 In antigen-stimulated B cells, cytokines such as IFN-γ and IL-27 trigger the activation of the Stat1 transcription factor and play a critical role in inducing germline transcription from the Iγ2a promoter that targets IgG2a to CSR.5-8 The T-box transcription factor T-bet, which was initially characterized as a transcriptional regulator of Th1 differentiation during the polarization of Th cells, has been shown to induce transcription from Iγ2a and promote switching to IgG2a.9-11 T-bet expression is increased by costimulation with either IFN-γ or IL-27 in mature B cells, and impaired IgG2a CSR in response to T cell–dependent stimulation is observed in T-bet–deficient mice.8,9,11 Although T-bet plays a major role in the regulation of IgG2a, the molecular mechanisms underlying the expression of this gene in activated B cells remains poorly understood.
Ets-1 is the prototype member of the ETS family of transcription factors, which all share a common winged helix-turn-helix DNA-binding domain.12,13 Ets-1 is involved in multiple biologic processes, such as differentiation, proliferation, and tumor progression; is highly expressed in the lymphoid lineages; and has been shown to regulate numerous genes involved in the development and function of B, T, and natural killer (NK) cells.14,15 Gene-targeted mutation of Ets-1 inhibits the polarization of CD4 Th cells into the Th1 lineage and increases the development of Th17 cells.16,17 In addition, inactivation of Ets-1 impairs the development and function of CD4 regulatory T cells.18 Therefore, Ets-1 appears to act as a critical regulator in several cytokine-mediated T-cell differentiation processes. Inactivation of Ets-1 also impairs B-cell differentiation at the early pro-B cell stage in the BM and during the maturation of splenocytes into Ab-secreting cells.19,20 John et al reported that Ets-1 regulates the activity of the Blimp1 transcription factor during plasma cell differentiation21 ; however, its role in B-cell activation and Ig isotype selection remains unknown.
In the present study, we investigated the role of Ets-1 in the response of B cells to Stat1-mediated IFN-γ receptor (IFNGR) signaling and the process of Ig isotype selection during CSR. Using Ets-1–deficient (Ets-1−/−) mice, we show that Ets-1 is required for IgG2a production in vivo and in vitro. Furthermore, we demonstrate that Ets-1 mediates the Stat1-dependent regulation of the T-bet locus in IFN-γ–activated B cells. These results identify Ets-1 as an essential factor for Stat1 function in B cells and for isotype selection during Ig CSR.
Methods
Mice
Mice carrying the inactivated ets-1 gene have been described previously.19,20 All mice used in this study, including wild-type C57Bl/6 mice, were maintained in our specific pathogen-free breeding facility (Departement d'Experimentation Animal, Hôpital Saint-Louis) and were killed for analysis at between 5 and 6 weeks of age. Rag2−/−/γc−/− chimera mice were in a C57Bl/6 background and were generated as described previously.18 In brief, fetal liver cells from Ets-1+/+ or Ets-1−/− day 18.5 embryos were injected intravenously into irradiated (3 Gy) RAG2−/−/γ-c−/− recipient mice. Chimeras were analyzed 6-12 weeks after grafting. All mouse experiments were subject to approval by the Institut Universitaire d'Hémathologie Institutional Animal Care and Use Committee.
Cell culture
Spleen cells were used for all experiments. B cells were enriched by depletion of CD43+ cells with MACS beads (Miltenyi Biotec; > 95% purity). Cells were maintained at 1.2 × 106 cells/mL in complete medium (RPMI 1640, 10% FCS, 25mM HEPES, 1mM sodium pyruvate, 2mM L-glutamine, 1 × nonessential amino acids, 5 × 10-5M 2-mercaptoethanol, and 1% antibiotics) with 25 μg/mL of lipopolysaccharide (LPS; serotype 0111:B4 from Escherichia coli; Sigma-Aldrich), 10μg/mL of anti-CD40 Abs (eBiosciences), 5 ng/mL of IL-4 (Sigma-Aldrich), 50 ng/mL of IFN-γ (PeproTech), and 100 ng/mL of IL-27 (PeproTech).
Detection of Ig
Concentrations of polyclonal IgG1, IgG3, IgG2a, and IgG2b were determined in sera and culture supernatants using isotype-specific ELISA (Southern Biotechnologies). The concentrations were determined by comparing test sample dilution series with isotype control standards (Southern Biotechnologies).
Flow cytometry
Reagents included anti–B220-APC (RA3-6B2), anti–mouse IgG2b-FITC (RMG2b-1), anti–mouse IgG3-FITC (R40-82), anti–mouse IgG1-PE (A85-1), anti–mouse IgG2a-biotin (Igh-b; 5.7; all BD Biosciences), and streptavidin-PerCy5.5 (eBiosciences). Cells were analyzed with a FACSCalibur (BD Biosciences) and FlowJo Version 8.8.6 software (TreeStar).
PCR assays
RT-PCR reactions were performed in a 50-μL reaction containing the indicated amounts of genomic DNA, 2 ng/μL of each primer, 0.2μM dNTP, 2mM MgCl2, 50mM KCl, 10mM Tris-HCl (pH 8.8), 0.1% Triton X-100, and 1 unit/50 μL of Taq polymerase. Reactions were performed for 4 minutes at 95°C; 32 cycles of 1 minute at 95°C, 1 minute at 57°C, and 1.5 minutes at 72°C; and 5 minutes at 72°C. Primers used for amplifications were: germline IgG2a: g2aF/g2aR; post-switch IgG2a: IμF/g2aR; germline IgG3: g3F/g3R; Hprt: HprtF/HprtR; and T-bet: T-betF/T-betR. The sequence-specific primers used for amplification are listed in supplemental Figure 4 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Real time RT-PCR was performed using the Fast SYBR Green Master Mix (Applied Biosystems) with the 7300 Real-Time PCR System (Applied Biosystems). Amounts of target DNA were calculated by reference to a log-linear standard regression curve. This curve was constructed from the number of cycles necessary to detect product accumulation after amplification of 2 μL of serial dilutions of a standard DNA solution. The calculation of the relative concentration was done using ABI 7300 software (Applied Biosystems).
Plasmid constructs
The minimal CNS IV region of the T-bet gene was PCR amplified with the primers CNS IVF and CNS IVF and cloned into pGL3-Promoter (Promega) to yield pGL3-CNS IV. The minimal Iγ2a promoter described by Collins et al5 was PCR amplified with the primers Iγ2aF and Iγ2aR and cloned into pGL3-basic (Promega) to yield pGL3b-Iγ2a. A site-directed mutagenesis kit (Stratagene) was used to mutate the GGA motif into CCA in the targeted ETS sites. The integrity of all regions cloned in the pGL3 vectors was established by sequencing. The PCR primers are listed in supplemental Figure 4.
