Abstract
The t(8;21)(q22;q22) is common in adult acute myeloid leukemia (AML). The RUNX1-ETO fusion protein that is expressed by this translocation is poorly leukemogenic and requires additional mutations for transformation. Loss of sex chromosome (LOS) is frequently observed in t(8;21) AML. In the present study, to evaluate whether LOS cooperates with t(8;21) in leukemogenesis, we first used a retroviral transduction/transplantation model to express RUNX1-ETO in hematopoietic cells from XO mice. The low frequency of leukemia in these mice suggests that the potentially critical gene for suppression of t(8;21) leukemia in humans is not conserved on mouse sex chromosomes. The gene encoding the GM-CSF receptor α subunit (CSF2RA) is located on X and Y chromosomes in humans but on chromosome 19 in mice. GM-CSF promotes myeloid cell survival, proliferation, and differentiation. To determine whether GM-CSF signaling affects RUNX1-ETO leukemogenesis, hematopoietic stem/progenitor cells that lack GM-CSF signaling were used to express RUNX1-ETO and transplanted into lethally irradiated mice, and a high penetrance of AML was observed in recipients. Furthermore, GM-CSF reduced the replating ability of RUNX1-ETO–expressing cells. These results suggest a possible tumor-suppressor role of GM-CSF in RUNX1-ETO leukemia. Loss of the CSF2RA gene may be a critical mutation explaining the high incidence of LOS associated with the t(8;21)(q22;q22) translocation.
Introduction
The t(8;21)(q22;q22) is one of the most common in adult de novo acute myeloid leukemia (AML), associated with nearly 40% of cases of FAB-M2 AML cases and in 8%-20% of AML cases, depending on the genetic background and geographic location of the population.1 The t(8;21)(q22;q22) translocation results from the fusion of the RUNX1 (AML1, CBFA2) gene, located on chromosome 21, with the ETO (MTG8, RUNX1T1) gene, located on chromosome 8.
Mouse models of the 8;21 translocation have been valuable for understanding the mechanisms of transformation in leukemia. RUNX1-ETO knock-in mice2,3 died at 12.5 days of embryogenesis and failed to establish definitive hematopoiesis. These results, similar to those seen in Runx1−/− mice,4,5 suggest that RUNX1-ETO has an inhibitory effect on the normal RUNX1 protein. To circumvent this early embryonic lethality, conditional and lineage-specific expression models of RUNX1-ETO have been established.6-10 Except for mice with Sca-1 locus-directed RUNX1-ETO expression, which developed a myeloproliferative disorder, all mice remained healthy with normal hematopoiesis during their lifespans. However, treatment with the DNA-alkylating mutagen N-ethyl-N-nitrosourea induced AML in MRP8-RUNX1-ETO transgenic mice7 and myeloid sarcoma in RUNX1-ETO conditional knock-in mice.9 Moreover, expression of RUNX1-ETO in human CD34+ cells induced expansion of stem cells in vitro and increased their survival,11 but xenograft models failed to develop leukemia.12 These results make clear the importance of additional mutations for the transformation of cells harboring the t(8;21)(q22;q22) translocation, a conclusion supported by the clinical observation that the RUNX1-ETO transcript is detectable in individuals without AML.13
Activating mutations of receptor tyrosine kinases are probably the best studied complementing mutations in t(8;21) AML. Experimental models have demonstrated the significance of TEL-PDGRFβR,14 FLT3-ITD,15 c-Kit(N822K),16 and NRAS17 in RUNX1-ETO leukemia. Cell-cycle arrest is observed on expression of RUNX1-ETO. Although the cell-cycle inhibitors p15Ink4b and p16Ink4a are not involved in leukemogenic transformation,18 p21Waf affects leukemogenic transformation by RUNX1-ETO directly.19 Dysfunction of ICSBP20 and overexpression of WT121 have also been found to favor the development of RUNX1-ETO leukemia.
The 8;21 translocation is often associated with additional cytogenetic abnormalities. Only approximately 30% of patients present with t(8;21)(q22;q22) as a sole abnormality. Loss of sex chromosome (LOS) is by far the most frequent abnormality found in association with t(8;21)(q22;q22) leukemia, accounting for 32%-59% of cases22-24 (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), in contrast to the inv(16)/t(16;16) and other types of AML, in which LOS occurs in less than 5% of patients. X and Y chromosomes are highly divergent in structure, except for short segments of homology called pseudoautosomal regions (PARs). However, in t(8;21) patients, X and Y chromosome loss have been observed at similar frequencies (supplemental Table 1). Therefore, we hypothesized that certain genes located on the PARs of human sex chromosomes may inhibit leukemogenesis by RUNX1-ETO.
In the present study, we first examined the effect of LOS on RUNX1-ETO–induced leukemogenesis using a retrovirus-mediated RUNX1-ETO expression and transplantation assay on XO hematopoietic cells from the offspring of XPafY* mice.25 Mice transplanted with XO and XY cells showed the same incidence of leukemia, suggesting that the loss of one mouse sex chromosome is not a cooperative factor. Comparing the genes located in the PARs of human and mouse sex chromosomes, we observed that CSF2RA, the gene for the GM-CSF receptor α subunit (CD116), is located on human PARs but on mouse chromosome 19. GM-CSF is a key cytokine in the control of the proliferation, survival, and differentiation of myeloid cells. Previous studies have suggested that t(8;21)(q22;q22) blasts are hyporesponsive to GM-CSF.26,27 The expression of CSF2RA mRNA was specifically down-regulated in t(8;21)(q22;q22) patients,28 suggesting that deficiency of GM-CSF signaling may be a contributing factor to t(8;21)–induced leukemia. Accordingly, we investigated the significance of deficiency of GM-CSF signaling in RUNX1-ETO leukemia using hematopoietic cells from IL-3/IL-5/GM-CSF receptor common β subunit knock-out (βc−/−) mice, which completely lack GM-CSF receptor–induced signaling.29 In transplantation experiments AML occurred at high frequency in βc−/− cells expressing RUNX1-ETO. Moreover, the activation of GM-CSF signaling in vitro affected the viability of RUNX1-ETO–expressing cells. These results suggest a previously unacknowledged negative role of GM-CSF signaling in RUNX1-ETO leukemia, which would provide a molecular basis for the high incidence of LOS in t(8;21)(q22;q22) AML.
