Abstract
The BRAFV600E mutation was recently detected in hairy cell leukemia (HCL) by whole exome sequencing. To make use of this new marker for diagnosis and follow-up of HCL, we developed a BRAFV600Emut-specific quantitative real-time PCR assay and validated it in 117 HCL patients and 102 non-HCL/BRAFwt patients. The cut-off level to discriminate BRAFV600E-positive/-negative cases was set at 0.023% BRAFV600E/BRAFwt. A total of 115 of 117 HCL (98.3%) demonstrated percentage BRAFV600E/BRAFwt above the cut-off (mean, 29.6 ± 41.1). The remaining 2 of 117 HCL with lower percentage BRAFV600E/BRAFwt ratios were also BRAFwt by deep-sequencing technology. Sixteen HCL-variant patients showed percentage BRAFV600E/BRAFwt values corresponding to “non-HCL.” Follow-up studies in 19 HCL cases demonstrated a decrease of percentage BRAFV600E/BRAFwt during therapy. The log-reductions as determined by RT-PCR and immunophenotyping correlated significantly (P < .001). In conclusion, we confirmed BRAFmut as a useful marker in HCL, its absence in HCL variant, and developed an RT-PCR-based assay to monitor minimal residual disease in HCL.
Introduction
Hairy cell leukemia (HCL) is a clearly characterized disease that differs from other mature B-cell neoplasms by its clinical presentation, unique immunophenotype, and morphology, as well as the occurrence of bone marrow (BM) fibrosis, or therapy.1-4 So far, diagnosis of HCL was based on cytomorphology, immunophenotyping, and immunohistopathology. Recently, a V600E mutation of BRAF (v-raf murine sarcoma viral oncogene homolog B1) was detected in all 48 patients with HCL analyzed by whole-exome and Sanger sequencing.5 The BRAFV600E was restricted to typical HCL and was not observed in HCL-variant (HCL-v) or splenic marginal zone lymphoma.5 BRAF mutations have been described before in different cancers, including melanoma,6 papillary thyroid cancer, or lymphoblastic leukemia.7 Here, we developed a new quantitative real-time PCR assay to detect the BRAFV600E mutations. We evaluated this assay in 117 patients at diagnosis of HCL as confirmed by multiparameter flow cytometry (MFC) and performed follow-up studies compared with MFC in a proportion of these patients.
Methods
Patients
We investigated 117 patients at diagnosis of HCL as confirmed by MFC (28 females; 89 males; median age, 57.9 years; range, 24.3-88.4 years), 9 of whom had been part of the recently published sequencing study.5 In addition, 2 independent cohorts (1, 16 patients with HCL-v; and 2, 102 patients with different acute/chronic myeloid/lymphatic leukemias or nonmalignant diseases) were included for comparison studies. BM or peripheral blood (PB) samples were sent to the Munich Leukemia Laboratory between August 2005 and June 2011. All patients gave written informed consent with genetic analysis and research studies in accordance with the Declaration of Helsinki. The study was approved by the Munich Leukemia Laboratory Institutional Review Board.
After erythrocyte lysis of the BM/PB samples, all HCL and all HCL-v cases were investigated by 5-color flow cytometry.8 Classification as HCL was based on bright monotypic surface immunoglobulin, bright coexpression of CD20, CD22, and CD11c, and expression of CD103 and CD25.1 HCL-v was diagnosed in the absence of CD25.1
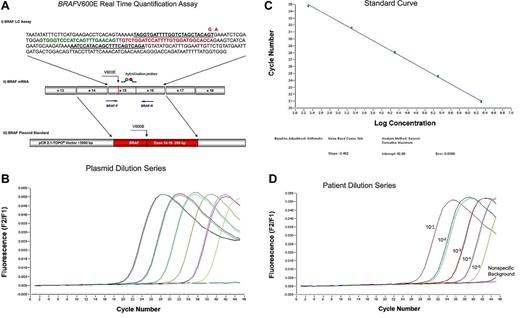
As the BRAFV600E is characterized by an invariant T > A nucleotide substitution, we decided to develop an mRNA-based reverse transcription allele-specific real-time quantification (RT-PCR) assay. Quantification of the BRAFV600E was performed by measuring the expression of BRAFV600E mRNA compared with BRAF wild-type (BRAFwt) mRNA. Values were given in percentage BRAFV600E/BRAFwt. RT-PCR was performed by LightCycler Version 1.5 System (Roche Diagnostics) with the application of hybridization probes as the detection format as detailed in Figure 1A. Standard curves for both assays were produced by 10-fold dilution series of 6 different plasmid concentrations. In all the following runs, a reference dilution was analyzed and the external standard curve was loaded compared with a reference sample. To analyze the efficiencies and sensitivities, we performed serial dilution experiments of the plasmids and BRAFmut patient samples in BRAFwt cDNA. Efficiencies were calculated from the slope of the standard curves and ranged from 1.93 to 1.96, and were similar in the BRAFwt and BRAFmut assays (Figure 1B-C). Limiting dilution series of cDNA of 2 cases with highly infiltrated HCL in cDNA of a normal control revealed a sensitivity of 10−4 to 10−5 (Figure 1D).