Transient transfections and luciferase assays
WEHI cells were transiently transfected with 1 μg of the indicated pGL3-luciferase plasmids using electroporation with the Amaxa Cell Line Nucleofector KitV (Lonza). For each transfection, 0.5 μg of β-galactosidase–expressing plasmid (Invitrogen) was added for normalization. Transfected cells were stimulated overnight with 100 ng/mL of mouse IFN-γ, and luciferase assays were performed as described previously.18
Retroviral transduction
Transfections of MSCV-based retroviral constructs encoding T-bet–IRES-GFP (a gift of Dr L. Rogge, Pasteur Institute, Paris) or green fluorescent protein (GFP) empty vector controls were performed using Lipofectamine 2000 (Invitrogen) with DMEM medium (Invitrogen). Ecotropic retroviral supernatant was produced by cotransfecting HEK293T cells with the gag-pol-env–expressing plasmid pCL-Eco (Imgenex) and the indicated viral construct. Transfected cells were cultured in high-glucose DMEM (Invitrogen) with GlutaMAX containing 10% FBS, 100 IU/mL of penicillin, and 100 μg/mL of streptomycin (culture medium). The virus supernatants were harvested 48 hours after transfection, filtered through a 0.45-μm filter, and either frozen at −80°C or used immediately for infection. Purified B cells were activated with LPS for 12 hours and transduced by overnight culture with the indicated viral supernatant in culture medium containing 4 μg/mL of polybrene. Twenty-four hours after transduction, cells were washed and maintained in the indicated culture medium.
Coimmunoprecipitation and Western blot analyses
Transfected Cos7 cells were lysed in buffer A (1% Nonidet P-40, 50mM Tris HCl, pH 8, 150mM NaCl, 1mM PMSF, and a complete protease inhibitor mixture; Roche, Pasteur Institute, Paris Applied Sciences). Supernatants were precleared by incubation with protein-G-agarose beads (GE Healthcare) and then incubated with rabbit polyclonal anti–Ets-1 Ab (C-20; Santa Cruz Biotechnology). Immunoprecipitates were separated on SDS-PAGE gels, transferred onto nitrocellulose membranes, and blotted with a rabbit polyclonal anti-STAT1 or anti–phosphorylated-STAT1 Ab (Santa Cruz Biotechnology). For Western blot analysis, cells were lysed in buffer A and 40 μg of protein was separated on SDS-PAGE gels, transferred onto nitrocellulose membranes, and blotted with anti-actin and anti–T-bet Abs (Santa Cruz Biotechnology).
ChIP assays
ChIP assays were performed using the Imprint ChIP Kit (Sigma-Aldrich), as described previously.22 CD43-depleted cells were sorted from spleens and stimulated with 20 μg/mL of LPS and 100 ng/mL of IFN-γ for 1 hour. Formaldehyde (final concentration, 1%) was then added to cross-link proteins and DNA. The cell lysates were sonicated and immunoprecipitated with normal rabbit serum (Upstate Biotechnology), anti-STAT1 (Santa Cruz Biotechnology), and anti–Ets-1 (Santa Cruz Biotechnology). The immunoprecipitated DNA was eluted and amplified by real-time SYBR Green PCR using an ABI 7300 (Applied Biosystems). Values were normalized to serial dilution of corresponding input controls and are expressed as the fold enrichment relative to normal rabbit serum for each experiment. The sequence-specific primers used for amplification are listed in supplemental Figure 4.
Results
Secretion of IgG2a and expression of germline γ2a transcripts are inhibited in Ets-1−/− B cells
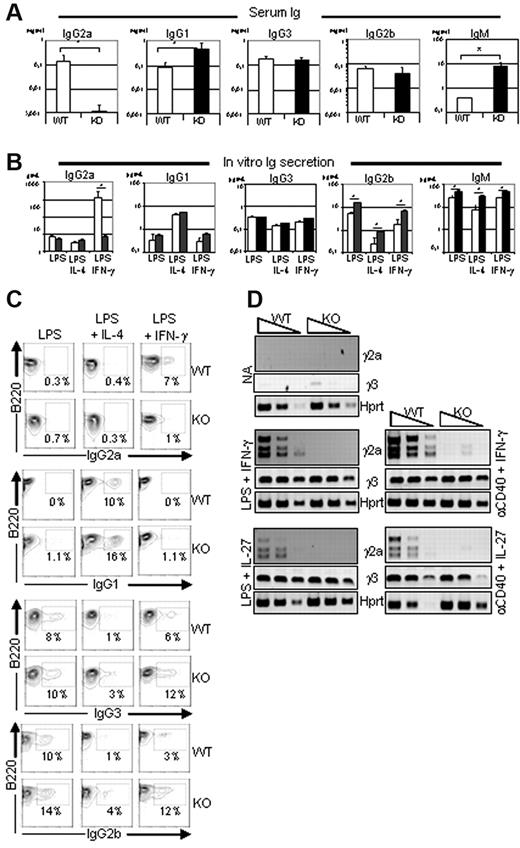
To investigate the role of Ets-1 in Ig production, we analyzed the serum of Ets-1−/− mice.19,23 As described previously,18,19 the levels of IgG1 and IgM were elevated in the sera of 5- to 6-week-old viable Ets-1−/− mice compared with controls, whereas IgG2b and IgG3 levels were normal (Figure 1A). In contrast, the serum concentration of IgG2a was severely reduced in Ets-1−/− mice. To further explore this defect, and, given that Ets-1−/− mice have a poor viability,20 we generated chimera mice by injecting fetal liver cells from Ets-1−/− or wild-type embryos into irradiated RAG2−/−/γ-c−/− recipients (hereafter referred to as Ets-1 chimeras) and used these reconstituted mice as a source of B splenocytes. B cells isolated from spleens of wild-type and Ets-1−/− chimeras secreted high levels of IgG1, IgG2b, IgG3, and IgM after in vitro activation with LPS or LPS plus IL-4 (Figure 1B). In contrast, stimulation with LPS plus IFN-γ induced very low levels of IgG2a secretion in Ets-1−/− B cells compared with controls (Figure 1B). Consistent with this, surface expression of IgG2a was not induced in Ets-1−/− B cells, but was detected on approximately 7% of controls in response to LPS plus IFN-γ (Figure 1C). Conversely, induction of IgG2b, IgG3, and IgG1 occurred at normal or slightly elevated levels on Ets-1−/− cells after stimulation with LPS or LPS plus IL-4, respectively (Figure 1C). Defective IgG2a production was observed in Ets-1−/− B cells, although these cells responded to LPS plus IFN-γ stimulation by proliferation, up-regulation of Aicda (supplemental Figures 1-2), and expression of IgG2b and IgG3 (Figure 1C). Therefore, Ets-1−/− B cells have a defect in IgG2a production both in vivo and in vitro.