Methods
Animals
Retroviral transduction and hematopoietic cell transplantation
For details on retroviral transduction and hematopoietic cell transplantation, see supplemental Methods.
Statistical analysis
For details on the statistical analysis, see supplemental Methods.
Flow cytometric analysis
For details on the flow cytometric analysis, see supplemental Methods.
Cytology and histology
For details on cytology and histology, see supplemental Methods.
Methylcellulose colony-forming assay
For analysis of βc−/− cells, 2 × 104 BM cells were plated in 35-mm dishes in 1 mL of base methylcellulose (M3134) medium (StemCell Technologies) prepared according to the instructions of the manufacturer. Recombinant mouse (rm) IL-3, rmIL-5, or rmGM-CSF (PeproTech) was added at 10 ng/mL.
For replating assays, lineage-negative BM cells from C57B6/J mice were separated using magnetic beads (Miltenyi Biotec). Cells were cultured in StemSpan SFEM (StemCell Technologies) 15% FBS, 10 ng/mL of rmIL-3, 50 ng/mL of rmSCF, 50 ng/mL of rmFLT3L, 1% penicillin-streptomycin, and 1% glutamine at a concentration of 2 × 105 cells/mL, and prestimulated overnight. Retroviral vectors MIP and RE are similar to Mig and MigRE except that the enhanced green fluorescent protein (EGFP) gene is replaced with the puromycin N-acetyl transferase gene (Figure 1A). For retroviral transduction, viral supernatants from MIP or RE vectors were added at 20% vol/vol with 4 μg/mL of polybrene to the cells, and spinoculated. The procedure was repeated on the next day. The transduction efficiency of control MigR1-infected cells was 40%-60%. MIP- and RE-transduced cells were collected in IMDM with 2% FBS, and 2 × 104 cells were plated in 35-mm plates in 1 mL of M3434 medium (StemCell Technologies) with 1 μg/mL of puromycin and 1% penicillin-streptomycin in duplicate. When applicable, 10 ng/mL of rmGM-CSF or rmIL-5 was added. Cells were kept at 37°C and 4% CO2 in a humidified chamber. After 7 days, colonies were counted and replated following the procedure described for the initial plating.
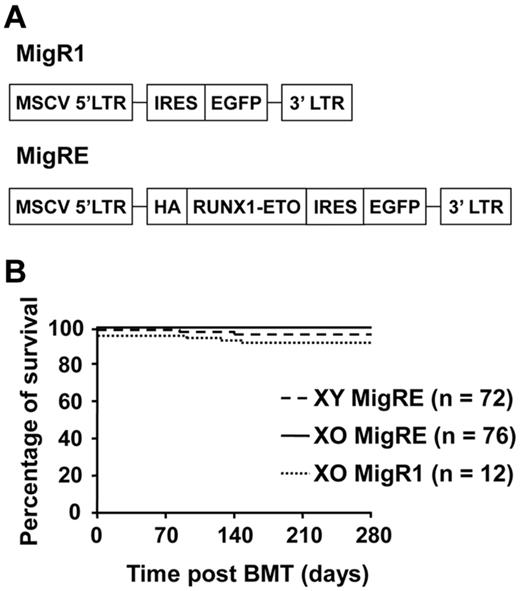
Loss of 1 X chromosome in mice does not promote RUNX1-ETO–induced leukemia. (A) Schematic representation of retroviral constructs. MSCV indicates murine stem cell virus; LTR, long-terminal repeat; HA, hemagglutinin tag; and IRES, internal ribosome entry site. (B) Kaplan-Meier survival curve of animals transplanted with MigRE-transduced XO or XY cells. Pooled results from 6 different transplantation experiments are shown.
Loss of 1 X chromosome in mice does not promote RUNX1-ETO–induced leukemia. (A) Schematic representation of retroviral constructs. MSCV indicates murine stem cell virus; LTR, long-terminal repeat; HA, hemagglutinin tag; and IRES, internal ribosome entry site. (B) Kaplan-Meier survival curve of animals transplanted with MigRE-transduced XO or XY cells. Pooled results from 6 different transplantation experiments are shown.
CSF2RA expression in Kasumi-1 cell line
For details on CSF2RA expression in the Kasumi-1 cell line, see supplemental Methods.
Immunoblotting
For analysis of RUNX1-ETO expression in leukemic animals, protein was extracted from frozen pellets of spleen cells directly on sample buffer (62.5mM Tris-HCl, pH 6.8, 2% wt/vol SDS, 10% glycerol, 50mM DTT, and 0.01% wt/vol bromophenol blue). Samples were separated by electrophoresis and transferred into nitrocellulose membranes (Amersham Hybond-ECL; GE Healthcare), blocked, and stained overnight with HA.11 Ab (1:1000; clone 16B12; Covance) in 5% wt/vol skim milk in PBST 0.1% vol/vol Tween-20. Membranes were washed and incubated with ECL anti–mouse IgG HRP (1:1000; GE Healthcare) for 4 hours before chemiluminescence reaction and exposure to autoradiographic films.
For analysis of phosphoproteins, lineage-negative cells were prepared as described in “Methyl cellulose colon-forming assay.” After retroviral transduction, cells were incubated with 2 μg/mL of puromycin for 48 hours, washed, and starved in IMDM at 37°C and 4% CO2 for the indicated times. Stimulations were performed with 50 ng/mL of rmGM-CSF with or without 20% FBS. After the indicated incubation times, cells were collected, washed, and lysed with RIPA buffer (50mM Tris-HCl, pH 7.6, 150mM NaCl, 1% vol/vol Triton X-100, 0.5% wt/vol sodium deoxycholate, and 1mM EDTA) and added to a 0.2mM PMSF, protease and phosphatase inhibitor cocktail (Roche) and 1mM DTT. Lysates were mixed by pipetting with sample buffer for loading and storage. Lysates from approximately 1 × 105 cells were loaded per lane. Samples were separated in 10% polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were blocked and stained overnight in TBST 0.1% vol/vol Tween-20 solution with 5% wt/vol BSA with the following Ab dilutions: 1:1000 of pStat5 (Tyr694; D47E7) or Stat5 (3H7), 1:2000 of pAkt (Ser473; D9E) or Akt, and 1:8000 of pErk1/2 (Thr202/Tyr204)(13.14.4E) or Erk1/2 (MK12; BD Pharmingen). All Abs were from Cell Signaling Technology unless noted otherwise. Membranes were washed and incubated with ECL anti–mouse or anti–rabbit IgG HRP (1:2000) for 2 hours before chemiluminescence reaction and exposure to autoradiographic films. Densitometric analysis was performed using CS Analyzer Version 3 software (Atto).