RT-PCR assay for the detection of BRAFV600E. (A) BRAFV600E real-time quantification assay. (i) Assays for BRAFwt and BRAFmut were almost identical, with the exception of the BRAFV600E mutation-specific primer, which includes the nucleotide substitution of the T > A at the 3′end and an additional mismatch nucleotide (A > G at the third position from the 3′end) that further enhances the specific detection of mutated transcripts. Forward and reverse primer are underlined. Mutated nucleotide and mismatch nucleotide in the forward primer are given in red. Green represents fluorescein-labeled detection probe; and red, LC640-labeled probe. In detail, the reaction was performed in a final volume of 20 μL by use of 2 μL mastermix (LightCycler Fast Start DNA Master Hybridization Probes; Roche Diagnostics), 4mM MgCl2, 0.5μM of each forward (BRAFwt-F: TAGGTGATTTTGGTCTAGCTACAGT or BRAFmut-F: TAGGTGATTTTGGTCTAGCTACGGA) and reverse primer (BRAF-R: TCTGACTGAAAGCTGTATGGATT; Metabion), 0.25μM of each of the 2 fluorescent hybridization probes (BRAF-Fl: GTGGGTCCCATCAGTTTGAACAG-fluorescein and BRAF-LCred640: LC640-GTCTGGATCCATTTTGTGGATGGCACC-phosphate), and 2 μL cDNA (accounting for an equivalent of mRNA of 200 000 cells). Amplification was performed after initial incubation at 95°C for 10 minutes in a 3-step cycle procedure (denaturation 95°C, 1 second, ramp rate 20°C/s, annealing temperature 64°C, 10 seconds, ramp rate 20°C/s, and extension 72°C, 26 seconds, ramp rate 2°C/s) for 45 cycles. (ii) Position of primers (BRAF-F and BRAF-R), V600E mutation, and hybridization probes at the mRNA are indicated. (iii) For plasmid standards, exons 14 to 16 of BRAFwt and BRAFmut each were cloned into a pCR2.1TOPO vector. (B) Ten-fold dilution series of the plasmid carrying the BRAFV600E: 200 000, 2 plasmid copies. (C) Standard curve showing efficiency of the BRAFV600E specific PCR of 1.93. (D) Ten-fold dilution of a diagnostic BRAFV600E mutated patient sample in cDNA of an unmutated patient showing a sensitivity of 1 of 100 000. BRAFwt signal is indicated as “nonspecific background.”
RT-PCR assay for the detection of BRAFV600E. (A) BRAFV600E real-time quantification assay. (i) Assays for BRAFwt and BRAFmut were almost identical, with the exception of the BRAFV600E mutation-specific primer, which includes the nucleotide substitution of the T > A at the 3′end and an additional mismatch nucleotide (A > G at the third position from the 3′end) that further enhances the specific detection of mutated transcripts. Forward and reverse primer are underlined. Mutated nucleotide and mismatch nucleotide in the forward primer are given in red. Green represents fluorescein-labeled detection probe; and red, LC640-labeled probe. In detail, the reaction was performed in a final volume of 20 μL by use of 2 μL mastermix (LightCycler Fast Start DNA Master Hybridization Probes; Roche Diagnostics), 4mM MgCl2, 0.5μM of each forward (BRAFwt-F: TAGGTGATTTTGGTCTAGCTACAGT or BRAFmut-F: TAGGTGATTTTGGTCTAGCTACGGA) and reverse primer (BRAF-R: TCTGACTGAAAGCTGTATGGATT; Metabion), 0.25μM of each of the 2 fluorescent hybridization probes (BRAF-Fl: GTGGGTCCCATCAGTTTGAACAG-fluorescein and BRAF-LCred640: LC640-GTCTGGATCCATTTTGTGGATGGCACC-phosphate), and 2 μL cDNA (accounting for an equivalent of mRNA of 200 000 cells). Amplification was performed after initial incubation at 95°C for 10 minutes in a 3-step cycle procedure (denaturation 95°C, 1 second, ramp rate 20°C/s, annealing temperature 64°C, 10 seconds, ramp rate 20°C/s, and extension 72°C, 26 seconds, ramp rate 2°C/s) for 45 cycles. (ii) Position of primers (BRAF-F and BRAF-R), V600E mutation, and hybridization probes at the mRNA are indicated. (iii) For plasmid standards, exons 14 to 16 of BRAFwt and BRAFmut each were cloned into a pCR2.1TOPO vector. (B) Ten-fold dilution series of the plasmid carrying the BRAFV600E: 200 000, 2 plasmid copies. (C) Standard curve showing efficiency of the BRAFV600E specific PCR of 1.93. (D) Ten-fold dilution of a diagnostic BRAFV600E mutated patient sample in cDNA of an unmutated patient showing a sensitivity of 1 of 100 000. BRAFwt signal is indicated as “nonspecific background.”
For confirmation of the RT-PCR results, 15 cases with and 15 without BRAFV600E were verified by Sanger sequencing. In addition, 2 cases with BRAFV600wt, as assessed by RT-PCR, were investigated by deep-sequencing (454 Life Sciences).9 Correlation between PCR and MFC results was performed according to Spearman, and expression levels of BRAF were compared between groups using Student t test.
Results and discussion
First, we defined the nonspecific background of the assay that can be caused by the normal wild-type allele in the 102 “non-HCL” controls. This cohort was composed of the following: acute myeloid leukemia, n = 9; myelodysplastic syndrome, n = 7; myeloproliferative neoplasm, n = 13; chronic myeloid leukemia, n = 4; chronic myeloid leukemia in major molecular response, n = 10; chronic myelomonocytic leukemia, n = 4; chronic eosinophilic leukemia, n = 2; T-acute lymphoblastic leukemia, n = 1; B-acute lymphoblastic leukemia, n = 2; follicular lymphoma: n = 3; mantle cell lymphoma, n = 8; splenic marginal zone cell lymphoma, n = 1; chronic lymphocytic leukemia, n = 11; other mature B-non-Hodgkin lymphoma, 20; nonmalignant diseases, n = 7. Unspecific percentage BRAFV600E/BRAFwt values of “non-HCL” controls were very low (median, 0.003; range, 0.000-0.030; mean ± SD, 0.005 ± 0.006). The cut-off level for discrimination of BRAFV600E-positive and -negative cases was therefore defined as 0.023 percentage BRAFV600E/BRAFwt (3 SD above the mean). In addition, the 16 patients with HCL-v showed percentage BRAFV600E/BRAFwt values in the range of the “non-HCL” controls (median, 0.006; range, 0.000-0.012; mean, 0.006 ± 0.004).
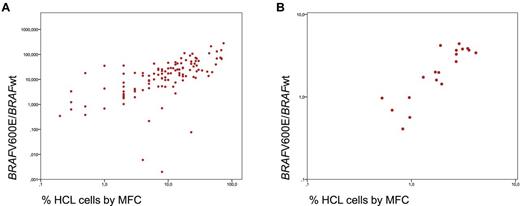
Subsequently, the 117 patients with a proven diagnosis of HCL were analyzed. In 115 of 117 HCL (98.3%), the percentage BRAFV600E/BRAFwt expression was above the cut-off (median, 17.2; range, 0.077-280.3; mean, 29.6 ± 41.1). Expression levels measured in PB samples (n = 37) were slightly higher (median percentage BRAFV600E/BRAFwt, 13.89; range, 0.138-280.3) than those in BM (median, 25.09; range, 0.077-131.6; P = .042). This higher expression correlates with a higher infiltration of pathologic cells in PB (23.9%) compared with BM (12.4%; P = .002). MFC detected a median of 10.0% leukemic cells (range, 0.2%-74.0%; mean, 16.1% ± 17.2%). BRAFV600E expression and the percentage of HCL cells by MFC at diagnosis showed a significant correlation (r = 0.741, P = .001; Figure 2A). Two of the 117 (1.7%) HCL patients had percentage BRAFV600E/BRAFwt values in the range of normal controls (0.000 and 0.006, respectively) and thus are considered as having BRAFwt. The percentage of HCL cells by MFC amounted to 4.0% and 8.0%, respectively, in these cases. We confirmed the absence of the V600E or a variant mutation by 454 deep sequencing of exon 15 with 198- and 398-fold coverage, respectively, which was corresponding to a detection limit of approximately 0.5% to 1% of cells. Both patients were females (age 52 and 61 years) and showed good response to cladribine (patient 1) and splenectomy (patient 2). Besides BM assessment by cytomorphology and MFC, the diagnosis of HCL was confirmed in patient 1 by BM immunohistochemistry and in patient 2 by immunohistochemistry after splenectomy. In a recently published paper,15 not only variant but also IGHV4-34–expressing HCL were reported to lack the BRAFV600E mutation (11 of 53 examined HCL cases, all with IGHV4-34 usage). However, both our cases did not reveal IGHV4-34 usage. Thus, it remains to be studied which HCLs are BRAFwt and whether this rare subset has a common still unknown genetic defect.
Correlation of PCR and MFC. (A) Correlation of BRAFV600E expression level and the percentage of HCL cells by MFC at first diagnosis. (B) The logarithmic reduction between diagnosis and follow-up assessment as determined for BRAFV600E expression level by RT-PCR and for HCL cells by MFC.
Correlation of PCR and MFC. (A) Correlation of BRAFV600E expression level and the percentage of HCL cells by MFC at first diagnosis. (B) The logarithmic reduction between diagnosis and follow-up assessment as determined for BRAFV600E expression level by RT-PCR and for HCL cells by MFC.
Follow-up studies including 1 to 4 samples per patient were performed in 19 HCL cases in parallel by MFC and RT-PCR. At diagnosis, the mean percentage BRAFV600E/BRAFwt was 38.0 ± 60.8, and was decreasing to a mean of 7.3 ± 19.8 by a mean log reduction of 2.0 ± 1.6 during therapy. MFC revealed a mean of 17.4% ± 22.0% leukemic cells at diagnosis, decreasing by a mean log reduction of 1.8 ± 1.2 to a mean of 3.0% ± 7.7% during follow-up. The log reduction as observed by RT-PCR and immunophenotyping again showed a significant correlation (r = 0.896; P < .001; Figure 2B). Only 1 of 7 patients who achieved immunologic CR during follow-up maintained a slightly increased percentage BRAFV600E/BRAFwt of 0.05, whereas 6 obtained also a molecular CR with percentage BRAFV600E/BRAFwt values in the range of negative controls. One patient showed an increasing percentage of HCL cells by MFC and in parallel an increasing BRAFV600E/BRAFwt (from 0.988 to 1.9) 36 days before the cytomorphologic relapse appeared.
In conclusion, our study confirms the high specificity of the BRAFV600E for HCL.10,11 In HCL patients, RT-PCR for the detection of BRAFV600E provides a new valid, rapid, and highly sensitive molecular minimal residual disease parameter, which may be clinically applied similarly to the settings in chronic lymphocytic leukemia and mantle cell lymphoma12,13 and provides a high sensitivity level, which is even higher compared with an allele-specific PCR applied to an independent cohort as published recently.14 Furthermore, this new RT-PCR assay facilitates the diagnosis of HCL by detection of the BRAFV600E in all mutation carriers and improves the discrimination of HCL from HCL variant cases. However, as shown here for the first time, single cases with classic HCL do not carry BRAF gene mutations. Furthermore, this RT-PCR assay may be used to monitor minimal residual disease with sensitivity comparable with and potentially higher than that achieved by flow cytometry. In addition, RT-PCR has the advantage to be applicable also retrospectively without the need for viable cells. In general, we would recommend use of RT-PCR and immunophenotyping in combination to further improve and strengthen the diagnosis and minimal residual disease monitoring in patients with HCL.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.S., T.H., C.H., and W.K. performed design of study; U.B. and W.K. analyzed data and wrote the first draft of the manuscript; S.S., N.W., M.U., F.D., and V.G. performed molecular studies; T.H. performed cytomorphology; W.K. was responsible for immunophenotyping; and all authors contributed to write the manuscript and approved the final version.
Conflict-of-interest disclosure: S.S., T.H., C.H., and W.K. declare part ownership of the Munich Leukemia Laboratory GmbH. F.D., N.W., M.U., and V.G. are employed by the Munich Leukemia Laboratory GmbH. The remaining author declares no competing financial interests.
Correspondence: Susanne Schnittger, MLL Munich Leukemia Laboratory, Max-Lebsche-Platz 31, 81377 Munich, Germany; e-mail: susanne.schnittger@mll.com.