Defective expression and secretion of IgG2a in Ets-1−/− B cells. (A) Serum Ig isotype levels. ELISA quantification of serum Ig in 5- to 6-week-old viable Ets-1−/− (KO, dark bar graphs) and littermate controls (WT, open bar graphs). Bar graphs represented the average of 7 mice of each genotype. Error bars indicate ±1 SD. The statistical significances were calculated with the Student t test. *P < .005. (B) In vitro Ig secretion after B-cell stimulation. ELISA quantification of IgG1, IgG2a, IgG2b, IgG3, and IgM produced in the supernatant of B splenocytes from Ets-1−/− (dark bar graphs) and wild-type (open bar graphs) chimeras cultured for 6 days. CD43− B cells were activated in medium containing 25 μg/mL of LPS, LPS plus 5 ng/mL of IL-4 (LPS-IL-4), or LPS plus 50 ng/mL of IFN-γ (LPS-IFN-γ). Bar graphs represent the average of 5 independent experiments. Error bars indicate ± 1 SD. *P < .005. (C) CD43− purified B cells from wild-type (WT) and Ets-1−/− (KO) were activated in the indicated conditions and CSR was analyzed by FACS. Numbers represent the percentages of cells falling in each gate. Results are representative of 3 independent experiments. (D) Semiquantitative RT-PCR analysis of γ2a and γ3 germline transcripts in activated B cells. RNA was isolated from B cells purified from spleens of Ets-1−/− (KO) and wild-type (WT) chimeras either untreated (NA, not activated) or after day 2 of in vitro stimulation. Cells were activated in medium containing 25 μg/mL of LPS, 10 μg/mL of anti-CD40 Abs, 5 ng/mL of IL-4, 50 ng/mL of IFN-γ, or 100 ng/mL of IL-27; 10-fold serial dilutions of template cDNA are indicated. Transcription of the Hprt gene was used as a control. Results are representative of 4 independent experiments.
Defective expression and secretion of IgG2a in Ets-1−/− B cells. (A) Serum Ig isotype levels. ELISA quantification of serum Ig in 5- to 6-week-old viable Ets-1−/− (KO, dark bar graphs) and littermate controls (WT, open bar graphs). Bar graphs represented the average of 7 mice of each genotype. Error bars indicate ±1 SD. The statistical significances were calculated with the Student t test. *P < .005. (B) In vitro Ig secretion after B-cell stimulation. ELISA quantification of IgG1, IgG2a, IgG2b, IgG3, and IgM produced in the supernatant of B splenocytes from Ets-1−/− (dark bar graphs) and wild-type (open bar graphs) chimeras cultured for 6 days. CD43− B cells were activated in medium containing 25 μg/mL of LPS, LPS plus 5 ng/mL of IL-4 (LPS-IL-4), or LPS plus 50 ng/mL of IFN-γ (LPS-IFN-γ). Bar graphs represent the average of 5 independent experiments. Error bars indicate ± 1 SD. *P < .005. (C) CD43− purified B cells from wild-type (WT) and Ets-1−/− (KO) were activated in the indicated conditions and CSR was analyzed by FACS. Numbers represent the percentages of cells falling in each gate. Results are representative of 3 independent experiments. (D) Semiquantitative RT-PCR analysis of γ2a and γ3 germline transcripts in activated B cells. RNA was isolated from B cells purified from spleens of Ets-1−/− (KO) and wild-type (WT) chimeras either untreated (NA, not activated) or after day 2 of in vitro stimulation. Cells were activated in medium containing 25 μg/mL of LPS, 10 μg/mL of anti-CD40 Abs, 5 ng/mL of IL-4, 50 ng/mL of IFN-γ, or 100 ng/mL of IL-27; 10-fold serial dilutions of template cDNA are indicated. Transcription of the Hprt gene was used as a control. Results are representative of 4 independent experiments.
We next analyzed transcripts initiated from the isotype-specific intronic promoters (I) that precede CSR during in vitro B-cell activation. Semiquantitative RT-PCR revealed similar inductions of Iγ3 germline transcripts in wild-type and Ets-1−/− B cells after LPS or anti-CD40 Ab (α-CD40) stimulation with either IFN-γ or IL-27 (Figure 1D). Similarly, PCR products corresponding to γ2a germline transcripts were readily detected in wild-type B cells in response to LPS or α-CD40 activation in IFN-γ– or IL-27–containing medium (Figure 1D). In contrast, γ2a transcripts were not detected in Ets-1−/− B cells under the same conditions. These data show that Ets-1 is required to activate transcription from the Iγ2a promoter in response to stimulations commonly used to induce switching to IgG2a.
To investigate the role of Ets-1 in the regulation of the γ2a gene, we cloned the Iγ2a promoter in the pGL3 luciferase (pGL3-Iγ2a) reporter vector and tested its function in Cos7 cells. Cotransfection of pGL3-Iγ2a with an Ets-1 expression vector did not increase the activity of Iγ2a (supplemental Figure 3). Given that the PU.1 transcription factor has been shown to regulate the IgH locus,24 including I promoters,25,26 we tested Iγ2a activity in PU.1-transfected Cos7 cells. Expression of PU.1 readily induced Iγ2a activity, whereas the addition of Ets-1 only slightly increased promoter function (supplemental Figure 3). In contrast, transfection of PU.1 with T-bet markedly enhanced the activity of Iγ2a, and, again, coexpression of the 2 factors with Ets-1 led to only a marginal increase in promoter function. These results indicate that Ets-1 is not necessary for the activity of the Iγ2a promoter in vitro, and suggest that it might play an indirect role in the regulation of the IgG2a gene.
Ets-1 interacts with Stat1 after activation of the INFGR/JAK2 pathway
Given the defective induction of IgG2a expression in response to IFN-γ and the probable indirect role of Ets-1 in this process, we investigated the function of the IFNGR signaling pathway in Ets-1 mutant B cells. Splenocytes isolated from Ets-1−/− or wild-type chimeras displayed equivalent levels of phosphorylated Stat1 after IFN-γ stimulation in vitro (supplemental Figure 4). This result indicates that IFNGR signaling remained functional in Ets-1−/− B cells and suggests that inactivation of Ets-1 affects more distal elements in the IgG2a secretion process.
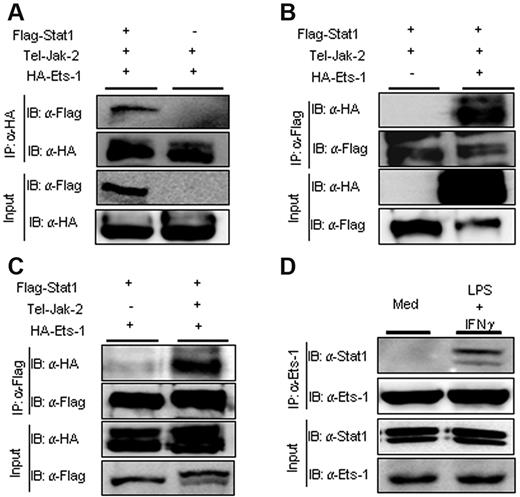
Because Ets-1 has been shown to interact with numerous transcription factors, we investigated whether activation of the Jak2/Stat1 pathway, which occurs after engagement of the IFNGR or IL-27 receptor, led to the formation of Ets-1/Stat1 complexes. Vectors expressing Flag-tagged Ets-1 or hemagglutinin (HA)–tagged Stat1 were transfected in Cos7 cells and the interaction between the 2 transcription factors was monitored by coimmunoprecipitation. A construct expressing the oncogenic TEL/JAK2 fusion protein was cotransfected with the Ets-1 and Stat1 expression vectors to give constitutive Stat1 phosphorylation. Immunoprecipitation with Ab to HA demonstrated that Ets-1 associated with Flag-Stat1 protein (Figure 2A). Similarly, Abs recognizing the Flag epitope on Stat1 readily precipitated the HA-tagged Ets-1 protein (Figure 2B). Furthermore, the complex between Ets-1 and Stat1 depended on expression of the TEL/JAK2 protein that triggered Stat1 phosphorylation (Figure 2C). To verify that Ets-1/Stat1 interactions occur under physiologic conditions, we performed coimmunoprecipitation experiments with anti–Ets-1 Abs on cellular extracts from LPS- and IFN-γ–stimulated B splenocytes. Our results show that Stat1 interacted specifically with Ets-1 after LPS and IFN-γ treatment (Figure 2D), indicating that Stat1 and Ets-1 interact upon activation of the JAK2-signaling pathway in B cells.
Ets-1 interacted physically with Stat1 after JAK2 activation in vivo. Expression plasmids of HA-tagged Ets-1 or Flag-tagged Stat1 was transfected into Cos7 cells with or without expression vector encoding the TEL-JAK2 protein. Lysates from transfected cells were immunoprecipitated (IP) with anti-HA Ab (A) or anti-Flag Ab (B-C). Total protein (30 μg) from each transfected sample was loaded as an input. Immunoblots (IB) were developed with anti-HA Ab (revealing Ets-1) or anti-Flag Ab (revealing Stat1) as indicated. (D) Lysates from CD43−–purified B splenocytes activated for 1 hour with 25 μg/mL of LPS and 100 ng/mL of IFN-γ or left in medium alone (Med) were immunoprecipitated (IP) with anti–Ets-1 Abs. Total protein (30 μg) from each sample was loaded as input controls. Immunoblots (IB) were developed with anti–Ets-1 or anti-Stat1 Abs as indicated.
Ets-1 interacted physically with Stat1 after JAK2 activation in vivo. Expression plasmids of HA-tagged Ets-1 or Flag-tagged Stat1 was transfected into Cos7 cells with or without expression vector encoding the TEL-JAK2 protein. Lysates from transfected cells were immunoprecipitated (IP) with anti-HA Ab (A) or anti-Flag Ab (B-C). Total protein (30 μg) from each transfected sample was loaded as an input. Immunoblots (IB) were developed with anti-HA Ab (revealing Ets-1) or anti-Flag Ab (revealing Stat1) as indicated. (D) Lysates from CD43−–purified B splenocytes activated for 1 hour with 25 μg/mL of LPS and 100 ng/mL of IFN-γ or left in medium alone (Med) were immunoprecipitated (IP) with anti–Ets-1 Abs. Total protein (30 μg) from each sample was loaded as input controls. Immunoblots (IB) were developed with anti–Ets-1 or anti-Stat1 Abs as indicated.
T-bet expression is impaired in Ets-1−/−–activated B cells
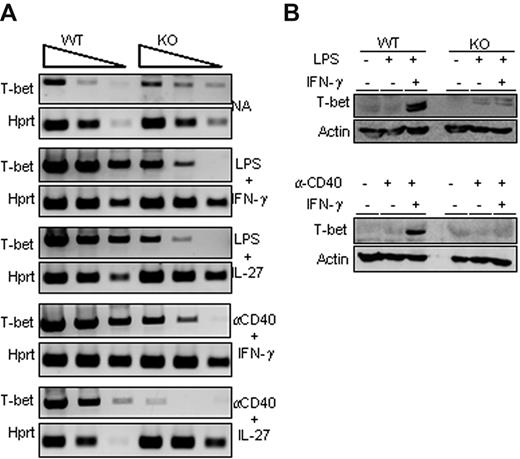
Because the T-bet transcription factor plays a major role in γ2a germline transcription and IgG2a secretion, we analyzed its expression status in activated B cells isolated from wild-type or Ets-1−/− chimeras. RT PCR analysis of wild-type B cells stimulated by LPS or α-CD40 in the presence of either IFN-γ or IL-27 showed a robust induction of T-bet transcription (Figure 3A). Strikingly, activation of Ets-1−/− B cells poorly up-regulated T-bet and did not induce expression of T-bet protein (Figure 3A-B). We further investigated T-bet expression in B cells stimulated under T cell–dependent conditions (with α-CD40 and either IFN-γ or IL-27) and, again, whereas T-bet was weakly induced in Ets-1−/− B cells, it was markedly enhanced in the wild-type control (Figure 3A). Moreover, activated wild-type B cells stimulated with LPS or α-CD40 in IFN-γ– or IL-27–containing medium expressed the T-bet protein, whereas this transcription factor was undetectable in Ets-1−/− B-splenocytes stimulated under the same conditions (Figure 3B). These data show that Ets-1 is required for activation of T-bet expression in in vitro activated B cells.
Impaired T-bet up-regulation in Ets-1−/− B cells. (A) Semiquantitative RT-PCR analysis of T-bet expression in in vitro–stimulated B cells purified from spleens of Ets-1−/− (KO) and wild-type (WT) chimera activated B cells. RNA were harvested from cells activated for 2 days under the indicated conditions or left untreated (NA, not activated). Shown are the 10-fold serial dilutions of template cDNA amplified with primers specific for the T-bet gene and Hprt gene used as a control. Results are representative of 5 independent experiments. (B) Western analyses of T-bet in activated B cells. Freshly isolated wild-type (WT) and Ets-1−/− (KO) B cells were treated for 48 hours under the following conditions: top panel, 25 μg/mL of LPS, 20 ng/mL of IFN-γ, or untreated; bottom panel, 10 μg/mL of anti-CD40 Abs, 20 ng/mL of IFN-γ, or untreated. Whole-cell lysates were subjected to Western analysis with Abs recognizing T-bet or actin as loading control. Results are representative of 3 independent experiments.
Impaired T-bet up-regulation in Ets-1−/− B cells. (A) Semiquantitative RT-PCR analysis of T-bet expression in in vitro–stimulated B cells purified from spleens of Ets-1−/− (KO) and wild-type (WT) chimera activated B cells. RNA were harvested from cells activated for 2 days under the indicated conditions or left untreated (NA, not activated). Shown are the 10-fold serial dilutions of template cDNA amplified with primers specific for the T-bet gene and Hprt gene used as a control. Results are representative of 5 independent experiments. (B) Western analyses of T-bet in activated B cells. Freshly isolated wild-type (WT) and Ets-1−/− (KO) B cells were treated for 48 hours under the following conditions: top panel, 25 μg/mL of LPS, 20 ng/mL of IFN-γ, or untreated; bottom panel, 10 μg/mL of anti-CD40 Abs, 20 ng/mL of IFN-γ, or untreated. Whole-cell lysates were subjected to Western analysis with Abs recognizing T-bet or actin as loading control. Results are representative of 3 independent experiments.
Forced expression of T-bet rescues transcription and secretion of IgG2a in in vitro stimulated Ets-1−/− B cells
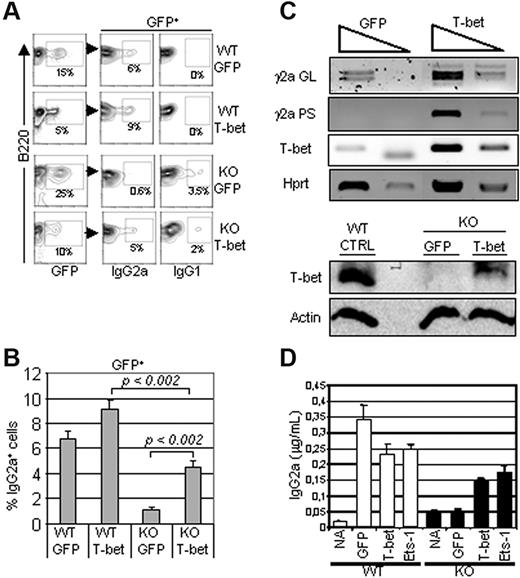
We next investigated whether T-bet down-regulation played a role in defective IgG2a production in Ets-1−/− B cells. B cells isolated from spleens of Ets-1−/− chimeras were activated with LPS and IFN-γ and transduced with a retrovirus expressing GFP alone or GFP plus T-bet. The expression levels of IgG2a and IgG1 were analyzed to investigate the efficiency and the specificity of Ig CSR among T-bet–infected B cells. FACS analysis of GFP-expressing cells revealed that expression of T-bet increased the expression of IgG2a in Ets-1−/− B cells in response to LPS and IFN-γ significantly (Figure 4A-B). However, as described previously,27 T-bet was not sufficient to inhibit switching to IgG1, and a low-level expression of this isotype remained in T-bet–transduced Ets-1−/− B cells (Figure 4A). Consistent with this, RT-PCR analysis of transduced Ets-1−/− B cells showed a marked increase in γ2a germline transcripts compared with control cells (Figure 4C top panel). In addition, IgG2a post-switch transcripts were readily induced in T-bet–expressing Ets-1−/− B cells compared with controls (Figure 4C bottom panel). Furthermore, ELISA performed on the culture supernatants revealed that the expression of T-bet partially rescued IgG2a secretion in IFN-γ–treated activated Ets-1−/− B cells (Figure 4D). Therefore, expression of T-bet can rescue expression, germline transcription, and secretion of IgG2a in Ets-1 B−/− cells.
T-bet expression rescued IgG2a production in Ets-1−/− B cells. B cells from Ets-1−/− chimeras were stimulated in vitro with 25 μg/mL of LPS and 50 ng/mL of IFN-γ and simultaneously infected with GFP or GFP-T-bet retroviruses. Seventy-two hours after stimulation, cells were analyzed by FACS and RNA was harvested. (A) FACS analysis of IgG2a and IgG1 expression on wild-type (WT) and Ets1−/− (KO) cells transduced with GFP alone (GFP) or GFP-T-bet (T-bet). The percentages of cells falling in the gates are indicated. Results are representative of 3 experiments. (B) The bar graphs show the mean percentages of IgG2a-expressing cells in 3 independent experiments as in panel A. The statistical significance was calculated with the Student t test and the P values are shown. (C) Top panel, semiquantitative RT-PCR of 10-fold serial dilutions of template cDNA from cells infected with the indicated viruses. Transcription of T-bet, γ2a germline (GL), and post-switch (PS) were analyzed. The Hprt gene was used as a control. Bottom panel, Western blot analysis of Ets-1−/− B cells transduced with the empty (GFP) or T-bet–expressing viruses (T-bet) in the same experiment as in panel A. Whole-cell lysates were subjected to Western analysis with Abs recognizing T-bet or actin as loading control. (B) B cells from the spleens of Ets-1−/− (KO) or wild-type (WT) chimeras were left untreated (NA) or stimulated with 25 μg/mL of LPS and 50 ng/mL of IFN-γ and infected with retrovirus expressing GFP alone, Ets-1-GFP (Ets-1), or T-bet-GFP (T-bet). Secretion of IgG2a was measured by ELISA after 5 days in culture. Results are representative of 3 independent experiments.
T-bet expression rescued IgG2a production in Ets-1−/− B cells. B cells from Ets-1−/− chimeras were stimulated in vitro with 25 μg/mL of LPS and 50 ng/mL of IFN-γ and simultaneously infected with GFP or GFP-T-bet retroviruses. Seventy-two hours after stimulation, cells were analyzed by FACS and RNA was harvested. (A) FACS analysis of IgG2a and IgG1 expression on wild-type (WT) and Ets1−/− (KO) cells transduced with GFP alone (GFP) or GFP-T-bet (T-bet). The percentages of cells falling in the gates are indicated. Results are representative of 3 experiments. (B) The bar graphs show the mean percentages of IgG2a-expressing cells in 3 independent experiments as in panel A. The statistical significance was calculated with the Student t test and the P values are shown. (C) Top panel, semiquantitative RT-PCR of 10-fold serial dilutions of template cDNA from cells infected with the indicated viruses. Transcription of T-bet, γ2a germline (GL), and post-switch (PS) were analyzed. The Hprt gene was used as a control. Bottom panel, Western blot analysis of Ets-1−/− B cells transduced with the empty (GFP) or T-bet–expressing viruses (T-bet) in the same experiment as in panel A. Whole-cell lysates were subjected to Western analysis with Abs recognizing T-bet or actin as loading control. (B) B cells from the spleens of Ets-1−/− (KO) or wild-type (WT) chimeras were left untreated (NA) or stimulated with 25 μg/mL of LPS and 50 ng/mL of IFN-γ and infected with retrovirus expressing GFP alone, Ets-1-GFP (Ets-1), or T-bet-GFP (T-bet). Secretion of IgG2a was measured by ELISA after 5 days in culture. Results are representative of 3 independent experiments.
Ets-1 directly regulates the T-bet enhancer
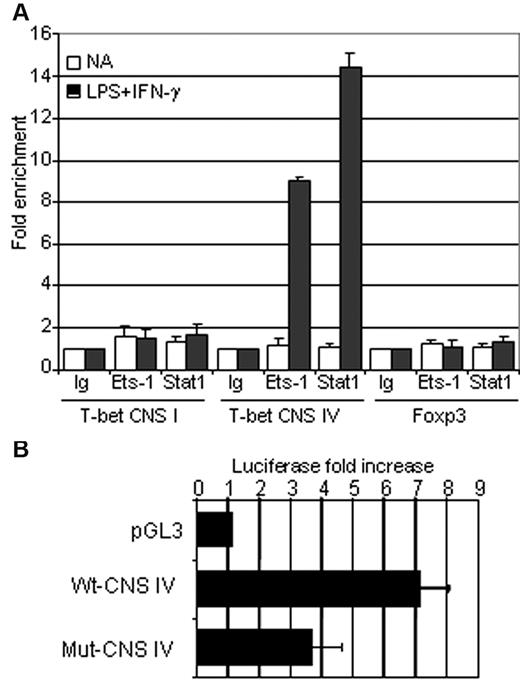
To examine the function of Ets-1 in the regulation of the T-bet locus, we sought to identify conserved nucleotide sequences (CNSs) that often correspond to transcriptional regulatory elements. Using the UCSC Genome Bioinformatics Site (http://genome.ucsc.edu), we scanned a 14-kb region located upstream of the transcription start site and found 4 CNSs (supplemental Figure 5). Analysis of mouse and human CNS regions with TFSEARCH software (www.cbrc.jp/research/db/TFSEARCH.html) revealed the presence of 2 conserved ETS-binding sites located near STAT-interacting motifs within the CNS IV sequence, but failed to identify similar motifs in CNS I, II, and III (supplemental Figure 5). The CNS IV region has been reported previously to function as an enhancer in NKT and T cells and corresponds to a DNAse-hypersensitive site with STAT-binding sequences in activated human T cells.28,29 Given the potential role of Ets-1 and Stat1 in the regulation of T-bet in activated B cells, the in vivo binding of these transcription factors was assessed by ChIP analysis in purified B splenocytes using anti–Ets-1, anti-Stat1, or irrelevant (Ig) Abs. These experiments showed significant Stat1 and Ets-1 binding to the CNS IV region in LPS- and IFN-γ–activated B cells (Figure 5A). Furthermore, although Ets-1 was expressed before and after activation (supplemental Figure 6), no binding was detected in resting cells. As expected from the computerized sequence analysis, we did not observe Stat1 or Ets-1 binding to the CNS I region that corresponds to the T-bet promoter (Figure 5A). As a control, we assessed a DNA segment of the Foxp3 gene that is not expressed in B cells and found neither Stat1 nor Ets-1 binding.
Ets-1 is involved in the regulation of the T-bet enhancer in vivo and in vitro. (A) B cells were purified from the spleens of wild-type C57Bl/6 mice and either stimulated for 1 hour with 25 μg/mL of LPS and 50 ng/mL of IFN-γ (dark histograms) or left untreated (open histograms). ChIP analysis was performed using normal rabbit serum (Ig), anti-STAT1 (Stat1), or anti–Ets-1 (Ets-1) Abs. Quantification of immunoprecipitated DNA fragments was performed by real-time PCR using primers for the promoter region (CNS I), the enhancer (CNS IV), and the irrelevant Foxp3 gene (Foxp3). Values were normalized to corresponding input control and are expressed as the fold enrichment relative to normal rabbit serum for each experiment. Results are representative of 3 individual experiments. (B) The ETS-binding sites in CNS IV are essential for T-bet enhancer activity. Luciferase activity of pGL3 empty vector (pGL3), wild-type T-bet CNS IV (Wt-CNS IV), or CNS IV carrying mutated ETS-binding sites (Mut-CNS IV). Constructs were transfected into the WEHI B-cell line and normalized luciferase activities were measured in nonactivated and overnight IFN-γ (50 ng/mL)–stimulated cells. Bar graphs show the average fold increase in luciferase activity after activation of 4 independent experiments. Errors bars indicate SDs.
Ets-1 is involved in the regulation of the T-bet enhancer in vivo and in vitro. (A) B cells were purified from the spleens of wild-type C57Bl/6 mice and either stimulated for 1 hour with 25 μg/mL of LPS and 50 ng/mL of IFN-γ (dark histograms) or left untreated (open histograms). ChIP analysis was performed using normal rabbit serum (Ig), anti-STAT1 (Stat1), or anti–Ets-1 (Ets-1) Abs. Quantification of immunoprecipitated DNA fragments was performed by real-time PCR using primers for the promoter region (CNS I), the enhancer (CNS IV), and the irrelevant Foxp3 gene (Foxp3). Values were normalized to corresponding input control and are expressed as the fold enrichment relative to normal rabbit serum for each experiment. Results are representative of 3 individual experiments. (B) The ETS-binding sites in CNS IV are essential for T-bet enhancer activity. Luciferase activity of pGL3 empty vector (pGL3), wild-type T-bet CNS IV (Wt-CNS IV), or CNS IV carrying mutated ETS-binding sites (Mut-CNS IV). Constructs were transfected into the WEHI B-cell line and normalized luciferase activities were measured in nonactivated and overnight IFN-γ (50 ng/mL)–stimulated cells. Bar graphs show the average fold increase in luciferase activity after activation of 4 independent experiments. Errors bars indicate SDs.
To investigate the functional significance of ETS-binding sites on the activity of CNS IV, this DNA region was cloned into the pGL3-promoter vector and luciferase activity was measured in transfected WEHI B cells. Whereas the native enhancer exhibited an approximately 8-fold increase in activity in response to IFN-γ treatment, mutation of the 2 ETS-binding sites reduced the enhancer function of the CNS IV region markedly (Figure 5B). These results demonstrated that Ets-1 and Stat1 bind to CNS IV element in IFN-γ–stimulated B cells and further established the critical role of ETS-binding sites for the enhancer function of this region.
Ets-1 regulates histone modifications and Stat1 binding at the T-bet enhancer
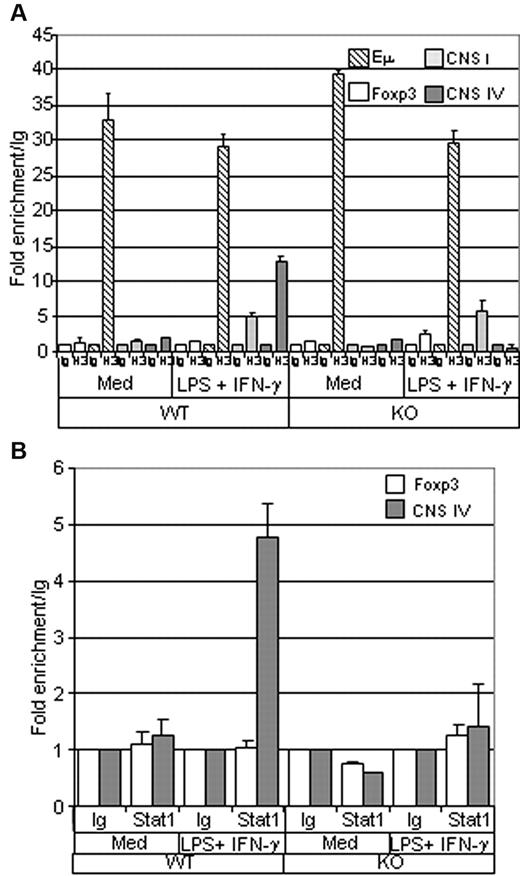
Histone modifications play an important role in the regulation of the genes activated during the differentiation of many cell types. To further investigate the function of Ets-1 in the regulation of the T-bet locus, we analyzed histone H3 acetylation (ac-H3) at the CNS IV enhancer region in B cells isolated from spleens of wild-type or Ets-1−/− chimeras. As expected, unstimulated wild-type and Ets-1−/− B cells had low levels of ac-H3 at the CNS IV region and at the Foxp3 enhancer-negative control (Figure 6A). Conversely, the transcribed IgH locus was enriched in ac-H3 at the intronic IgH enhancer (Eμ) in the cells of both genotypes. Strikingly, whereas stimulation with LPS and IFN-γ triggered efficient ac-H3 at CNS IV in wild-type B cells, H3 remained unacetylated in activated Ets-1−/− B splenocytes (Figure 6A). In contrast, H3 acetylation at the T-bet promoter (CNS I) occurred readily in activated wild-type and mutant B cells (Figure 6A). Therefore, Ets-1 is required for H3 acetylation of the T-bet enhancer in in vitro–activated B cells.
Histone H3 acetylation and Stat1 binding at the T-bet enhancer are impaired in Ets-1−/− B cells. (A) ChIP analysis of histone H3 acetylation. B cells were purified from the spleens of Ets-1−/− (KO) or wild-type (WT) chimeras and either stimulated for 1 hour with 25 μg/mL of LPS and 50 ng/mL of IFN-γ (LPS + IFN-γ) or left untreated (Med). ChIP was performed using either normal rabbit serum (Ig) or anti-acetylated histone H3 (H3) Abs. Quantification of immunoprecipitated DNA fragments was performed by real-time PCR using primers for the IgH intronic enhancer Eμ (striped), CNS I promoter region (light gray), the CNS IV enhancer (dark gray), and the irrelevant Foxp3 gene (white). Bar graphs show the values normalized to the corresponding input control and are expressed as the fold enrichment relative to normal rabbit serum. Results are representative of 3 individual experiments. (B) ChIP analysis of Stat1 binding. B cells were treated as in panel A and ChIP was performed using either normal rabbit serum (Ig) or ant-Stat1 (Stat1) Abs. Quantification of immunoprecipitated DNA fragments was performed by real-time PCR with primers for the CNS IV enhancer (dark gray) or the irrelevant Foxp3 gene (white). Bar graphs show the values normalized to corresponding input control and are expressed as the fold enrichment relative to normal rabbit serum. Results are representative of 3 individual experiments.
Histone H3 acetylation and Stat1 binding at the T-bet enhancer are impaired in Ets-1−/− B cells. (A) ChIP analysis of histone H3 acetylation. B cells were purified from the spleens of Ets-1−/− (KO) or wild-type (WT) chimeras and either stimulated for 1 hour with 25 μg/mL of LPS and 50 ng/mL of IFN-γ (LPS + IFN-γ) or left untreated (Med). ChIP was performed using either normal rabbit serum (Ig) or anti-acetylated histone H3 (H3) Abs. Quantification of immunoprecipitated DNA fragments was performed by real-time PCR using primers for the IgH intronic enhancer Eμ (striped), CNS I promoter region (light gray), the CNS IV enhancer (dark gray), and the irrelevant Foxp3 gene (white). Bar graphs show the values normalized to the corresponding input control and are expressed as the fold enrichment relative to normal rabbit serum. Results are representative of 3 individual experiments. (B) ChIP analysis of Stat1 binding. B cells were treated as in panel A and ChIP was performed using either normal rabbit serum (Ig) or ant-Stat1 (Stat1) Abs. Quantification of immunoprecipitated DNA fragments was performed by real-time PCR with primers for the CNS IV enhancer (dark gray) or the irrelevant Foxp3 gene (white). Bar graphs show the values normalized to corresponding input control and are expressed as the fold enrichment relative to normal rabbit serum. Results are representative of 3 individual experiments.
Given the key function of Stat1 in the regulation of the T-bet locus and the role of Ets-1 in the activity of its enhancer, we investigated whether the binding of Stat1 to CNS IV relied on Ets-1 by performing ChIP analysis with anti-Stat1 Abs on wild-type and Ets-1−/− splenocytes. As expected, Stat1 binding was not detected in resting B cells; however, LPS plus IFN-γ treatment triggered recruitment of Stat1 at the CNS IV region in wild-type B cells (Figure 6B). In contrast, binding of Stat1 to CNS IV enhancer was not observed in Ets1−/− B cells activated under the same conditions (Figure 6B). Therefore, Ets-1 is required for Stat1 binding to the T-bet enhancer in IFN-γ–treated activated B cells.
Discussion
Secretion of different Ig isotypes contributes greatly to the efficiency of the immune response. In the present study, we explored the molecular mechanisms that regulate CSR at the Cγ2a region and the subsequent secretion of the γ2a Ig subclass. We demonstrate herein that inactivation of the Ets-1 transcription factor leads to a severe deficit in IgG2a serum levels that is not due to impaired proliferation of activated B cells or decreased CSR per se. This defect appears to be cell autonomous, because Ets-1−/− B cells failed to produce IgG2a in response to in vitro activation in the context of proinflammatory cytokines, namely IFN-γ and IL-27. We have provided evidence that the function of the Iγ2a promoter does not require the activity of Ets-1 in reporter assays, suggesting an indirect role for this factor in the regulation of the γ2a locus. Indeed, expression of the T-bet transcription factor, which is a known regulator of IgG2a secretion, is markedly impaired in activated Ets-1−/− B cells. Furthermore, our results reveal a direct role of Ets-1 in the regulation of the T-bet transcriptional enhancer, and suggest that Ets-1 cooperates with Stat1 to induce epigenetic changes associated with T-bet expression. These results identify Ets-1 as an essential factor in the control of CSR targeting to particular Ig isotypes via transcriptional regulation of T-bet.
Nevertheless, Ets-1 deficiency may affect both T-bet–dependent and T-bet–independent regulation of the IgG2a region. Indeed, induction of γ2a germline transcripts and subsequent switching on Cγ2a occurs on various stimulations, only some of which also up regulate T-bet expression in B cells.8,11,30 Consistent with this, gene-targeted inactivation of T-bet inhibits the production of IgG2a in response to T cell–independent but not T cell–dependent stimulations.8,11 In contrast, our data show that inactivation of Ets-1 inhibits Iγ2a induction in response to both T cell–dependent (anti-CD40) and T cell–independent (LPS) stimulations. Furthermore, Ets-1−/− mice have no detectable serum IgG2a, suggesting that the major pathways controlling the secretion of this isotype require Ets-1 activity. Signaling through type I IFN is thought to induce class switching to IgG2a by IFN-γ– and T-bet–independent regulation of the Iγ2a region.11 Interestingly, the receptor for IFN-αβ triggers activation of Stat1 and Stat2. One important finding reported herein is the interaction between Stat1 and Ets-1 after JAK2 activation. The formation of an Ets-1/Stat5 complex has been described previously in T cells in response to IL-2 stimulation, indicating that Ets-1 can interact with multiple Stat factors.31 These data suggest that Ets-1 might be required for the function of several Stat proteins by acting downstream of Th1 cytokines to control IgG2a induction directly or indirectly.
We found that serum levels or in vitro induction of IgG1, IgG2b, and IgG3 were increased in the Ets-1−/− mice (Figure 1). ETS-binding sites have been found at several cis-acting regulatory sequences throughout the IgH locus.24,32 One possibility is that Ets-1 could inhibit the production of these classes of Ig directly. However, several lines of evidence suggest that this function, if it exists, would not be responsible for the defect of IgG2a in Ets-1−/− mice. First, elevated serum levels of IgG1 and IgE have been shown to occur through a systemic process that involves Ets-1−/− T cells and that can be corrected by functional regulatory T cells.18 Second, Eμ and HS1,2, which contain ETS-binding sites, are dispensable for efficient IgH CSR.33 In addition, deletion of critical regulatory sequences such as HS3b, HS4 inhibits the production of IgG isotypes that are secreted efficiently in Ets-1−/− mice. Third, forced T-bet expression rescued IgG2a production in Ets-1−/− B cells, indicating that Ets-1 per se is dispensable for this process. Therefore, Ets-1 is likely to play an indirect function in the control of IgG2a secretion. In this context, the increase in vitro production of IgG2b and IgG3 in response to LPS and IFN-γ might have been due to a compensatory mechanism in response to the inhibition of IgG2a.
In the present study, we have shown that Ets-1 controls the expression of T-bet in activated B cells through its binding to an enhancer located 13 kb upstream of the coding sequence. The transcriptional regulation of T-bet has been studied mainly in activated cytotoxic CD8 T cells and during Th1 polarization of CD4 helper cells. The cis-acting sequences that we characterized herein as having an enhancer function in B cells also act as an enhancer in T and NKT cells, where its activity depends on Stat1 and Stat4 binding after IFN-γ and IL-12 stimulation, respectively.29 Our results are consistent with these previous studies, but further demonstrate that, in B cells, Ets-1 acts as a necessary cofactor in the regulation of the T-bet enhancer by Stat proteins. Considering the conserved function of the enhancer in T and B lineages, one could speculate that Ets-1 could also be required for T-bet expression in T cells. Interestingly, gene-targeted mutation of Ets-1 has been shown to impair the development of Th1 cells and to reduce T-bet expression in activated CD4+ T cells.16 However, unlike in B cells, T-bet is still expressed in the Th1 cells of Ets-1–mutated mice, suggesting different regulation pathways for this locus in B versus T cells. Indeed, in human T cells, whereas the enhancer corresponds to an IFN-γ–dependent DNase, I–hypersensitive site, T-BET expression also occurs after TCR engagement alone.34 These data support the idea that Ets-1 is required for the enhancer function of T-bet in T and B lymphocytes, but also that a cytokine/Ets-1–independent pathway allows low T-bet expression in the T-cell lineage.
Finally, the data reported here support a model in which Ets-1 associates with Stat1 and binds to the T-bet enhancer, causing chromatin remodeling and activation of the T-bet gene, in response to B-cell activation in the context of proinflammatory cytokines. Increased T-bet expression subsequently targets the γ2a region for CSR by activating transcription of this region from the Iγ2a promoter. Our results identify a novel function for Ets-1 in the regulation of terminal B-cell differentiation and Ig-isotype selection.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank J. C. Brouet, M. Goodhardt, L. Rogge, and E. Pinaud for helpful discussions and critical reading of the manuscript.
This work was supported by a grant from La Ligue Nationale Contre le Cancer (RS 06/75-16). H.N. was supported by fellowships from the Ministere de l'Education Nationale de la Recherche et de Technologie and from La Société Française d'Hématologie. E.M. was supported by fellowships from the Fondation pour la Recherche Médicale and from the Stem Cell Network, Canéropole Ile de France.
Authorship
Contribution: H.V.N. designed and performed the experiments and analyzed the data; E.M. and K.C. designed and performed the experiments, analyzed the data, and wrote the manuscript; R.D. and R.L. performed the experiments; J.-P.F. and B.A. designed the experiments and wrote the manuscript; and J.-C.B. conceived the study, performed and oversaw the data analysis, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jean-Christophe Bories, EA 3963, Institut Universitaire d'Hématologie, Hôpital Saint Louis, 1 avenue Claude Vellefaux, 75475 Paris Cedex 10, France; e-mail: jc.bories@univ-paris-diderot.fr.