Results
Loss of one mouse sex chromosome does not cooperate with RUNX1-ETO to induce leukemia
Previous studies have shown that the expression of the t(8;21) fusion product RUNX1-ETO is involved in, but not sufficient for, induction of leukemia in mouse models. To evaluate the significance of LOS in t(8;21)–related leukemogenesis, hematopoietic cells from XO mice25 were used in retrovirus-mediated RUNX1-ETO expression and transplantation assays. Hematopoietic cells collected from XO mice or control XY littermates were transduced with control (MigR1) or RUNX1-ETO (MigRE) retrovirus (Figure 1A). During the observation period of more than 9 months, only 3 of 76 mice transplanted with XO cells expressing RUNX1-ETO developed AML. AML was also observed in 3 of 72 mice transplanted with XY cells expressing RUNX1-ETO, but in none of the XO cells transduced with MigR1 (Figure 1B). These results demonstrate that in mice, the loss of one sex chromosome does not provide additional advantage for the development of RUNX1-ETO–related leukemia.
The CSF2RA gene is located on human, but not mouse, sex chromosomes
Our experiments using mouse XO hematopoietic cells did not support the hypothesis that LOS is a cooperating mutation in RUNX1-ETO–induced leukemia. We next shifted our attention to the difference between human and mouse sex chromosomes. Because LOS in t(8;21)(q22;q22) patients involves both X and Y chromosomes at similar frequencies (supplemental Table 1), the gene(s) affecting RUNX1-ETO leukemia may be shared between X and Y chromosomes. The PARs are regions of homology that contain genes in common between sex chromosomes (supplemental Figure 1). CSF2RA is located on PAR1 of human X and Y chromosomes but on chromosome 19 in mice. CSF2RA encodes the α subunit of the GM-CSF receptor. Signaling from the GM-CSF cytokine promotes survival, proliferation, and differentiation in hematopoietic cells.30 Previous studies have shown that t(8;21)(q22;q22) blasts are hyporesponsive to GM-CSF.26,27 Jahns-Streubel et al observed the highest spontaneous proliferation but the lowest response to GM-CSF in cells from AML patients belonging to the favorable karyotype group, which includes t(8;21)(q22;q22).26 In addition, as shown in public databases, the expression of CSF2RA in t(8;21)(q22;q22) patients was lower than that in AML patients with other common translocations (supplemental Figure 2A-C), and in FAB-M2 leukemias, CSF2RA was specifically down-regulated in patients with the t(8;21)(q22;q22) translocation (supplemental Figure 3A-B).28 Moreover, the down-regulation of CSF2RA seemed to occur independently of other examined gene mutations, but was correlated positively with LOS (supplemental Table 3). We also confirmed the reduced expression of CSF2RA in t(8;21)(q22;q22) patients with real-time quantitative RT-PCR analysis (supplemental Figure 3C). These findings suggest that GM-CSF signaling may be involved in the pathogenesis of t(8;21)(q22;q22) AML.
Deficiency of GM-CSF signaling enhances the development of RUNX1-ETO–induced leukemia
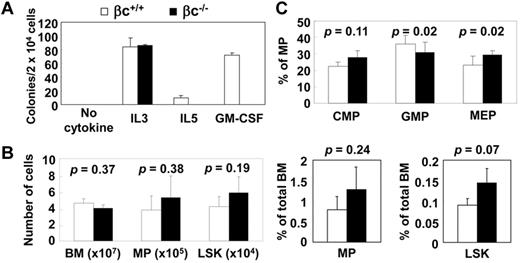
The receptor for GM-CSF is composed of α and β subunits, which heterodimerize to transmit signals in the cell. In humans, the GM-CSF receptor α subunit is specific to GM-CSF signaling, whereas the β subunit (βc) is shared among IL-3, IL-5, and GM-CSF. In contrast to humans, IL-3 signals in mice are transmitted both by βc and by an IL-3–specific β subunit (βIL-3).29 Accordingly, hematopoietic cells from βc−/− mice did not respond to GM-CSF or IL-5, but their response to IL-3 was unaffected (Figure 2A). Analysis of the βc−/− mice revealed no overt abnormalities in the hematopoietic system except for a lower number of eosinophils than in βc+/+ mice, which results from deficiency of IL-5 signaling.29 The stem cell and myeloid progenitor compartments of the βc−/− mice were also not affected (Figure 2B-C and supplemental Figures 4-5). Our data support the idea that deficiency of GM-CSF signaling does not significantly affect steady-state hematopoiesis in mice.
GM-CSF is not necessary for maintenance of hematopoiesis. (A) Response of wild-type (βc+/+) and βc−/− cells to single cytokines in colony assay. (B) Flow cytometric analysis of myeloid progenitor (MP) and LSK of βc+/+ and βc−/− mice. Absolute numbers and percentage of total BM are shown. MP, PI−/Lin−/IL-7Rα−/c-Kit+/Sca-1−; LSK, PI−/Lin−/IL-7Rα−/c-Kit+/Sca-1+ (n = 3; C) Flow cytometric analysis of common myeloid progenitor (CMP), PI−/Lin−/IL-7Rα−/c-Kit+/Sca-1−/FcγRlo/CD34+; granulocyte-macrophage progenitor (GMP), PI−/Lin−/IL-7Rα−/c-Kit+/Sca-1−/FcγRhi/CD34+; and megakaryocyte-erythrocyte progenitor (MEP), PI−/Lin−/IL-7Rα−/c-Kit+/Sca-1−/FcγRlo/CD34− in βc+/+ and βc−/− animals. (n = 6).
GM-CSF is not necessary for maintenance of hematopoiesis. (A) Response of wild-type (βc+/+) and βc−/− cells to single cytokines in colony assay. (B) Flow cytometric analysis of myeloid progenitor (MP) and LSK of βc+/+ and βc−/− mice. Absolute numbers and percentage of total BM are shown. MP, PI−/Lin−/IL-7Rα−/c-Kit+/Sca-1−; LSK, PI−/Lin−/IL-7Rα−/c-Kit+/Sca-1+ (n = 3; C) Flow cytometric analysis of common myeloid progenitor (CMP), PI−/Lin−/IL-7Rα−/c-Kit+/Sca-1−/FcγRlo/CD34+; granulocyte-macrophage progenitor (GMP), PI−/Lin−/IL-7Rα−/c-Kit+/Sca-1−/FcγRhi/CD34+; and megakaryocyte-erythrocyte progenitor (MEP), PI−/Lin−/IL-7Rα−/c-Kit+/Sca-1−/FcγRlo/CD34− in βc+/+ and βc−/− animals. (n = 6).
To study the role of GM-CSF signaling in leukemogenesis by RUNX1-ETO, hematopoietic cells from βc−/− mice were transduced with control (MigR1) or RUNX1-ETO (MigRE) retrovirus and transplanted into wild-type recipients. All MigR1-transplanted animals remained healthy until the completion of the study. In the RUNX1-ETO group, 32 animals died of leukemia, with a median survival time of 230 days; 6 animals in the RUNX1-ETO group remained healthy until completion of the study (penetrance: 84.2%; Figure 3A). Immunoblotting with lysates from spleens of leukemic mice confirmed the expression of full-length RUNX1-ETO (Figure 3B and supplemental Figure 6). To confirm that the βc−/− genotype is favorable to leukemogenesis, MigRE-transduced βc−/− and βc+/+ cells were used in the same set of transplantation assays. Whereas only 2 of 7 animals in the βc+/+ RUNX1-ETO group died of leukemia, all 5 animals succumbed to leukemia in the βc−/− RUNX1-ETO group (Figure 3C). We also investigated whether deficiency of GM-CSF signaling would provide a further advantage to the leukemogenic ability of RUNX1-ETO9a, an isoform of RUNX1-ETO that does not require additional exogenous mutations to induce leukemia.31 Animals transplanted with RUNX1-ETO9a–expressing βc−/− cells had significantly shorter survival times than βc+/+–transplanted animals (Figure 3D), demonstrating that a deficiency of GM-CSF signaling is favorable for both RUNX1-ETO– and RUNX1-ETO9a–induced leukemogenesis.
Deficiency of GM-CSF signaling is favorable for the development of RUNX1-ETO–induced leukemia. (A) Kaplan-Meier survival curve of animals transplanted with βc−/− cells transduced with MigR1 or MigRE. Pooled results from 4 different transplantation experiments are shown. (B) Immunoblotting analysis of spleen cells from βc−/− MigRE leukemia animals using anti-HA tag (RUNX1-ETO) Ab. MigRE is a positive control. Wild-type (WT) is a negative control. (C) Kaplan-Meier survival curve of animals transplanted with βc−/− or βc+/+ cells transduced with MigR1 or MigRE. (D) Kaplan-Meier survival curve of animals transplanted with βc−/− or βc+/+ cells transduced with MigR1 or MigRE9a vectors.
Deficiency of GM-CSF signaling is favorable for the development of RUNX1-ETO–induced leukemia. (A) Kaplan-Meier survival curve of animals transplanted with βc−/− cells transduced with MigR1 or MigRE. Pooled results from 4 different transplantation experiments are shown. (B) Immunoblotting analysis of spleen cells from βc−/− MigRE leukemia animals using anti-HA tag (RUNX1-ETO) Ab. MigRE is a positive control. Wild-type (WT) is a negative control. (C) Kaplan-Meier survival curve of animals transplanted with βc−/− or βc+/+ cells transduced with MigR1 or MigRE. (D) Kaplan-Meier survival curve of animals transplanted with βc−/− or βc+/+ cells transduced with MigR1 or MigRE9a vectors.
AML observed in βc−/− RUNX1-ETO–transplanted animals
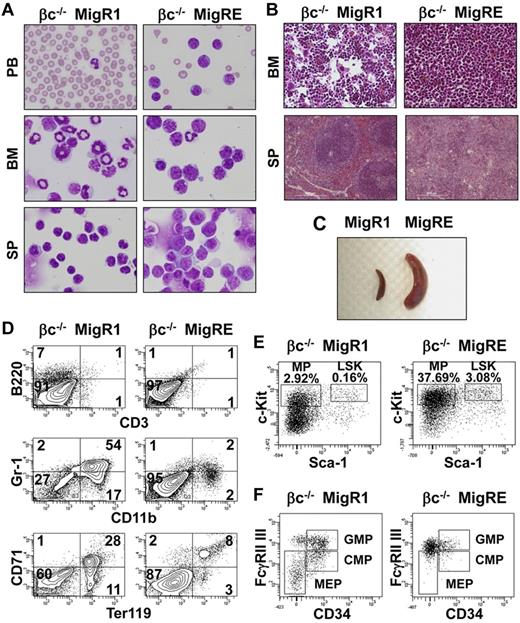
Moribund animals in the βc−/− RUNX1-ETO group were euthanized and examined for abnormalities in hematopoietic tissues according to the recommendations of the Bethesda Proposal.32 Clinical findings for these animals are described in detail in Table 1. In the peripheral blood, anemia, thrombocytopenia, and an increase in total WBC numbers and myeloid blasts were common findings. Cytological analysis of the BM showed an elevated percentage of blasts. The typical morphology of leukemic blasts in βc−/− RUNX1-ETO cells is shown in Figure 4A. Heterogeneity in size, basophilic cytoplasm, and a prominent Golgi zone were common; the nucleolus was not always evident. Azurophilic granules, although not uniformly evident, were detected in some cases. Histological analysis of the BM showed a massive infiltration of mononuclear cells (Figure 4B). Splenomegaly was pronounced in all animals (Figure 5C and Table 1). Histologically, the spleens of leukemic animals were characterized by loss of normal splenic architecture and infiltration of mononuclear cells (Figure 4B). Flow cytometric analysis of the BM of leukemic animals showed decreased expression of lineage markers in GFP-positive cells (Figure 4D). Analysis of the GFP-positive/lineage-negative compartment revealed a larger percentage of the leukemic cells than in the control population expressing the c-Kit marker (Figure 4E). The normal myeloid progenitor compartment in healthy animals can be divided into common myeloid progenitor, granulocyte-macrophage progenitor, and megakaryocyte-erythrocyte progenitor compartments, according to the expression of FcγR and CD34. In leukemic βc−/− RUNX1-ETO animals, this pattern was completely lost, with a single FcγRhi and CD34lo-dim population dominating this compartment (Figure 4F); this pattern is similar to that previously observed for the RUNX1-ETO9a mouse model.31 Oncogene activation through retroviral insertion is a concern when retroviral vectors are used for expression of the gene of interest. Southern blot analysis of spleens of leukemic animals showed that the transformed cells were mainly oligoclonal, ruling out the possibility of insertional mutagenesis–induced leukemia in our experiments (supplemental Figure 7). Furthermore, leukemic cells from these animals were able to replicate the disease in secondary transplanted recipients (data not shown). These results demonstrate that deficiency of GM-CSF signaling is a cooperating factor in the development of RUNX1-ETO–induced AML.
Characteristics of AML in βc−/− MigRE animals
| . | WBCs, 103/mL . | Hb, g/dL . | Plt, 103/mL . | BM blasts,% . | BM GFP, % . | Sp, mg . | SP GFP, % . |
|---|---|---|---|---|---|---|---|
| Mean | 24.5 | 5 | 198.6 | 45.5 | 60.7 | 456.1 | 64.8 |
| Range | 2.84-164.8 | 2.4-7.6 | 25-448 | 24-82.5 | 44-86.6 | 170-888 | 39-87.4 |
| Reference range | 1.8-10.7 | 11.0-15.1 | 792-2972 | < 5 | NA | < 100 | NA |
| . | WBCs, 103/mL . | Hb, g/dL . | Plt, 103/mL . | BM blasts,% . | BM GFP, % . | Sp, mg . | SP GFP, % . |
|---|---|---|---|---|---|---|---|
| Mean | 24.5 | 5 | 198.6 | 45.5 | 60.7 | 456.1 | 64.8 |
| Range | 2.84-164.8 | 2.4-7.6 | 25-448 | 24-82.5 | 44-86.6 | 170-888 | 39-87.4 |
| Reference range | 1.8-10.7 | 11.0-15.1 | 792-2972 | < 5 | NA | < 100 | NA |
Hb indicates hemoglobin levels; Plt, platelet number; Sp, spleen; SP GFP, GFP-positive cells cells in spleen; and NA, not applicable.
AML is observed in animals transplanted with βc−/− RUNX1-ETO cells. (A) Cytological analysis of a representative leukemic βc−/− MigRE animal. Peripheral blood (PB) smears, BM, and spleen (SP) cytospin samples stained with Wright-Giemsa are shown (original magnification, 400×). (B) Histopathological analysis of a representative leukemic βc−/− MigRE animal. H&E-stained sections of the BM (original magnification, 200×) and SP (original magnification, 100×) are shown. (C) Splenomegaly observed in leukemic βc−/− MigRE animal. (D) Flow cytometric analysis of the BM from a representative leukemic βc−/− MigRE animal; numbers represent percentage in GFP+BM cells. CD3/B220 are lymphocyte markers; CD11b/Gr-1, granulocyte markers; Ter119/CD71, erythroid markers. (E) Flow cytometric analysis of myeloid progenitor (MP) or LSK from a representative leukemic βc−/− MigRE animal. Respective percentages in GFP+BM are shown. (F) Flow cytometric analysis of MP populations from a representative leukemic βc−/− MigRE animal. CMP indicates common myeloid progenitor; GMP, granulocyte-macrophage progenitor; and MEP, megakaryocyte-erythrocyte progenitor.
AML is observed in animals transplanted with βc−/− RUNX1-ETO cells. (A) Cytological analysis of a representative leukemic βc−/− MigRE animal. Peripheral blood (PB) smears, BM, and spleen (SP) cytospin samples stained with Wright-Giemsa are shown (original magnification, 400×). (B) Histopathological analysis of a representative leukemic βc−/− MigRE animal. H&E-stained sections of the BM (original magnification, 200×) and SP (original magnification, 100×) are shown. (C) Splenomegaly observed in leukemic βc−/− MigRE animal. (D) Flow cytometric analysis of the BM from a representative leukemic βc−/− MigRE animal; numbers represent percentage in GFP+BM cells. CD3/B220 are lymphocyte markers; CD11b/Gr-1, granulocyte markers; Ter119/CD71, erythroid markers. (E) Flow cytometric analysis of myeloid progenitor (MP) or LSK from a representative leukemic βc−/− MigRE animal. Respective percentages in GFP+BM are shown. (F) Flow cytometric analysis of MP populations from a representative leukemic βc−/− MigRE animal. CMP indicates common myeloid progenitor; GMP, granulocyte-macrophage progenitor; and MEP, megakaryocyte-erythrocyte progenitor.
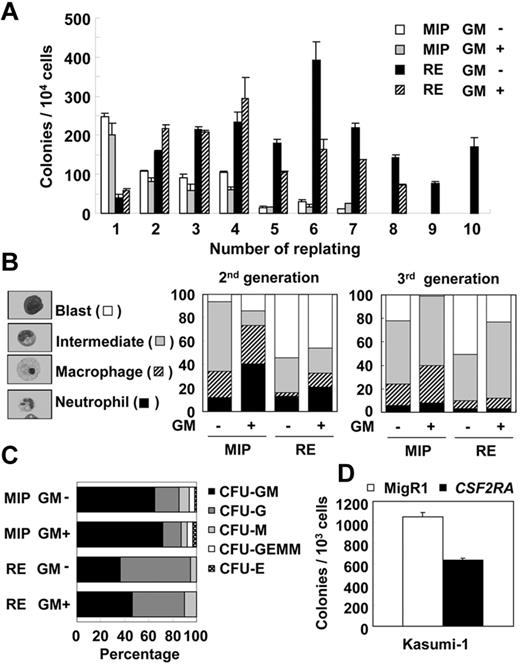
The GM-CSF cytokine negatively affects the self-renewal of RUNX1-ETO cells. (A) Representative result of 3 independent replating assays using BM cells transduced with MIP vector (MIP) or MIP-RE (RE), with or without addition of 10 ng/mL of GM-CSF. (B) Differential counts of cytospin samples from colony cells. Representative results of 2 independent experiments are shown. (C) Differential count of CFUs at the first replating. Representative results of 2 independent experiments. CFU-GM indicates granulocyte macrophage CFU; CFU-G, granulocyte CFU; CFU-M, macrophage CFU; CFU-GEMM, granulocyte, erythroid, macrophage, megakaryocyte CFU; and CFU-E, erythroid CFU. (D) Colony-forming efficiency of Kasumi-1 cells infected with MigR1 (MigR1) or MigR1-CSF2RA (CSF2RA) retrovirus. A representative result of average colony numbers of duplicate samples of 2 independent experiments is shown.
The GM-CSF cytokine negatively affects the self-renewal of RUNX1-ETO cells. (A) Representative result of 3 independent replating assays using BM cells transduced with MIP vector (MIP) or MIP-RE (RE), with or without addition of 10 ng/mL of GM-CSF. (B) Differential counts of cytospin samples from colony cells. Representative results of 2 independent experiments are shown. (C) Differential count of CFUs at the first replating. Representative results of 2 independent experiments. CFU-GM indicates granulocyte macrophage CFU; CFU-G, granulocyte CFU; CFU-M, macrophage CFU; CFU-GEMM, granulocyte, erythroid, macrophage, megakaryocyte CFU; and CFU-E, erythroid CFU. (D) Colony-forming efficiency of Kasumi-1 cells infected with MigR1 (MigR1) or MigR1-CSF2RA (CSF2RA) retrovirus. A representative result of average colony numbers of duplicate samples of 2 independent experiments is shown.
GM-CSF signaling negatively affects self-renewal of RUNX1-ETO cells
RUNX1-ETO, although not leukemogenic in vivo, is known to confer infinite replating ability to primary BM cells in vitro, as demonstrated in replating assays.3 In vivo, deficiency of GM-CSF signaling was favorable for the development of leukemia by RUNX1-ETO, and the replating assay of βc−/− cells showed that βc−/− cells were as efficient as βc+/+ cells in acquiring infinite replating ability after expression of RUNX1-ETO. Moreover, colony formation and proliferation were higher in βc−/− cells than in βc+/+ cells (supplemental Figure 8).
Analysis of blast cells from t(8;21)(q22;q22) patients showed that leukemic blasts were hyporesponsive to the GM-CSF cytokine in proliferation,26,27 suggesting that GM-CSF signaling may inhibit the development of RUNX1-ETO–induced leukemia. To investigate this possibility, we used the replating assay to observe the effects of GM-CSF signaling on the replating ability of RUNX1-ETO cells. As shown in Figure 5A, cells transduced with the control vector (MIP) ceased to form colonies after 7 generations, and the addition of GM-CSF to the medium did not affect the replating ability of these cells. In contrast, the addition of GM-CSF was found to inhibit the replating ability of RUNX1-ETO–expressing cells. With GM-CSF, RUNX1-ETO cells lost the ability to form colonies after 8 generations; RUNX1-ETO cells without GM-CSF continued to replate until termination of the experiment.
Because IL-5 signaling is also lost in βc−/− cells (Figure 2A), we further examined whether IL-5 signaling, similar to GM-CSF signaling, could negatively affect the replating of RUNX1-ETO cells. No such effect was seen with IL-5 (data not shown).
To examine whether GM-CSF affects the differentiation of RUNX1-ETO cells, differential counts were performed on cytospin samples from colony cells (Figure 5B). In the first generation, it was not possible to prepare cytospin samples from RUNX1-ETO cells because of insufficient cell number (see colony numbers at the first generation in Figure 5A). In the second generation, a clear effect of GM-CSF on the differentiation of granulocytes and macrophages was observed in control cells. In RUNX1-ETO cells, the addition of GM-CSF slightly increased the percentage of mature cells (macrophages and neutrophils). However, in the third and subsequent generations, the differentiation effect of GM-CSF was no longer evident in RUNX1-ETO cells. A similar effect was observed on colony types. In the presence of rmSCF, rmIL-3, recombinant human IL-6, and recombinant human EPO, myeloid, erythroid, and multipotent colonies are observed in control cells, whereas only myeloid colonies are observed in RUNX1-ETO cells. The addition of GM-CSF slightly increased the percentage of granulocyte-macrophage colonies in control cells and in RUNX1-ETO cells, but did not rescue the erythroid or multipotent colonies in RUNX1-ETO cells (Figure 5C).
Because the activation of GM-CSF signaling inhibited the immortalization of RUNX1-ETO–expressing primary murine BM cells, we also examined the effect of GM-CSF on the Kasumi-1 cell line derived from a t(8;21)(q22;q22) patient with LOS.33 Because blasts from t(8;21) patients and Kasumi-1 cells are hyposensitive to the GM-CSF cytokine,26,27 GM-CSF signaling was enhanced by transducing cells with retrovirus MigR1-CSF2RA (supplemental Figure 9). In colony assays with the presence of GM-CSF, CSF2RA-expressing cells had 40% fewer colonies than vector control-transduced cells (Figure 5D). This result indicates that the inhibitory effect of GM-CSF signaling is also observed in a human t(8;21) leukemia cell line.
Constitutively elevated phosphorylation of Erk1/2 in cells expressing RUNX1-ETO
Deficiency of GM-CSF signaling is favorable for development of leukemia by RUNX1-ETO in vivo. Activation of GM-CSF signaling in vitro inhibited the immortalization of primary cells by RUNX1-ETO and colony formation in a t(8;21) cell line. These results indicate that the activation of GM-CSF signaling may have a negative effect on the self-renewal of RUNX1-ETO–expressing cells. GM-CSF signaling is initiated on ligand binding and formation of a receptor complex, which activates major phosphorylation-dependent signaling pathways, including the Jak/Stat, Ras/MAPK, and PI3K pathways.34 To analyze the effect of GM-CSF–induced signaling in primary cells expressing RUNX1-ETO, lineage-negative BM cells were retrovirally infected with the control (MIP)– or RUNX1-ETO (RE)–expressing retrovirus, selected with puromycin for 48 hours, and analyzed by immunoblotting. In accordance with previously published protocols, starvation was performed for 2 hours and GM-CSF and serum were used for stimulation.35 The down-regulation of Stat5 phosphorylation was impaired in RUNX1-ETO cells. In addition, higher amounts of the 2 homologous proteins, Stat5a and Stat5b (92 and 94kDa), were observed. No significant abnormalities were found in phosphorylation of Akt. Notably, the phosphorylation of Erk 1 (p44MAPK) and Erk2 (p42MAPK; Erk1/2) was found to be constitutively elevated after 2 hours of serum and growth factor starvation (supplemental Figure 10A).
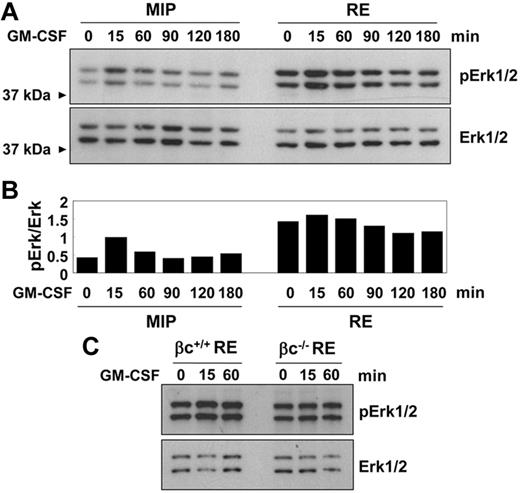
To examine whether the phosphorylation of Erk1/2 could be down-regulated after longer starvation times, the cells were starved for 6 hours before analysis. Moreover, comparison of stimulation conditions in our samples demonstrated that for the experimental conditions used in this study, serum was not a requirement for the detection of proteins phosphorylated after GM-CSF treatment (supplemental Figure 10B). Constitutively elevated Erk1/2 phosphorylation was still observed after 6 hours of serum starvation in the RUNX1-ETO samples (Figure 6A). Densitometry analysis of Erk1/2 bands on immunoblotting demonstrated that although the basal levels of phosphorylation were increased, the kinetics of response after the addition of GM-CSF were not affected (Figure 6B). Immunohistochemical analysis showed normal nuclear translocation of phosphorylated Erk1/2 in RUNX1-ETO cells (supplemental Figure 10C). The constitutive phosphorylation of Erk1/2 in RUNX1-ETO cells was not dependent on the presence of the GM-CSF receptor, as shown by the equal phosphorylation of βc−/− cells after 6 hours of starvation, although, as expected, phosphorylation of Erk1/2 after stimulation with GM-CSF increased in βc+/+ cells but not in βc−/− cells (Figure 6C).
Constitutively elevated phosphorylation of Erk1/2 in RUNX1-ETO cells. (A) Representative immunoblot showing lineage-negative BM cells transduced with MIP or RE and drug selected for 48 hours. Cells were serum and growth factor starved for 6 hours. GM-CSF was added at 50 ng/mL. (B) Quantification of band intensities by densitometry of the blots shown in panel A. Intensities of bands from phosphorylated Erk1/2 were normalized to Erk1/2. (C) Representative immunoblot of βc+/+ and βc−/− cells transduced with RE. Cells were starved and stimulated as in panel A.
Constitutively elevated phosphorylation of Erk1/2 in RUNX1-ETO cells. (A) Representative immunoblot showing lineage-negative BM cells transduced with MIP or RE and drug selected for 48 hours. Cells were serum and growth factor starved for 6 hours. GM-CSF was added at 50 ng/mL. (B) Quantification of band intensities by densitometry of the blots shown in panel A. Intensities of bands from phosphorylated Erk1/2 were normalized to Erk1/2. (C) Representative immunoblot of βc+/+ and βc−/− cells transduced with RE. Cells were starved and stimulated as in panel A.
Discussion
The t(8;21)(q22;q22) translocation is frequently detected in adult AML patients. However, whereas the RUNX1-ETO protein can block differentiation and prolong the survival of hematopoietic cells, it is weakly oncogenic by itself, requiring a second hit for transformation to full-blown leukemia. Because the t(8;21)(q22;q22) is frequently associated with LOS,22-24 we tested the hypothesis that LOS is an additional event required for leukemogenic transformation and identified the reduction of GM-CSF signaling as a likely molecular basis for the link between LOS and t(8;21)(q22;q22) AML.
Human X and Y chromosome deletions occur at similar frequencies in patients with t(8;21), suggesting that the candidate genes that affect RUNX1-ETO leukemia are located on the common regions of these chromosomes known as PARs. Considerable evidence from t(8;21) AML patients was found to suggest a possible negative function of GM-CSF signaling in t(8;21) AML, which led us to examine GM-CSF signaling in more detail. However, other genes located on PARs, namely IL3RA, CRLF2, and IL9R, have been reported to be involved in hematopoietic malignancies and may also cooperate in the pathogenesis of t(8;21). Moreover, because the entire sex chromosome is deleted, as opposed to isolated genes, there is also a possibility of cooperation between multiple genes on the PAR with the t(8;21).
We sought to determine whether the down-regulation of the GM-CSF signaling could contribute to the development of RUNX1-ETO leukemia. Because Csf2ra–knockout mice are not available and GM-CSF–knockout cells still respond to the cytokine produced by wild-type recipients, the βc–knockout mouse29 was chosen as the model for GM-CSF signaling–deficient cells. βc is shared among IL-3, IL-5, and GM-CSF receptors in both humans and mice. However, an extra mouse-specific βIL-3 restores signaling from IL-3, and the βc−/− cells are therefore deficient for GM-CSF and IL-5 signaling. Because the IL-5 requirement is limited to the eosinophil and basophil lineages, loss of IL-5 signaling would not likely affect leukemia development in early myeloid progenitors.
The animals transplanted with βc−/− cells expressing RUNX1-ETO developed leukemia with high penetrance but relatively long latency. Mono- to oligo-clones of leukemia cells detected by Southern blot analysis support the suggestion that further mutations may be necessary for leukemogenesis. Moreover, because in patients the lesion occurs only in one chromosome, and no deletion of CSF2RA on the other sex chromosome was reported in gene copy number studies,36,37 it is possible that CSF2RA gene dosage may also affect the outcome of leukemogenesis by t(8;21).
In contrast to t(8;21), high CSF2RA expression has been detected in inv(16) AML patients (supplemental Figure 2A). The inversion of chromosome 16 results in the fusion of CBFB with MYH11. CBFβ is a binding partner of RUNX1 known to stabilize the RUNX1-DNA interaction. The present study's focus on the possible contribution of deficient GM-CSF signaling to leukemogenesis is superficially at odds with a reported observation of high CSF2RB expression in inv(16) AML patients and mouse models.38 The study reporting this association, however, found Csf2rb− cells to be enriched for preleukemic progenitors compared with Csf2rb+ cells, although the bulk of leukemia cells were represented by the abnormal population expressing Csf2rb. This report suggests that in inv(16) AML, GM-CSF signaling may be involved in a different mechanism of leukemogenesis from that which we considered. It is also worth mentioning that in addition to Kasumi-1 cells, another cell line, SKNO-1, was also derived from a t(8;21)(q22;q22) patient with LOS.39 However, leukemia cells from this patient were originally cultured in the presence of GM-CSF and established as a GM-CSF–dependent cell line. It is possible that the negative effect of GM-CSF on RUNX1-ETO–involved leukemia is blocked by additional mutations in SKNO-1 cells.
The GM-CSF cytokine was first detected in a lung-conditioned medium as a factor that efficiently induced granulocyte-macrophage colonies.40 Apart from the effects on later stages of cell differentiation, GM-CSF was also shown to affect the biology of hematopoietic stem/progenitor cells. Seminal works using multipotent murine stem cell lines (FDC-Pmix) provided grounds for the theoretical delineation of the fate decision between self-renewal and committed differentiation of hematopoietic stem cells. FDC-Pmix cells cultured with IL-3 self-renewed continuously, but activation of GM-CSF signaling induced initial proliferation, followed by differentiation and a decrease in clonogenicity.41 In agreement with these observations, GM-CSF induces granulopoiesis in vivo initially through mobilization of granulocyte-macrophage progenitors and megakaryocyte-erythrocyte progenitors,42 and the expansion of stem/progenitor cells by GM-CSF seems to occur at the expense of more primitive cells.43
The activation of GM-CSF signaling initiates major tyrosine phosphorylation–dependent signaling pathways, including Jak/Stat, Ras/MAPK, and PI3K pathways.34 Because signaling from GM-CSF was able to counteract the effects of RUNX1-ETO expression in mouse BM cells, we sought to examine the pathways involved in this process. Analysis of primary BM cells at the early time points of RUNX1-ETO expression revealed the constitutively elevated phosphorylation of Erk1/2 in RUNX1-ETO cells. This activation was not dependent on the presence of the GM-CSF receptor, because βc−/− cells also showed constitutive phosphorylation in response to RUNX1-ETO expression. The mechanisms of the constitutive activation of Erk1/2 in RUNX1-ETO cells are not known. One possible explanation is the transcriptional repression of Neurofibromatosis-1 (NF1) by RUNX1-ETO.44 NF1 encodes neurofibromin, and mutation of NF1 is frequent in juvenile myelomonocytic leukemia.45 Neurofibromin is a GTPase-activating protein that converts activated Ras/GTP to the inactive Ras/GDP; neurofibromin dysfunction results in hyperactivation of Ras and, as a consequence, hyperactivation of the MAPK pathway.
Erk1/2 belongs to the MAPK pathway, which is made up of a conserved family of protein kinases that are activated in response to virtually all mitogenic factors. Deregulation of the Ras/MAPK pathway is frequent in myeloid leukemia.46 However, contradictory to this finding, sustained expression of MEK in CD34+ cells results in apoptosis,47 and expression of mutated RAS induces proliferation and differentiation of CD34+ cells.48 These observations demonstrate that the pathway to oncogenesis is not a straightforward one, especially considering the role of Ras/MAPK signaling in this process. In CD34+CD133+ cells, activation of Erk1/2 is involved in the maintenance of self-renewal. However, robust activation of Erk1/2 was less efficient than moderate activation in the maintenance of the repopulating ability of these cells.49 Accordingly, moderate activation of Erk1/2 by RUNX1-ETO may favor the self-renewal of stem/progenitor cells, whereas Erk1/2 phosphorylation strengthened by the addition of GM-CSF may instruct those cells to reduce stemness and differentiate. Erk1/2 phosphorylates and activates several transcription factors, such as Elk1, Creb1, Fos, Jun, and Runx1.46 A constitutive active mutant of Stat5 can confer cytokine independence in Ba/F3 cells through expression of pim-1, but further cytokine activation of Stat5 leads to apoptosis and differentiation through expression of JAB and p21.50 By similar mechanisms, GM-CSF may restore the expression of differentiation-related genes in RUNX1-ETO cells by further activation of Erk1/2.
The results of replating assays suggest that, at the initial immortalization phase, RUNX1-ETO–expressing cells are susceptible to the effects of GM-CSF. Furthermore, the enhancement of GM-CSF signaling in a t(8;21) AML cell line through expression of CSF2RA showed a negative effect on colony formation. Those results indicate that the susceptibility of t(8;21) blasts to GM-CSF is a treatment possibility to be explored further.
In summary, the present study demonstrates that deficiency of GM-CSF signaling is favorable for the development of leukemia by RUNX1-ETO in mouse models. Moreover, GM-CSF was able to inhibit the self-renewal of RUNX1-ETO–expressing primary murine BM cells and human t(8;21) Kasumi-1 cells. These findings suggest an unexpected tumor-suppressor role of GM-CSF in t(8;21) leukemias. Loss of the CSF2RA gene may be a critical mutation explaining the high incidence of LOS in t(8;21)(q22;q22). Future studies to identify downstream effectors of GM-CSF signaling in blocking such leukemogenesis may provide valuable insight into clinical treatment possibilities.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Paul Burgoyne for providing XPafY* mice, DNAX Research Institute for providing βc−/− mice, Ruud Delwel for AML patient information, and Dr Theo Ross and all members of the Zhang laboratory, especially Luke F. Peterson and Joseph R. Biggs, for valuable suggestions.
This work was supported by funding from the National Institutes of Health (grant R01CA096735 to D.-E.Z.). S.M. is a recipient of the Ruth L. Kirschstein National Research Service Award (F32HL091641).
National Institutes of Health
Authorship
Contribution: S.M. performed most of the experiments, analyzed the results, produced the figures, and wrote the manuscript; M.Y. performed the XO transplantations and Southern blot analyses; M-C.L. analyzed CSF2RA in FAB-M2 patients;, E-Y.A., S.W., and D.D. performed and advised on Western blot experiments; M.M. assisted with the experiments; T.H. performed immunofluorescence;G-S.F. assisted in experimental design and manuscript preparation; and D-E.Z. designed the experimental approach, analyzed the data, and supervised manuscript preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dong-Er Zhang, Moores UCSD Cancer Center, University of California, San Diego, 3855 Health Sciences Dr, Mail Stop 0815, Rm 5328, La Jolla, CA 92093; e-mail: d7zhang@ucsd.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal