Abstract
There are many examples of transcription factor families whose members control gene expression profiles of diverse cell types. However, the mechanism by which closely related factors occupy distinct regulatory elements and impart lineage specificity is largely undefined. Here we demonstrate on a genome wide scale that the hematopoietic GATA factors GATA-1 and GATA-2 bind overlapping sets of genes, often at distinct sites, as a means to differentially regulate target gene expression and to regulate the balance between proliferation and differentiation. We also reveal that the GATA switch, which entails a chromatin occupancy exchange between GATA2 and GATA1 in the course of differentiation, operates on more than one-third of GATA1 bound genes. The switch is equally likely to lead to transcriptional activation or repression; and in general, GATA1 and GATA2 act oppositely on switch target genes. In addition, we show that genomic regions co-occupied by GATA2 and the ETS factor ETS1 are strongly enriched for regions marked by H3K4me3 and occupied by Pol II. Finally, by comparing GATA1 occupancy in erythroid cells and megakaryocytes, we find that the presence of ETS factor motifs is a major discriminator of megakaryocyte versus red cell specification.
Introduction
GATA1 and GATA2 control a large number of developmental processes by directing transcription of critical target genes and affecting the regulatory activity of other transcription regulators and cofactors.1 These 2 GATA family members have homologous zinc fingers and bind similar DNA sequences in vitro.2,3 In addition, they are essential for blood cell development, and mice lacking either factor are not viable. How these homologous proteins bind to distinct loci in chromatin to regulate different sets of target genes in various tissues, however, is unclear.
GATA2 maintains hematopoietic stem and progenitor cells; mice lacking GATA2 die around embryonic day 11.5 because of defective hematopoiesis.4 Although GATA2 is most highly expressed in proliferating progenitors, its expression persists in mast cells where it is required for terminal maturation.5 In other hematopoietic lineages, such as erythroid cells, GATA2 expression is down-regulated during differentiation, and this decrease is required for terminal maturation.6 Mutations in GATA2 are associated with chronic myeloid leukemia, and GATA2 overexpression is seen in several subtypes of acute myeloid leukemia,7 further illustrating the central role of GATA2 in the control of hematopoietic development.
In contrast, GATA1 is essential for terminal differentiation of a subset of hematopoietic cells. In maturing erythrocytes and megakaryocytes, GATA1 activates many of the functional effectors of differentiation while repressing the proliferative transcriptional program.8,9 Mice that lack GATA1 in all cells (Gata1null) die of anemia in mid-gestation.10 In contrast, mice engineered to lack a DNaseI hypersensitive site between the 2 Gata1 promoters express only marginally reduced levels of GATA1 in the erythroid lineage and thus survive beyond birth.11,12 However, these mice fail to express detectable GATA1 in megakaryocytes and show prominent defects in this lineage. In humans, mutations in GATA1 occur in both acquired malignancies (eg, acute megakaryocytic leukemia) and inherited blood disorders (eg, dyserythropoietic anemia and thrombocytopenia).13 In murine models of GATA1 dysfunction, defective GATA1 function is accompanied by overexpression of GATA2, underscoring the critical interaction between these 2 factors in normal and aberrant hematopoiesis.9,14,15
Elegant studies in models of red blood cell differentiation have shown that GATA2 binding to key cis-regulatory elements in proliferating progenitors is displaced by GATA1 as differentiation progresses. This process, known as the GATA switch, involves exchange of one GATA factor for another on erythroid gene regulatory elements.16 Targets of the GATA switch include GATA2 and Kit, which are both strongly repressed during erythroid differentiation,17,18 as well as miR-144/451, which is highly up-regulated by GATA1 during erythroid differentiation.19 The extent of the GATA switch, and whether it operates in cells other than red blood cells, however, remains unexplored.
Several studies have characterized the genome-wide occupancy of GATA2 and GATA1 on chromatin in hematopoietic progenitors, erythrocytes, and megakaryocytes.1 However, when these studies examined both GATA1 and GATA2 occupancy, they used heterogeneous populations and were unable to track the dynamic interplay between GATA2 and GATA1 on chromatin. Here, we define the GATA1 and GATA2 binding patterns in developing megakaryocytes and, for the first time, demonstrate the existence of a GATA switch on a genome-wide scale. In addition, we characterize the chromatin landscape of GATA factor bound sites and show that the ETS1 transcription factor is a key determinant of GATA site selection and is associated with the H3K4me3 chromatin mark and GATA target activation. Finally, we reveal that co-occurrence of GATA and ETS motifs appears to be a major discriminator of megakaryocyte versus erythroid gene expression.
Methods
Cell culture
G1ME cells were cultured as described20 in 1% thrombopoietin-conditioned medium and differentiated by transduction with an MIGR1 retrovirus expressing HA-GATA1.
ChIP and sequencing
ChIP was performed as described previously21 using 5 to 10 × 107 G1ME cells and antibodies against GATA2 (sc-9008, Santa Cruz Biotechnology), H3K4me3 (07-473; Millipore), H3K27me3 (07-449; Millipore), or ETS1 (sc-350; Santa Cruz Biotechnology). GATA1 ChIPs were performed using 5 × 107 MIGR1-HA-GATA1–transduced G1ME cells at 48 hours after transduction and an antibody against the HA tag (sc-7392; Santa Cruz Biotechnology). Purified ChIP DNA or pre-IP control DNA was processed as described,22 and biologic replicates were sequenced using a GAII (Illumina) and mapped to the mouse (mm9) genome. Sequencing data were deposited in the Gene Expression Omnibus under accession number GSE31331.
ChIP-Seq binding site identification
Binding sites for transcription factors and histone marks were identified using MACS23 (Version 1.3.7.1) and SICER,24 respectively, and mapped to nearest genes using the ChIP-Seq Tool Set25 or a custom Perl script. Binding site overlaps between factors were determined with BEDTools,26 and statistical significance was calculated using the genome structure correction (GSC) test.27,28 Detailed methods are available in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Gene expression profiling
Biologic triplicates of MIGR1 or MIGR1-HA-GATA1–transduced G1ME cells were sorted for GFP on a MoFlo high-speed sorter (DakoCytomation) 72 hours after transduction. RNA was isolated using the RNeasy kit (QIAGEN), processed, and hybridized to Illumina mouse arrays. Gene expression data were deposited in the Gene Expression Omnibus under accession number GSE35695.
Results
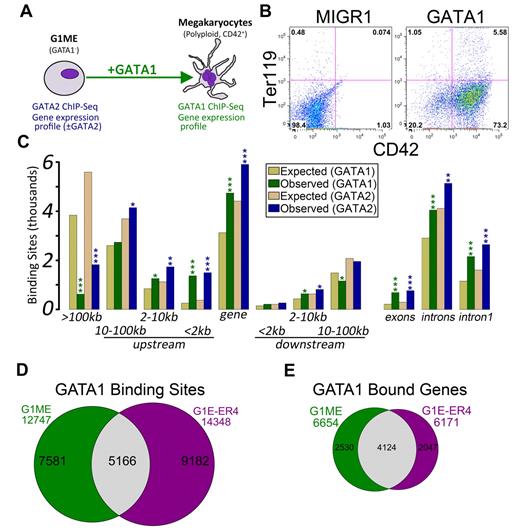
We set out to characterize the transcriptional regulatory programs controlled by GATA2 and GATA1 in a murine tissue culture model of megakaryocyte development, the Gata1-null megakaryocyte progenitor cell line, G1ME.20 Following restoration of GATA1 in the presence of thrombopoietin, these cells undergo terminal differentiation and exhibit hallmarks of mature megakaryocytes, including increased DNA content and expression of late markers, such as CD42 (Figure 1A-B; supplemental Figure 1). This approach allows us to draw conclusions that cannot be predicted from static extracts of mature erythroid cells or megakaryocytes.
Characterization of GATA-1 and GATA-2 binding sites in megakaryocytic cells. (A) Schematic of the model system used in this study, the murine erythromegakaryocytic progenitor cell line, G1ME. Gene expression profiles from GATA-2 knockdown conditions were previously published.21 (B) Flow cytometric plots showing the erythroid (Ter119) and megakaryocytic (CD42) characteristics of G1ME cells 72 hours after infection with GATA1 virus or GFP alone. (C) Distribution of GATA factor binding sites relative to genes. Gold and tan represent the average results of similar analyses performed on 10 randomly generated background BED files (expected) with identical chromosomal distribution and binding site size as the foreground sets (observed). P values from χ2 tests against the background control were all significant at P < .004. More significant values are as follows: *P < 10−10; **P < 10−50; and ***P < 10−100. (D) Venn diagram showing the intersection of GATA-1 binding sites in GATA-1–restored G1ME cells with the GATA-1 binding sites in estradiol-induced G1E-ER4 cells. A total of 40% of G1ME sites and 36% of G1E sites are bound in the opposite cell type. (E) Venn diagram showing the intersection of GATA-1–bound genes in GATA-1–restored G1ME cells with the GATA-1–bound genes in estradiol-induced G1E-ER4 cells. A total of 62% of G1ME occupied genes and 67% of G1E-ER4 occupied genes are bound in the opposite cell type.
Characterization of GATA-1 and GATA-2 binding sites in megakaryocytic cells. (A) Schematic of the model system used in this study, the murine erythromegakaryocytic progenitor cell line, G1ME. Gene expression profiles from GATA-2 knockdown conditions were previously published.21 (B) Flow cytometric plots showing the erythroid (Ter119) and megakaryocytic (CD42) characteristics of G1ME cells 72 hours after infection with GATA1 virus or GFP alone. (C) Distribution of GATA factor binding sites relative to genes. Gold and tan represent the average results of similar analyses performed on 10 randomly generated background BED files (expected) with identical chromosomal distribution and binding site size as the foreground sets (observed). P values from χ2 tests against the background control were all significant at P < .004. More significant values are as follows: *P < 10−10; **P < 10−50; and ***P < 10−100. (D) Venn diagram showing the intersection of GATA-1 binding sites in GATA-1–restored G1ME cells with the GATA-1 binding sites in estradiol-induced G1E-ER4 cells. A total of 40% of G1ME sites and 36% of G1E sites are bound in the opposite cell type. (E) Venn diagram showing the intersection of GATA-1–bound genes in GATA-1–restored G1ME cells with the GATA-1–bound genes in estradiol-induced G1E-ER4 cells. A total of 62% of G1ME occupied genes and 67% of G1E-ER4 occupied genes are bound in the opposite cell type.
We and others have recently shown that reduced expression of GATA2 in Gata1-deficient megakaryocyte progenitors leads to increased expression of myeloid lineage genes and reprogramming to functional macrophages.21,29 To extend our previous studies on GATA2 transcriptional targets, we performed ChIP followed by massively parallel sequencing (ChIP-Seq) using antibodies against GATA2 in proliferating undifferentiated G1ME cells and against GATA1 in differentiating cells. We obtained approximately 20 and 23 million mappable unique reads for GATA1 and GATA2, respectively, and identified 12 747 GATA1 binding sites and 18 149 GATA2 binding sites (Table 1). To improve the biologic power of our ChIP-Seq datasets, we integrated our findings with our previously published gene expression profiles from GATA2 knockdown21 G1ME cells and a newly generated profile from GATA1-restored G1ME cells.
Numbers of ChIP-Seq reads, binding sites, and bound genes
| IP antibody . | Sequencing reads . | Binding sites . | Bound genes . | ||||
|---|---|---|---|---|---|---|---|
| Mappable . | Unique . | A . | B . | C . | A ∩ B ∩ C . | ||
| GATA1 | 35 278 756 | 19 589 030 | 14 216 | 14 253 | 14 286 | 12 747 | 6654 |
| GATA2 | 24 006 095 | 22 661 685 | 20 982 | 20 909 | 20 895 | 18 149 | 7912 |
| ETS1 | 35 678 697 | 33 111 083 | 26 059 | 26 014 | 26 056 | 22 847 | 9005 |
| H3K4me3 | 16 069 369 | 13 486 369 | 36 911 | 36 913 | 36 982 | 36 277 | 10 749 |
| H3K27me3 | 18 861 924 | 18 230 420 | 45 436 | 45 477 | 45 475 | 42 631 | 4091 |
| INPUT | 55 715 331 | 47 452 331 | |||||
| IP antibody . | Sequencing reads . | Binding sites . | Bound genes . | ||||
|---|---|---|---|---|---|---|---|
| Mappable . | Unique . | A . | B . | C . | A ∩ B ∩ C . | ||
| GATA1 | 35 278 756 | 19 589 030 | 14 216 | 14 253 | 14 286 | 12 747 | 6654 |
| GATA2 | 24 006 095 | 22 661 685 | 20 982 | 20 909 | 20 895 | 18 149 | 7912 |
| ETS1 | 35 678 697 | 33 111 083 | 26 059 | 26 014 | 26 056 | 22 847 | 9005 |
| H3K4me3 | 16 069 369 | 13 486 369 | 36 911 | 36 913 | 36 982 | 36 277 | 10 749 |
| H3K27me3 | 18 861 924 | 18 230 420 | 45 436 | 45 477 | 45 475 | 42 631 | 4091 |
| INPUT | 55 715 331 | 47 452 331 | |||||
To gain insights into the mechanisms of GATA factor regulation in developing megakaryocytes, we first asked where occupied sites were located relative to annotated transcription start sites (TSS). In both datasets, we found a highly significant enrichment of binding sites within genes (GATA1: P = 8.2 × 10−242; GATA2: P = 5.4 × 10−146) and within the 2-kb promoter region (GATA1: P < 2.2 × 10−308; GATA2: P < 2.2 × 10−308), and a significant depletion of binding sites located more than 100 kb from the nearest TSS (GATA1: P < 2.2 × 10−308; GATA2: P < 2.2 × 10−308). Moreover, the binding sites occurred within the first intron significantly more often than expected by chance (GATA1: P = 1.9 × 10−209; GATA2: P = 2.7 × 10−162; Figure 1C). Even within the proximal promoter regions, the sites tended to localize within the 500-bp upstream of the TSS (supplemental Figure 2).
Comparison of GATA factor occupancy in megakaryocytes versus erythroid cells
Next, we sought to determine the extent of similarity between GATA1 binding in megakaryocytes and erythroid cells. Recently, Cheng et al identified 14 348 occupied segments corresponding to 6171 genes that were occupied by GATA1 during terminal red blood cell differentiation.30 We compared the locations of the erythroid GATA1 binding sites with those identified in our megakaryocytes using a base-wise overlap. Although the number of common binding sites was much greater than would be expected by chance (5166 common sites; Z-score = 349.3, P < 10−16, GSC test), the preponderance of GATA1 binding sites was specific to one of the 2 cell types (Figure 1D). A comparison of GATA1-bound genes showed that nearly 70% of genes bound by GATA1 in erythroid cells are also bound in megakaryocytes (P < 2.2 × 10−16; Figure 1E).
The genetic programs controlled by GATA1 and GATA2 are largely overlapping
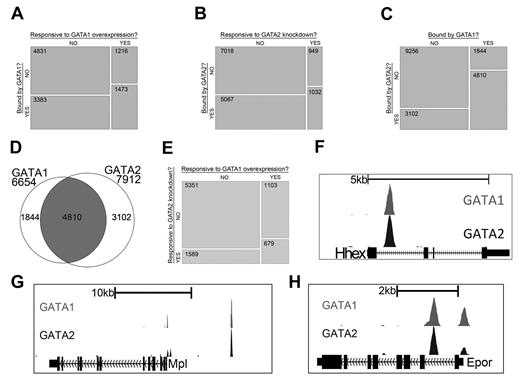
Gene expression studies have shown that GATA1 and GATA2 control overlapping sets of genes and that each factor can activate and repress target genes. By integrating gene expression data with our ChIP-Seq data, we next asked to what extent regulated genes are bound by each factor. We found that genes with significant changes in expression after restoration of GATA1 are significantly enriched for those bound by GATA1 (P < 2.2 × 10−16) and genes with significant changes in expression after knockdown of GATA2 are significantly enriched for those bound by GATA2 (P < 2.2 × 10−16; Figure 2A-B). Moreover, the list of genes that is bound by GATA1 significantly overlaps with the list of genes bound by GATA2 (P < 2.2 × 10−16), suggesting that many genes are being regulated by both factors (Figure 2C-D). We also observed that the list of genes that are differentially expressed following GATA1 restoration is significantly enriched for genes that are differentially expressed by knockdown of GATA2 (P < 2.2 × 10−16), supporting the idea that GATA1 and GATA2 directly regulate a common set of genes (Figure 2E).
GATA-1 and GATA-2 directly regulate many of the same genes during megakaryocytic development. (A) Mosaic plots showing that GATA-1–bound genes are significantly enriched for genes that are differentially expressed after restoration of GATA-1 in G1ME cells. (B) GATA-2–bound genes are significantly enriched for genes that are differentially expressed after shRNA-mediated down-regulation of GATA-2 in G1ME cells. (C) GATA-2–bound genes are significantly enriched for genes bound by GATA-1. The mosaic plots show the relative numbers of genes in each category as the area of the corresponding rectangle. If the whitespace between rows in each column are perfectly aligned, the response of the genes to the condition on the y-axis is independent of their categorization according to the condition on the x-axis. Deviations from perfect alignment represent enrichment or depletion as a condition of the y-axis category. The total number of genes in each category is shown inside the boxes. (D) Venn diagram showing the intersection of gene lists bound by GATA-1 and GATA-2. A total of 72% of GATA-1–occupied genes and 61% of GATA-2–occupied genes are bound by both factors. (E) Genes differentially expressed after shRNA-mediated down-regulation of GATA-2 are significantly enriched for genes differentially expressed after restoration of GATA-1 in G1ME cells. (F-H) GATA1 (top) and GATA2 (bottom) binding profiles at the Hhex, Mpl, and Epor loci. Peaks represent sequencing tag counts aligning to that genomic position.
GATA-1 and GATA-2 directly regulate many of the same genes during megakaryocytic development. (A) Mosaic plots showing that GATA-1–bound genes are significantly enriched for genes that are differentially expressed after restoration of GATA-1 in G1ME cells. (B) GATA-2–bound genes are significantly enriched for genes that are differentially expressed after shRNA-mediated down-regulation of GATA-2 in G1ME cells. (C) GATA-2–bound genes are significantly enriched for genes bound by GATA-1. The mosaic plots show the relative numbers of genes in each category as the area of the corresponding rectangle. If the whitespace between rows in each column are perfectly aligned, the response of the genes to the condition on the y-axis is independent of their categorization according to the condition on the x-axis. Deviations from perfect alignment represent enrichment or depletion as a condition of the y-axis category. The total number of genes in each category is shown inside the boxes. (D) Venn diagram showing the intersection of gene lists bound by GATA-1 and GATA-2. A total of 72% of GATA-1–occupied genes and 61% of GATA-2–occupied genes are bound by both factors. (E) Genes differentially expressed after shRNA-mediated down-regulation of GATA-2 are significantly enriched for genes differentially expressed after restoration of GATA-1 in G1ME cells. (F-H) GATA1 (top) and GATA2 (bottom) binding profiles at the Hhex, Mpl, and Epor loci. Peaks represent sequencing tag counts aligning to that genomic position.
To further validate our ChIP-Seq data, we examined the genomic regions surrounding several genes that are differentially expressed in our gene expression datasets and are bound by GATA1 and GATA2 in other hematopoietic cell types. For example, Hhex, which encodes a transcription factor that is critical for blood and endothelial cell development,31 is bound by GATA2 in hematopoietic progenitor cells32 and megakaryocytes.21 Our ChIP-Seq data identified a site bound by both GATA2 and GATA1 in the first intron of the Hhex gene centered between 2 suspected regulatory elements identified by bioinformatics approaches21,32 (Figure 2F). Moreover, we confirmed that GATA2 and GATA1 bind to previously identified GATA binding sites within the promoter regions of Epor33 and Mpl,34 which encode the erythropoietin and thrombopoietin receptors, respectively (Figure 2G-H). We also detected dual occupancy at the proximal promoter and −19-kb binding sites of the Sfpi1 (PU.1) locus29 (supplemental Figure 3A) as well as at the +9.5-kb switch site of the Gata2 locus35,36 (supplemental Figure 3B).
Analysis of GATA1 and GATA2 binding site locations reveals the existence of a GATA switch in megakaryocytic development
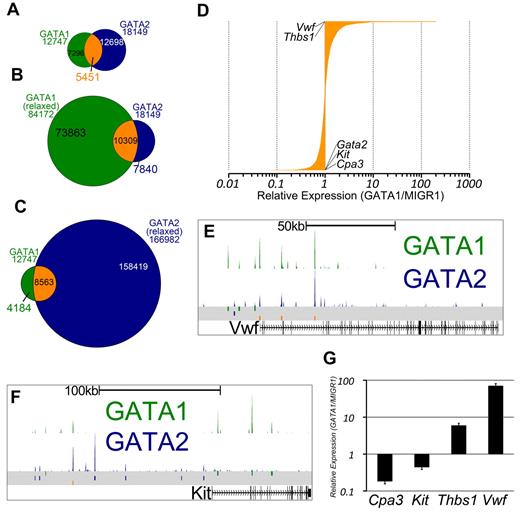
GATA1 and GATA2 participate in a chromatin occupancy switch at several critical genes during erythroid development.16 However, only a handful of GATA switch target genes have been reported, and the role of the GATA switch in lineages besides red blood cells is not clear. Thus, we asked whether, and to what extent, megakaryocytes exhibit a switch in GATA factor occupancy as has been shown for the erythroid lineage.17 We observed that nearly one-third of sites bound by GATA2 in undifferentiated G1ME cells were also occupied by GATA1 in differentiating cells (Z-score = 337.7, P < 10−16, GSC test; Figure 3A). To address the issue of whether GATA2 is truly replaced by GATA1, we reconstituted G1ME cells with GATA1, sorted for transduced cells, and performed ChIP-PCR for GATA2 and GATA1. At all genomic sites examined, we observed a substantial reduction in GATA2 occupancy following restoration of GATA1 (supplemental Figure 3C). Thus, we have identified, for the first time, a genome-wide GATA factor switch in megakaryocyte development.
The intersection of GATA1 and GATA2 binding site sets reveals GATA1-selective, GATA2-selective, and GATA switch binding sites. (A) Venn diagram showing the intersection of binding sites between the sets of GATA1 and GATA2 binding sites. A total of 43% of GATA1-bound sites and 30% of GATA2-bound sites are GATA “switch sites.” (B-C) Intersection of GATA1 and GATA2 ChIP-Seq binding sites using relaxed binding site identification parameters (P < .01) for one of the factors to allow for identification of a high confidence set of GATA1-selective or GATA2-selective binding sites. (D) Bar graph showing the expression profiles of 3321 GATA switch-bound genes. Data are depicted as relative average expression in GATA1-restored condition compared with the MIGR1 vector control condition. Two of the most strongly induced genes (Vwf and Thbs1) are indicated on the plot. Cpa3, Kit, and Gata2 are strongly repressed by GATA1 restoration and are also indicated on the plot. (E-F) GATA1 and GATA2 binding profiles at the Vwf and Kit loci as in Figure 2F through H. (G) Real-time quantitative PCR to confirm down-regulation of Cpa3 and Kit and the induction of Thbs1 and Vwf following GATA1 restoration in G1ME cells. PCRs were performed in triplicate from biologic duplicates 72 hours after infection. Bars represent the mean relative expression in GATA1-restored condition compared with the MIGR1 vector control condition; error bars represent SD.
The intersection of GATA1 and GATA2 binding site sets reveals GATA1-selective, GATA2-selective, and GATA switch binding sites. (A) Venn diagram showing the intersection of binding sites between the sets of GATA1 and GATA2 binding sites. A total of 43% of GATA1-bound sites and 30% of GATA2-bound sites are GATA “switch sites.” (B-C) Intersection of GATA1 and GATA2 ChIP-Seq binding sites using relaxed binding site identification parameters (P < .01) for one of the factors to allow for identification of a high confidence set of GATA1-selective or GATA2-selective binding sites. (D) Bar graph showing the expression profiles of 3321 GATA switch-bound genes. Data are depicted as relative average expression in GATA1-restored condition compared with the MIGR1 vector control condition. Two of the most strongly induced genes (Vwf and Thbs1) are indicated on the plot. Cpa3, Kit, and Gata2 are strongly repressed by GATA1 restoration and are also indicated on the plot. (E-F) GATA1 and GATA2 binding profiles at the Vwf and Kit loci as in Figure 2F through H. (G) Real-time quantitative PCR to confirm down-regulation of Cpa3 and Kit and the induction of Thbs1 and Vwf following GATA1 restoration in G1ME cells. PCRs were performed in triplicate from biologic duplicates 72 hours after infection. Bars represent the mean relative expression in GATA1-restored condition compared with the MIGR1 vector control condition; error bars represent SD.
Because we observed that GATA1 or GATA2 binding sites that are not at our 5451 switch sites often have tag counts for the other factor that are higher than background levels, we sought to identify a list of truly selective GATA1 or GATA2 binding sites. To that end, we used MACS to call binding sites at a 1000-fold less stringent P value cutoff and overlapped the relaxed binding site datasets with our high-confidence set of binding sites. Binding sites that are present in the GATA2 “stringent” list and not present in the GATA1 “relaxed” list are then considered to be GATA2-selective sites, as these sites are not occupied by GATA1, even under the most liberal peak-calling conditions. In this way, we have identified high-confidence sets of GATA1-selective (4184) and GATA2-selective (7840) binding sites (Figure 3B-C).
Because many of the GATA switch targets described in the literature are repressed by GATA1 (Gata2,17 Kit,18 Sfpi1,29 and Cbfa2t337 ), we asked how GATA switch genes are regulated in megakaryocytes. To address this, we assigned each GATA switch site to the nearest TSS and obtained a list of 3518 genes. We found that approximately equal numbers of GATA switch-regulated genes are up-regulated and down-regulated (Figure 3D). Moreover, we identified Vwf and Thbs1 as 2 of the genes most induced by the GATA switch and found that the GATA switch down-regulates Kit and Cpa3 in megakaryocytes (Figure 3E-G). Together, these data show that the GATA switch is prominent, robust, and directionally agnostic in megakaryopoiesis. Furthermore, occupancy by GATA2 at a specific genomic locus is neither necessary nor sufficient for subsequent occupancy by GATA1.
ETS motifs are significantly overrepresented in megakaryocytic GATA binding sites
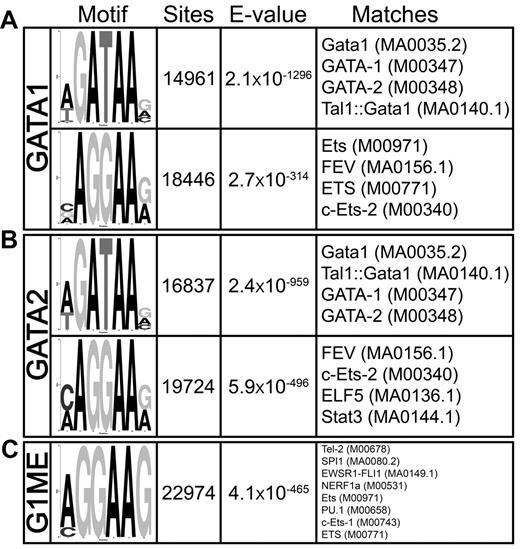
Previous studies in erythroid cells revealed that GATA1 binds to sites that also contain consensus motifs for SCL, RUNX1, LRF, KLF1, and to a lesser extent ETS factors.30,37-39 We used DREME40 to search for overrepresented sequence motifs in and around GATA2 and GATA1 binding sites in megakaryocytes. The most enriched motif was the GATA consensus [A/T]GATAA[G/A/C], with nearly 15 000 sites detected within the GATA1 occupied sites (E-value = 2.1 × 10−1296) and almost 17 000 distributed throughout the GATA2-bound regions (E = 2.4 × 10−959; Figure 4A). Intriguingly, the second-most significantly enriched motif was a core ETS factor binding motif AGGAA[G/A], and more than 18 000 and 20 000 ETS motifs were found within the GATA1 (E = 2.7 × 10−314) and GATA2 (E = 5.9 × 10−496) bound regions, respectively (Figure 4B). We also identified a statistically significant enrichment of other transcription factor motifs, including those that resemble KLF (E = 2.1 × 10−233), SMAD (E = 2 × 10−92), SCL (E = 5.6 × 10−25), and PPARG (E = 1.8 × 10−6) motifs. Of these motifs, ETS sites were by far the most prominent and significant in megakaryocytes.
GATA-1 and GATA-2 occupied genomic sites are highly enriched for GATA and ETS motifs in megakaryocytes. DREME motif identification of 500-bp sequences surrounding each of the (A) 12 747 GATA1 and (B) 18 149 GATA2 binding sites relative to a shuffled background control. (C) Most enriched motif identified by DREME in megakaryocytic GATA1 binding sites compared with the erythroid GATA1 binding sites as background sequence. The “Motif” column displays the sequence logo generated from the position-weight matrix of the overrepresented motif. The “Sites” column is a count of the number of times a sequence matching the motif appears within the collection of binding site genomic regions. Note that a motif may appear more than one time within a single binding region. “E-value” is a statistical measure of the overrepresentation of the motif; values closer to zero are more statistically significant. The “Matches” column shows the 4 best matches to the motif position-weight matrix from TOMTOM. In parentheses are the unique identifiers for the transcription factor motifs from the Jaspar or Transfac databases.
GATA-1 and GATA-2 occupied genomic sites are highly enriched for GATA and ETS motifs in megakaryocytes. DREME motif identification of 500-bp sequences surrounding each of the (A) 12 747 GATA1 and (B) 18 149 GATA2 binding sites relative to a shuffled background control. (C) Most enriched motif identified by DREME in megakaryocytic GATA1 binding sites compared with the erythroid GATA1 binding sites as background sequence. The “Motif” column displays the sequence logo generated from the position-weight matrix of the overrepresented motif. The “Sites” column is a count of the number of times a sequence matching the motif appears within the collection of binding site genomic regions. Note that a motif may appear more than one time within a single binding region. “E-value” is a statistical measure of the overrepresentation of the motif; values closer to zero are more statistically significant. The “Matches” column shows the 4 best matches to the motif position-weight matrix from TOMTOM. In parentheses are the unique identifiers for the transcription factor motifs from the Jaspar or Transfac databases.
Given that GATA1 often binds at distinct sites within erythroid cells and megakaryocytes, we suspected that different cofactors would be responsible for recruiting GATA factors to chromatin in different cell types. Thus, we asked what motifs were enriched in megakaryocytic GATA1 binding sites compared with erythroid GATA1 binding sites.30 We used DREME to identify enriched motifs, using the GATA1 bound regions from G1E-ER4 cells as the background set. We found a highly significant enrichment of an ETS motif sequence within the megakaryocytic GATA binding sites relative to the erythroid binding sites (E = 4.1 × 10−465; Figure 4C) and failed to identify any motifs enriched in erythroid GATA1 binding sites relative to the megakaryocytic sites. These findings suggest that ETS factor cooperation with GATA binding may be a key determinant of lineage specific site selection by GATA factors.
ETS1 co-occupies a portion of GATA1 and GATA2 sites
Because of the high incidence of ETS motifs recovered from our ChIP-Seq data and the striking enrichment of ETS motifs in megakaryocytic GATA1 binding sites compared with erythroid sites, we sought to identify the ETS factor that occupies these binding sites. We performed ChIP-PCR across a panel of GATA2 binding sites using antibodies against several ETS family transcription factors expressed in megakaryocytes (supplemental Figure 4). These experiments suggested that the ETS1 transcription factor binds at or near a subset of GATA2-occupied regions in G1ME cells.
During development, ETS1 is expressed in many mesodermal lineages, and it has a well-established role in lymphoid development.41 In addition, ETS1 is up-regulated during megakaryocyte development, and its overexpression in CD34+ hematopoietic progenitor cells drives megakaryopoiesis at the expense of erythropoiesis. Gel shift, luciferase reporter, and ChIP experiments in CD34+ cells point to a direct activating role for ETS1 at the GATA2 promoter.42 To investigate the relationship of these factors on chromatin, we performed ChIP-Seq for ETS1 and identified 22 847 binding sites throughout the genome (Table 1). Only 1857 (8.1%) of these binding sites overlap with GATA2 binding sites (Z-score = 36.2, P < 10−16,GSC) and 1713 (7.5%) occupy a genomic site that is later occupied by GATA1 (Z-score = 77.4, P < 10−16,GSC); 901 (3.9%) of the ETS1 occupied regions overlap with GATA switch sites (supplemental Figure 5A-B). The ETS1 binding sites are enriched for several ETS family motifs (supplemental Figure 5C) and are associated with 9005 genes (Table 1). More than 25% (2316) of the ETS1-bound genes also contain a GATA switch site (P < 2.2 × 10−16, χ2 test).
Emergent patterns of multifactor occupancy and histone methylation marks
To gain additional information about the chromatin state surrounding GATA1 and GATA2 binding sites, we performed ChIP-Seq using antibodies directed against a histone methylation mark associated with active chromatin, histone 3 trimethyl-lysine 4 (H3K4me3); a mark associated with silenced chromatin, histone 3 trimethyl-lysine 27 (H3K27me3); and reanalyzed a publicly available dataset from a ChIP-Seq that used an antibody against RNA polymerase II (Pol II) in G1ME cells.43 We used SICER to identify chromatin domains marked by the trimethylated histones and found 36 277 H3K4me3 domains with a median width of 2200 bp and 42 631 H3K27me3 domains with a median width of 4200 bp (supplemental Figure 6). In proliferating G1ME cells, approximately 4% of the genome is covered by H3K4me3 and 10% is covered by H3K27me3.
To gain insights into the patterns of GATA factor occupancy and histone methylation marks in the vicinities of the GATA2 binding sites, we used the HOMER Version 2.6 software package44 to create heatmaps. For each of the 18 149 GATA2 binding sites, we plotted sequencing tag density (ChIP-Seq signal intensity) for each ChIP-Seq dataset within 25-bp bins across a 6-kb region centered on the GATA2 binding site. This allowed us to visualize patterns of occupancy by comparing multiple adjacent heatmaps (Figure 5A). Several patterns emerged from this analysis.
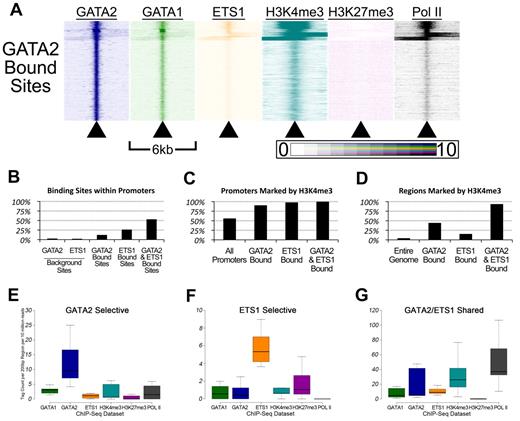
Genomic sites bound by GATA-2 and ETS-1 are marked by heavy H3K4 trimethylation and occupied by RNA Pol II. (A) Heatmaps depicting the tag density of GATA2 (dark blue), GATA1 (green), ETS1 (orange), H3K4me3 (teal), H3K27me3 (purple), and Pol II (black) across 6-kb genomic regions centered on the locations of each of the 18 149 GATA2 binding sites ordered by k-means clustering. Each row represents a 6-kb genomic region that surrounds a single GATA2 binding site. Columns represent 25-bp bins that are colored according to tag density. Bins were colored on a linear scale where those with zero tags were colored white and bins with 10 or more tags were colored most intensely. (B) Percentage of GATA2-selective, ETS1-selective, and GATA2/ETS1 shared sites that are located within gene proximal promoters, defined as the 2 kb immediately upstream of an annotated TSS. (C) Percentage of promoters bound by GATA2 and/or ETS1 that are also marked by trimethylation on lysine 4 of histone 3. (D) Percentage of non-promoter genomic regions marked by H3K4me3 that are also bound by GATA2 and/or ETS1. (E-G) Box-and-whisker plots show the tag counts for genomic regions bound by GATA2 and/or ETS1, normalized to 200-bp regions and 10 million total reads.
Genomic sites bound by GATA-2 and ETS-1 are marked by heavy H3K4 trimethylation and occupied by RNA Pol II. (A) Heatmaps depicting the tag density of GATA2 (dark blue), GATA1 (green), ETS1 (orange), H3K4me3 (teal), H3K27me3 (purple), and Pol II (black) across 6-kb genomic regions centered on the locations of each of the 18 149 GATA2 binding sites ordered by k-means clustering. Each row represents a 6-kb genomic region that surrounds a single GATA2 binding site. Columns represent 25-bp bins that are colored according to tag density. Bins were colored on a linear scale where those with zero tags were colored white and bins with 10 or more tags were colored most intensely. (B) Percentage of GATA2-selective, ETS1-selective, and GATA2/ETS1 shared sites that are located within gene proximal promoters, defined as the 2 kb immediately upstream of an annotated TSS. (C) Percentage of promoters bound by GATA2 and/or ETS1 that are also marked by trimethylation on lysine 4 of histone 3. (D) Percentage of non-promoter genomic regions marked by H3K4me3 that are also bound by GATA2 and/or ETS1. (E-G) Box-and-whisker plots show the tag counts for genomic regions bound by GATA2 and/or ETS1, normalized to 200-bp regions and 10 million total reads.
First, GATA2 bound sites are associated with regions marked by H3K4me3 and occupied by Pol II (Figure 5A). We found that 2.5% of randomly selected GATA2 background sites (and 1.9% of randomly selected ETS1 background sites) were located within 2 kb of a TSS, compared with 12% of GATA2 binding sites (P < 2.2 × 10−308), 26% of ETS1 binding sites (P < 2.2 × 10−308), and 53% of shared GATA2/ETS1 binding sites (P < 2.2 × 10−308; Figure 5B). Among these promoter-associated binding sites, 90.2% of the GATA2-bound sites were marked by H3K4me3, 97.8% of ETS1-bound sites were marked by H3K4me3, and 99.9% of shared GATA2/ETS1 binding sites at promoters were marked by H3K4me3, whereas only 55.8% of all promoters in G1ME cells were marked by H3K4me3 (Figure 5C). Moreover, among the GATA2, ETS1, and shared GATA2/ETS1 binding sites that were situated outside of proximal promoter regions, 44.1%, 15.2%, and 93.4%, respectively, were marked by H3K4me3 (Figure 5D). Overall, GATA2 and ETS1 sites were significantly enriched for H3K4me3, both within and outside of promoters. In addition, whereas only 53.3% of shared GATA2/ETS1 binding sites were located within promoters, H3K4me3 marked approximately 97% of all shared sites, suggesting that GATA2 and ETS1 associate almost exclusively at actively transcribed genes in megakaryocytes (Figure 5B and data not shown). Moreover, we observed that regions bound by both GATA2 and ETS1 had significantly higher H3K4me3 and Pol II tag counts than regions bound by only one of those factors (Figure 5E-G; supplemental Figure 7).
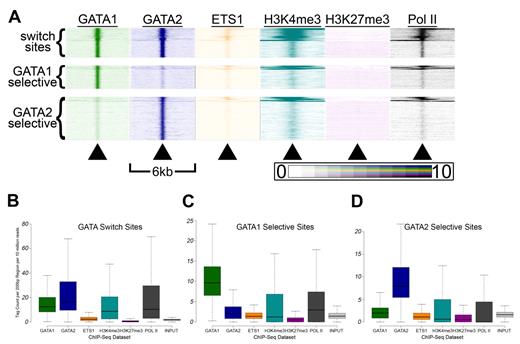
Second, we suspected that GATA switch sites may exhibit distinct patterns of histone modifications and ETS1 binding compared with single-factor-selective sites. Indeed, when we generated heatmaps for each class of GATA occupied sites, we observed distinct patterns (Figure 6A). Thus, we examined more closely the distribution of tag densities at GATA switch and GATA selective sites and found that GATA switch sites had higher mean and median GATA1 and GATA2 tag counts than sites selectively bound by only one GATA factor (Figure 6B-D). In addition, GATA switch sites had significantly higher mean and median H3K4me3 and Pol II tag counts than single-factor-selective sites (Figure 6B). These findings suggest that single-factor–selective binding sites may be enriched for false positives and that the GATA switch may be a more prevalent mode of GATA-mediated regulation than our estimates suggest. Indeed, we find that the list of genes that contain at least one GATA switch site (and no GATA1- or GATA2-selective binding sites) is significantly enriched (P = 7.7 × 10−6, χ2 test) for genes that are differentially expressed after GATA1 restoration. In contrast, the list of genes that contain at least 1 GATA1-selective binding site (and no GATA switch sites) is not significantly enriched (P = .17, χ2 test) for genes that are differentially expressed after restoration of GATA1.
GATA switch sites have higher H3K4me3 and Pol II signals than single-factor bound sites. (A) Tag density heatmaps as in Figure 5A for each of the 5451 GATA switch sites (top), the 4184 GATA1 selective binding sites (middle), and the 7840 GATA2 selective binding sites (bottom). (B-D) Box-and-whisker plots show the tag counts for genomic regions bound by GATA1 and/or GATA2, as in Figure 5E through G.
GATA switch sites have higher H3K4me3 and Pol II signals than single-factor bound sites. (A) Tag density heatmaps as in Figure 5A for each of the 5451 GATA switch sites (top), the 4184 GATA1 selective binding sites (middle), and the 7840 GATA2 selective binding sites (bottom). (B-D) Box-and-whisker plots show the tag counts for genomic regions bound by GATA1 and/or GATA2, as in Figure 5E through G.
Third, GATA2 binding sites were in chromatin regions that had low H3K27me3 tag densities. This was somewhat unexpected given that GATA2 has a known role as a direct transcriptional repressor. Thus, we examined the H3K27me3-marked domains identified by SICER for overlap with the GATA binding sites. We found that 17% of GATA2 binding sites (P = 1.63 × 10−87 vs random background) and 15.5% of GATA1 binding sites (P = 2.32 × 10−25 vs random background) were localized within H3K27me3 domains. This contrasts with our findings regarding the co-occurrences of GATA binding sites and H3K4me3 domains. Specifically, we observed that 33.4% of H3K4me3 domains contained a binding site for GATA1 (Z-score = 1625.8, P < 10−16, GSC) and/or GATA2 (Z-score = 1103.0, P < 10−16, GSC) compared with 4.7% of random background sites (P < 2.2 × 10−308), and 49.5% of GATA2 binding sites were located within an H3K4me3 domain. From these data, we conclude that, although GATA2 is more likely to be situated in a chromatin domain marked by the activating H3K4me3 mark, it does bind within H3K27me3-marked domains.
Fourth, our heatmaps suggest that regions enriched for both H3K4me3 and H3K27me3 were rarely associated with GATA binding sites (Figures 5A and 6A). Given the multipotent nature of our cell line model and the fact that bivalent chromatin domains are prominent and critical in embryonic stem cells,45,46 we asked whether bivalent domains are prevalent in megakaryocyte progenitors. We found more than 8500 H3K27me3 domains that were also marked by H3K4me3, of which 2866 (34%) overlap promoters (supplemental Table 1). However, only 176 of these bivalent promoters were occupied by ETS1, which indicates that the role of ETS1 in lineage fate decision occurs independently of bivalent chromatin marking. Next, we asked how the locations of bivalent domains, which generally mark developmentally poised chromatin regions, are related to the locations of the dynamically bound GATA switch sites. We found that approximately 11% of GATA switch sites were located within RefSeq gene promoters and less than 10% of switch sites were located within bivalent chromatin domains. In addition, we observed that only 37 (0.7%) GATA switch sites overlapped a promoter and a bivalent chromatin domain (supplemental Table 2), suggesting that the GATA switch is unlikely to control lineage-specific gene expression by regulating bivalent chromatin marking of promoters as is prevalent in pluripotent cells.
GATA1 and GATA2 orchestrate broad transcriptional programs across hematopoiesis
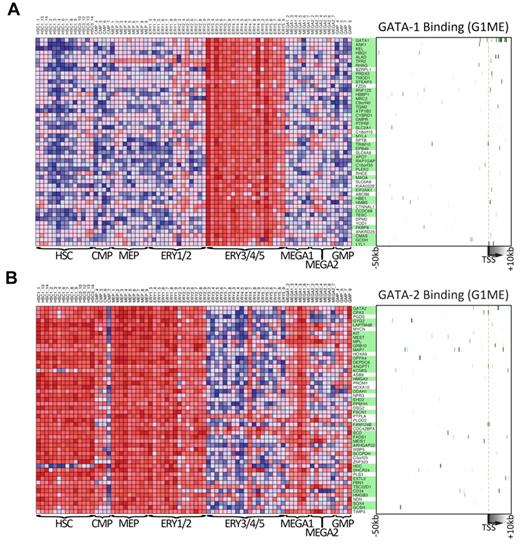
To obtain further insights about the general functions of GATA1 and GATA2 during hematopoietic development, we took advantage of data from a recently published study that profiled gene expression across 211 prospectively isolated human hematopoietic samples.47 Using the Differentiation Map Portal, we obtained the gene names and expression profiles of the 50 “nearest neighbor” genes for GATA1 and GATA2, those whose expression profiles most closely resembled the global expression pattern of the queried gene. As expected, GATA1 and its nearest neighbors are strongly expressed during erythropoiesis but expressed at relatively low levels in hematopoietic progenitors, early erythroid cells, and early megakaryocytes (Figure 7A). In contrast, GATA2 and its neighbors are highly expressed in hematopoietic progenitors as well as in erythroid and megakaryocyte progenitors but expressed at much lower levels in more mature erythroid cells (Figure 7B). These gene expression data clearly demonstrate a switch in GATA factor expression during hematopoiesis and a corresponding switch in the expression of the nearest neighbor genes. Given that these neighbors were tightly coexpressed with GATA1 and GATA2 across many lineages, we asked whether they were direct targets of GATA factors in G1ME cells. Indeed, we found that 37 of 50 GATA1 nearest neighbors (P = 7.4 × 10−9, χ2 test) and 34 of 50 GATA2 nearest neighbors (P = 1.5 × 10−4, χ2 test) are bound by their respective GATA factor (Figure 7A-B green boxes). Together, these data show that genes identified by expression profile patterns in human hematopoietic cells can provide critical information about the network of direct targets of GATA1 and GATA2. These findings put GATA1 and GATA2 at the top of the complex regulatory hierarchy controlling hematopoietic differentiation.
Genes that are expressed similarly to either GATA1 or GATA2 across human hematopoiesis are often bound by that factor in G1ME cells. (A) The expression levels across primary human hematopoietic cell types for the 50 GATA-1 “nearest neighbor” genes from DMAP47 are shown in the heatmap on the left. On the right, a heatmap depicts GATA-1 binding at each gene in G1ME cells, where intensity of green represents tag density at that position relative to the model gene depicted below the heatmap. In the center, gene names highlighted in green have binding sites within this potential regulatory region (−50 kb to +10 kb relative to the TSS) or were assigned a binding site by the nearest TSS (within 50 kb) criteria. (B) Heatmaps of GATA-2 “nearest neighbor” genes showing gene expression in primary human hematopoietic (left) and GATA-2 binding sites in G1ME cells (right) as in panel A.
Genes that are expressed similarly to either GATA1 or GATA2 across human hematopoiesis are often bound by that factor in G1ME cells. (A) The expression levels across primary human hematopoietic cell types for the 50 GATA-1 “nearest neighbor” genes from DMAP47 are shown in the heatmap on the left. On the right, a heatmap depicts GATA-1 binding at each gene in G1ME cells, where intensity of green represents tag density at that position relative to the model gene depicted below the heatmap. In the center, gene names highlighted in green have binding sites within this potential regulatory region (−50 kb to +10 kb relative to the TSS) or were assigned a binding site by the nearest TSS (within 50 kb) criteria. (B) Heatmaps of GATA-2 “nearest neighbor” genes showing gene expression in primary human hematopoietic (left) and GATA-2 binding sites in G1ME cells (right) as in panel A.
Discussion
DNA sequences containing a match to the GATA binding motif are prevalent throughout the genome. However, not all GATA motifs are bound by GATA2 or GATA1 during development, and little information exists about how the binding sites are chosen. In erythroid cells, several studies have shed some light on the requirements for GATA1 occupancy, but a full list of binding determinants has not yet been identified. In addition, these studies did not explore the genome-wide dynamic binding patterns of GATA2 and GATA1 across a developmental timeline. Here, we describe the full complement of sites bound by GATA2 or GATA1 in 2 stages of megakaryocyte maturation. These data have allowed us to identify thousands of genomic sites that are targets of the GATA switch as well as thousands of other sites that are bound selectively by GATA2 or GATA1 during megakaryocyte differentiation.
Our new GATA1 binding dataset from megakaryocytes has allowed us to answer questions about the similarities and differences in GATA1 occupancy patterns between 2 closely related lineages. Interestingly, we found that GATA1 binds to many of the same genes in erythroid and megakaryocytic cells, although it uses different binding sites in the 2 lineages. This finding provides new insights into how transcription factors can have qualitatively different effects on the same genes in different lineages. Because we identified no differences between the GATA motifs found in GATA1-bound sites in erythroid versus megakaryocytic cells, we suspected that cofactors associated with GATA1 play an instructive role in determining lineage-selective GATA1 occupancy. Consistent with the established role of ETS family proteins in megakaryopoiesis, we identified ETS motifs substantially more frequently in megakaryocytic GATA binding sites than in erythroid GATA sites. The ETS family members GABPα and FLI1 have complementary early and late roles, respectively, in megakaryopoiesis, and they can potentiate GATA1/FOG1-mediated transcriptional activation.48 Somewhat surprisingly, we were unable to confirm GABPα or FLI1 occupancy by ChIP-PCR in G1ME cells at previously characterized sites and instead found that ETS1 co-occupies many GATA2 sites at critical megakaryocytic genes. Previous studies in human CD34 cells have established a role for ETS1 in megakaryocyte differentiation and show that ETS1 promotes megakaryopoiesis at the expense of erythropoiesis.42 More recent work demonstrates that ETS1 is a target of miR-155 and implicates this regulatory axis as a potential player in the erythromegakaryocytic lineage fate choice.49 Together, these data led us to perform ChIP-Seq for ETS1 in megakaryocyte progenitors to determine the full range of interplay between GATA factors and ETS1.
Despite the relatively low levels of coincident binding between GATA2 and ETS1, there is a strong relationship between the 2 factors in megakaryocytes. In particular, ETS1 binding overlaps with GATA2 occupancy at a subset of sites. These co-occupied sites have very strong H3K4me3 and Pol II signals. In general, Pol II ChIP-Seq signals strongly correlate with gene expression levels. Thus, we propose that our data point to the existence of 2 distinct types of GATA binding sites: (1) those that are coincidently occupied by GATA2 and ETS1 and are highly transcriptionally active; and (2) those that are occupied by GATA2 and not ETS1 and may be active at a low level, poised, or repressed through a mechanism that is not likely to involve H3K27me3. Furthermore, we suspect that ETS1 also binds 2 distinct classes of sites in megakaryocytes: (1) one class that is co-occupied by GATA2 (or GATA1 during differentiation) and highly transcriptionally active; and (2) the other class is bound only by ETS1 (and not by GATA factors) and may represent sites that are (1) transcriptionally activated by ETS1 in earlier hematopoietic lineages, (2) transcriptionally activated later during megakaryocyte development by ETS1, (3) actively repressed by ETS1 in G1ME cells, and/or (4) not functionally relevant binding sites.
Bivalent chromatin domains
One mechanism through which differentiating cells have attained the ability to rapidly alter transcription after making a lineage choice is suspected to involve bivalent marking of chromatin.45,46 In this situation, histones near the promoters of developmentally dynamic genes are modified by 2 opposing covalent modifications: H3K4me3 (a mark of transcriptional activation) and H3K27me3 (a mark of transcriptional repression). In embryonic stem cells, PRC and MLL complexes maintain the bivalent mark and the presence of both modifications is critical for maintaining pluripotency. In hematopoietic stem cells, bivalent chromatin often marks hematopoietic regulator genes and a majority of bivalent domains resolve to a single trimethylation mark at the onset of differentiation. However, some bivalent domains persist beyond the hematopoietic stem cell stage, and the full extent to which bivalent chromatin domains act in maturing multipotent progenitor cells of the hematopoietic system remains unclear. Here we demonstrate that bivalent domains are fairly common in G1ME cells but that these regions are strikingly underrepresented at key GATA switch sites. These results suggest that bivalent chromatin domains are not directly controlled by GATA factors or that GATA-regulated bivalent domains are resolved before the initiation of terminal differentiation.
Factor switching and hematopoietic lineage commitment
Our study reveals that the GATA switch occurs in both erythroid cells and megakaryocytes. However, it is important to note that expression of Gata2, a key target of the GATA switch, is different within the 2 lineages. Gata2 expression is rapidly down-regulated in G1E-ER4 cells induced to differentiate to erythroid cells: Gata2 mRNA declines by more than 100-fold within 3 hours and is undetectable by 24 hours.8 In contrast, in G1ME cells Gata2 mRNA is reduced 2.4-fold by 42 hours and 3.7-fold by 72 hours after GATA1 expression, respectively.29 These differences are recapitulated in primary human cells.50 Thus, despite the exchange of GATA factors on the −77, −1.8, and +9.5 sites of the GATA2 locus, GATA2 expression is not diminished to the same degree. This difference may be a consequence of differential cofactor recruitment or different kinetics of GATA1 displacement of GATA2. Nevertheless, we predict that the increased level of GATA2 in megakaryocytes directly contributes to specification and the differential gene expression program of the 2 closely related cell types.
The G1ME system offers the distinct advantage of rapid induction of megakaryocyte maturation through GATA1 complementation within an arrested progenitor cell and, thus, provides a simple system to study the complex actions of GATA1 in megakaryopoiesis. However, complete absence of GATA1 in this MEP-like cell may not be entirely physiologically accurate. Consequently, GATA2 levels may be artificially high in uninfected cells, and GATA1 may be artificially high in transduced cells; these potentially higher protein levels may lead to increased occupancy at otherwise weak binding sites or promiscuous binding at otherwise unoccupied sites. Future studies to precisely define how GATA1 and GATA2 select their binding sites will provide additional insights into lineage selection and hematopoietic cell differentiation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jindan Yu for critical review of the manuscript.
This work was supported in part by the National Institutes of Health (NIH, P50 award GM081892 to the Chicago Center for Systems Biology), National Cancer Institute (awards CA101774 to J.D.C. and CA143869 to the Physical Sciences-Oncology Center at Northwestern University), the Samuel Waxman Cancer Research Foundation (J.D.C.), NIH (T32-CA080621, L.C.D.), Malkin Family Scholar Awards (L.C.D. and T.M.C.), a National Science Foundation Graduate Research Fellowship (T.M.C.), and the Chicago Biomedical Consortium (Scholar Award), supported by the Searle Funds at the Chicago Community Trust (L.C.D.).
A portion of the data analysis was performed on the QUEST High Performance Computing System at Northwestern University.
National Institutes of Health
Authorship
Contribution: L.C.D. and T.M.C. generated ChIP-Seq datasets; L.C.D. analyzed data and interpreted results; C.D.B., K.P.W., and J.D.C. assisted in analyzing data and interpreting results; J.D.C. supervised the study; and L.C.D. and J.D.C. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John Crispino, Northwestern University, 303 East Superior St, Lurie 5-113, Chicago, IL 60611; e-mail: j-crispino@northwestern.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal