Abstract
Fanconi anemia (FA) is an inherited genetic disorder associated with BM failure and cancer predisposition. In the present study, we sought to elucidate the role of microRNAs (miRNAs) in the hematopoietic defects observed in FA patients. Initial studies showed that 3 miRNAs, hsa-miR-133a, hsa-miR-135b, and hsa-miR-181c, were significantly down-regulated in lymphoblastoid cell lines and fresh peripheral blood cells from FA patients. In vitro studies with cells expressing the luciferase reporter fused to the TNFα 3′-untranslated region confirmed in silico predictions suggesting an interaction between hsa-miR-181c and TNFα mRNA. These observations were consistent with the down-regulated expression of TNFα mediated by hsa-miR-181c in cells from healthy donors and cells from FA patients. Because of the relevance of TNFα in the hematopoietic defects of FA patients, in the present study, we transfected BM cells from FA patients with hsa-miR-181c to evaluate the impact of this miRNA on their clonogenic potential. hsa-miR-181c markedly increased the number and size of the myeloid and erythroid colonies generated by BM cells from FA patients. Our results offer new clues toward understanding the biologic basis of BM failure in FA patients and open new possibilities for the treatment of the hematologic dysfunction in FA patients based on miRNA regulation.
Introduction
microRNAs (miRNAs) are 20- to 24-nucleotide RNAs that regulate gene expression mainly by destabilizing target mRNAs through partial base pairing complementary sites.1 Since their original description, multiple miRNAs have been shown to regulate the development and differentiation of different tissues,2 including the hematopoietic system.3-5 In addition, miRNAs have been shown to act as tumor suppressors6 or oncogenes in the development of hematologic malignancies.7-9
Because Fanconi anemia (FA) is a complex disease mainly associated with BM failure and cancer predisposition,10 we investigated whether specific miRNAs are deregulated and play a role in the hematopoietic dysfunction of FA patients. Among the hematologic dysfunctions of FA patients, macrocytosis is often the first detected, followed by thrombocytopenia and neutropenia. A high incidence of cancers, principally acute myeloid leukemia and squamous cell carcinoma, is also associated with FA.11 Data from the International Fanconi Anemia Registry showed that in these patients, the actuarial risk of developing BM failure and hematologic and nonhematologic malignancies by 40 years of age is 90%, 33%, and 28%, respectively.10 Differences in the clinical symptoms of FA patients are difficult to interpret. Nevertheless, because FA is a chromosomal instability disorder, FA cells accumulate DNA damage at an increased rate. Unrepaired DNA damage may activate apoptotic pathways, thus potentially leading to BM failure, or may induce additional mutations and translocations that may finally result in solid tumors or leukemias.12
Although the exact function of FA proteins is not clearly understood, 8 FA proteins (FANCA, FANCB, FANCC, FANCG, FANCF, FANCE, FANCL, and FANCM) interact to form an FA core complex that is responsible for the mono-ubiquitination of FANCD2 and FANCI, known as the ID complex.13,14 After mono-ubiquitination, both proteins migrate to sites of DNA damage, forming DNA repair foci in association with other proteins, including FANCJ/BRIP1, FANCD1/BRCA2, and FANCN/PALB2.15 An additional molecule, the nuclease FAN1, was shown recently to be an essential partner in the FA pathway because of its interaction with the ID complex and its further recruitment to sites of DNA damage.16-19 Finally, the observation that FANCP (SLX4)20,21 has endonuclease activity is allowing investigators to unravel the role of the FA/BRCA pathway in the repair of DNA interstrand cross-links during replication.22
In the present study, miRNA expression analyses in lymphoblastic cell lines (LCLs) and peripheral blood (PB) cells from FA patients and healthy donors (HDs) showed that 3 different miRNAs were specifically down-regulated in FA hematopoietic cells. In silico studies and in vitro experiments of gene interference showed that one of the miRNAs that is consistently down-regulated in FA samples is hsa-miR-181c. Moreover, our results demonstrate that this miRNA interacts with TNFα, playing an important role in the functional properties of FA hematopoietic progenitors.
Methods
Cell lines and primary cells
FA patients were diagnosed based on clinical symptoms and chromosome breakage tests of PB cells using DNA cross-linker drugs.23 Patients and HDs were encoded to protect their confidentiality, and informed consent was obtained in all cases according to institutional regulations. EBV-transformed LCLs from FA patients were grown in RPMI medium (GIBCO) supplemented with 15% FBS, glutamine, and antibiotics (0.5% penicillin and streptomycin).
PB mononuclear cells (MNCs) were obtained by Ficoll fractionation. BM samples were depleted from erythrocytes with hydroxyethyl starch as described previously24 and cultured in IMDM supplemented with 20% FBS and antibiotics (0.5% penicillin and streptomycin) in the presence of thrombopoietin (50 ng/mL; R&D Systems), SCF (150 ng/mL; Peprotech), and Flt3 ligand (50 ng/mL; Invitrogen).
The RKO cell line (a colon carcinoma cell line) was grown in DMEM supplemented with 10% FBS, glutamine, and antibiotics (0.5% penicillin and streptomycin).
RNA extraction, reverse transcription, and real-time PCR quantification of miRNAs
2-5.106 cells from either LCLs or PB from healthy and FA-A patients was used for RNA extraction using the TRIzol total RNA isolation reagent (Molecular Probes). cDNA was synthesized using gene-specific primers designed by Applied Biosystems (http://www5.appliedbiosystems.com/tools/mirna/) according to the TaqMan miRNA assay protocol (PE Applied Biosystems). The reaction was prepared as described previously.25 Real-time quantitative PCR (qPCR) was performed using an Applied Biosystems 7300 Sequence Detection system and a Rotor Gene using 1.33 μL of RT product, 1× TaqMan Universal PCR master mix, and 1 μL of primers and probe mix of the TaqMan miRNA assay protocol (PE Applied Biosystems). The reaction was incubated at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 10 minutes. The Ct data were calculated using default threshold settings. Normalization of the data was done using the RNU6B gene as an endogenous control. Relative quantification of expression of analyzed miRNAs was calculated with the 2−ΔΔCt method (Applied Biosystems user bulletin number 2, P/N 4303859). Data are presented as log 2−ΔΔCt of the relative quantity of miRNAs, normalized and compared with the mean relative expression value of control cell lines or PB samples. A supervised analysis using the significant analysis of microarrays algorithm was performed to identify miRNAs with statistically significant changes in expression between groups.
DNA methylation analysis
DNA was extracted using the QIAamp DNA Mini Kit (QIAGEN). Methylation-specific PCR (MSP) was used to analyze the methylation status of the CpG sites located 5′ upstream of hsa-miR-181c and was performed as described previously. For the hsa-miR-181c-MSP, the following primers were used: hsa-miR-181c-MD (5′-GTTCGTAGATTTAGGTTAGGGGC-3′) and hsa-miR-181c-MR (5′-CAATAATCGCACAAAATTCGAC-3′), which amplify a 151-bp product, for the methylated reaction and primers hsa-miR-181c-UD (5′-GTTTGTAGATTTAGGTTAGGGGTGA-3′) and hsa-miR-181c-UR (5′-CTCCAATAATCACACAAAATTCAAC-3′), which amplify a 154 bp product, for the unmethylated reaction. PCR conditions for the hsa-miR-181c-MD/hsa-miR-181c-MR primers were 94°C for 10 minutes, followed by 35 cycles at 94°C for 1 minute, 60°C for 1 minute, and 72°C for 1 minute. PCR conditions for the hsa-miR-181c-UD/hsa-miR-181c-UR primers were 94°C for 10 minutes, followed by 35 cycles at 94°C for 1 minute, 58°C for 1 minute, and 72°C for 1 minute. The final extension was at 72°C for 10 minutes. The products were separated by electrophoresis on a 1.8% agarose gel.
The 4 CpG dinucleotides that are located in the genomic sequence that results in mature miRNA hsa-miR-181c were analyzed by pyrosequencing, as described previously.26 After bisulfite treatment of DNA, “hot-start” PCR was performed using hsa-miR-181c-PD (5′-TTTATGAGAGAAAAGGGGTTTTTTAT-3′) and hsa-miR-181c-PR (biotine-5′-AAAAATAACAATTCCAAACCTCAAA-3′). PCR conditions were 94°C for 10 minutes, followed by 40 cycles at 94°C for 1 minute, 58°C for 1 minute, and 72°C for 1 minute. The final extension was at 72°C for 10 minutes. The resulting biotinylated PCR products were immobilized to streptavidin Sepharose High Performance beads (GE Healthcare) and processed to yield high-quality ssDNA using the PyroMark Vacuum Prep Workstation (Biotage) according to the manufacturer's instructions. The pyrosequencing reactions were performed using Pyromark ID (Biotage) and sequence analysis was performed using the hsa-miR-181c-SEQ1 (5′-ATGTTTTTTGGTTTTTTGTTATTTATTTTA-3′) and hsa-miR-181c-SEQ2 (5′-TTTGTTAAGGGTTTGGGGGAATATTTAATTTG-3′) primers and PyroQ-CpG Version 1.0.9 analysis software (Biotage).
Human male genomic DNA universally methylated for all genes (Intergen) was used as a positive control for methylated alleles (not shown). Water blanks were included with each assay. The results were always confirmed by repeating the MSP assays or by pyrosequencing after an independently performed bisulfite treatment.
Cell transfection and transduction with miRNAs
Lymphoblast cell lines and primary BM samples from FA-A patients were transfected either with a negative-precursor miRNA control (Pre-miR Control #1; Applied Biosystems) or a Pre-miR-181c precursor molecule using siNeoFX reagent (Ambion). Briefly, retronectin-coated plates were used to transfect 2.5 × 105 LCLs or BM cells with Pre-miR molecules. Transfection was repeated after 24 hours and 48 hours later, cells were harvested for functional and expression assays. To determine the transfection efficiency in these experiments, a carboxyfluorescein dye–labeled Pre-miR negative control (Applied Biosystems) was used. The transfection efficiency was determined as the percentage of FITC-positive cells as measured by flow cytometry 2 days after transfection. Cell viabilities were determined by analyzing the proportion of propidium iodide–negative cells by flow cytometry (EPICS XL; Coulter Electronics).
To stably transduce BM cells from FA patients, infective supernatants containing lentiviral vectors carrying the Pre-miR-181c and enhanced green fluorescent protein (EGFP) genes (Pre-miR-181c-EGFP LV; System Biosciences) or a control vector (EGFP LV) were generated as described previously.27 Briefly, 5 × 105 FA-BM cells were cultured in IMDM supplemented with 20% FBS and 300 ng/mL of human SCF (Peprotech), 100 ng/mL of human thrombopoietin (R&D Systems), and 100 ng/mL of Flt3 ligand (Invitrogen) in retronectin-coated wells (Takara Shuzo). Two rounds of infection were conducted with either LV:Pre-miR-181c EGFP or LV:EGFP and the efficiency of transduction was analyzed by the percentage of EGFP-positive cells measured by flow cytometry in a FACS LSRFortessa flow cytometer (BD Biosciences)
mRNA expression analyses of target genes
BM samples transfected with Pre-miR control or Pre-miR-181c were harvested 48 hours after transfection and RNA was extracted with TRIzol reagent, as described previously. Total RNA was obtained and the expression of TNFα, IL-1β, RAD54B, and β-ACTIN was studied with SYBR Green Supermix (Applied Biosystems) using the following primers: TNFα-F: 5′-CAGCCTCTTCTCCTTCCTGAT-3′ and TNFα-R: 5′-GCCAGA GGGCTGATTAGAGA-3′; Il-1β-F: 5′-CTGTCCTGCGTGTTGAAAGA-3′ and IL-1β-R: 5′-TTGGGTAAT TTTTGGGATCTACA-3′; RAD54B-F: 5′-TCATGATCTGCTTGACTGTGAG-3′ and RAD54B-R: 5′-TTTTTCCAACGAATCACCTGT-3′; and hDNA-RNA-βACTIN-F: 5′-ATTGGCAATGAGCGGTTCC-3′ and hRNA-βACTIN-R:5′-CACAGG ACTCCATGCCCA-3′.
TNFα analyses
Intracellular levels of TNFα in erythrocyte-depleted BM cells previously transduced or transfected with miRNA constructs were determined by flow cytometry 48 hours after miRNA transfection. Samples were incubated for 5 hours with 50 ng/mL of phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich), lipopolysaccharide (LPS; 1μg/mL; Sigma-Aldrich), or resiquimod (R848;1 μg/mL; Alexis Biochemicals), together with 5 μg/mL of brefeldin A (Sigma-Aldrich), which inhibits the intracellular transport of cytokines into the Golgi complex, avoiding the extracellular release of the cytokines.28 Cells were washed in PBA (PBS + 0.1% BSA) and incubated for 30 minutes at 4°C with anti-CD45–FITC or anti-CD45–PC5 (Beckman Coulter). Stained cells were resuspended for 20 minutes at 4°C in Cytofix/Cytoperm fixation/permeabilization solution (BD Biosciences) according to the manufacturer's instructions. After permeabilization, cells were washed twice with washing buffer and stained for intracellular TNFα using an anti–TNFα-PE Ab (BD Biosciences). Flow cytometric analysis was conducted in a FACS LSRFortessa (BD Biosciences).
Luciferase reporter assays
For reporter assays, a region of wild-type 3′-untranslated region (3′UTR) from TNFα, the 3′UTR from mutated TNFα, 3′UTR from IL1β, and the target hsa-miR-181c (which is perfectly complementary to hsa-miR-181c sequence) were constructed annealing the following primers: 3′UTR-TNF-α.F: 5′-CTAGTATTATTTATTTATTTACAGATGAATGTATTTATTTG GGAGACCGGGGTA-3′ and 3′UTR-TNF-α.R: 5′-AGCTTACCCCGGTCTCCCAAATAAATACATTCATCTGTAAATAAATAAATAATA-3′; 3′UTR-TNF-α mutated. F: 5′-TCGAGATTATTTATTTATTTACAGATCTGCGTATTTATTTGGGAGACCGGGGTGC-3′ and 3′UTR-TNF-α mutated. R: 5′-GGCCGCACCCCGGTCTCCCAAATAAATACGCAGATCTGTAAATAAATAAATAATC-3′; 3′UTR-IL1-β.F: 5′-TCGAGGTACCCAG AGAGTCCTGTGCTGAATGTGGACTCAATCCCTAGGGCTG GGC-3′ and 3′UTR-IL1-β.R: 5′-GGCCGCCCAGCCCTAGGGATTGAGTCCACATTCAGCACAGGACTCTCTGGGTACC-3′; and 3′UTR-Target miRNA181c.F: 5′-TCGAGATTATTACTCACCGACAGGTTGAATGTT TTTATTTGGGAGACCGGGGTGC-3′ and 3′UTR-Target miRNA181c.R: 5′-GGCCGCACCCCGGTCTCCCAAATAAAAACATTCAACCTG TCGG TGAGTAATAATC-3′.
Annealing was conducted by incubating both primers for 4 minutes at 95°C and for 10 minutes at 70°C in annealing buffer (100mM potassium acetate, 30mM HEPES, pH 7.4, and 2mM magnesium acetate). Primers were then phosphorylated and cloned into the siCHECK2 vector from Promega digested with XhoI and NotI.
Fifty-thousand RKO cells were plated in DMEM containing 10% FBS. Twenty-four hours later, cells were transfected with the siCHECK vectors either with the Pre-miR control or the Pre-miR-181c using lipofectamine (Invitrogen) following the manufacturer's instructions. For inhibition experiments, an anti-miR control or an anti–miR-181c were transfected. For LCLs, nucleofection was conducted using the cell line optimization 96-well nucleofector kit from Amaxa according to the manufacturer's recommendations. Nucleofection was done by combining siCHECK-TNFα or siCHECK-mutTNFα with Pre-miR-181c. The luciferase reporter assay was performed 24-72 hours after transfection using the Dual-Glo luciferase kit (Promega).
The ratio between the firefly and the Renilla luciferase allows the normalization of luciferase values. Ratios were normalized against the control plasmid. RKO experiments with miRNA transfection were performed 5 times in duplicate, and anti-miR experiments were performed 3 times, also in duplicate. Three independent experiments were conducted in LCLs. In all cases, the expression of endogenous miRNA after transfection was analyzed by RT-qPCR. Statistical differences were determined using the Student t test.
In vitro culture of hematopoietic progenitors
For the assessment of colony forming cells, samples consisting of at least 150 000 mononuclear or erythrocyte-depleted BM cells were cultured at 37°C in 5% CO2 and 95% humidity in Methocult H4434 medium containing SCF, GM-CSF, IL-3, and erythropoietin as growth factors (StemCell Technologies) according to standardized procedures.29 Colonies were scored after 2 weeks in culture in an inverted microscope (Olympus IX70 WH10x/22) with a 4× objective.
Statistical analysis
Results are shown as the means ± SEM. Differences between groups were assessed using the 2-tailed Student t test. Statistical analysis of the data was performed using Prism Version 4.0 software (GraphPad).
Results
Down-regulated expression of specific miRNAs in LCLs and primary blood cells from FA-A patients
To study the involvement of miRNAs in FA, we initially determined the expression levels of 157 miRNAs in LCLs obtained from 6 HDs and 8 FA-A patients using the TaqMan miRNA assay kit. After normalizing the data using RNU6B as a control, we determined the mean expression level of each miRNA in the different FA cell lines and compared them with the mean values determined in control LCLs. These analyses showed a down-regulated expression of 8 miRNAs in FA LCLs: hsa-miR-99a, hsa-miR-133a, hsa-miR-135b, hsa-miR-139, hsa-miR-181c, hsa-miR-182, hsa-miR-183, and hsa-miR-199s (all of them P < .01 and significant analysis of microarrays score > 0.9; data not shown).
Fresh PB MNCs from 9 FA-A patients and 6 HDs were used to confirm data obtained in LCLs. In these experiments, the expression levels of 7 of the 8 miRNAs that were down-regulated in FA-LCLs were analyzed (hsa-miR-199s miRNA was not analyzed because it has been eliminated from the miRNA annotation list). As shown in Figure 1, no evident changes of expression were noted in 4 of 7 miRNAs, whereas a consistent down-regulated expression of 3 miRNAs, hsa-miR-133a, hsa-miR-135b, and hsa-miR-181c, was observed in fresh FA PB cells.
Comparative analysis of miRNA expression levels in primary blood cells from FA-A patients and HDs. The figure shows the relative expression of hsa-miR-99a, hsa-miR-133a, hsa-miR-135b, hsa-miR-139, hsa-miR-181c, hsa-miR-182, and hsa-miR-183 determined by qPCR in fresh MNCs obtained from HDs (■) and FA-A patients (▴). Top panels show miRNAs in which expression was not modified in samples from FA patients compared with those from HDs. Bottom panels show miRNAs significantly down-regulated in FA-A patients. Expression levels were related to the expression of the RNU6B gene and normalized to mean values corresponding to HD samples.
Comparative analysis of miRNA expression levels in primary blood cells from FA-A patients and HDs. The figure shows the relative expression of hsa-miR-99a, hsa-miR-133a, hsa-miR-135b, hsa-miR-139, hsa-miR-181c, hsa-miR-182, and hsa-miR-183 determined by qPCR in fresh MNCs obtained from HDs (■) and FA-A patients (▴). Top panels show miRNAs in which expression was not modified in samples from FA patients compared with those from HDs. Bottom panels show miRNAs significantly down-regulated in FA-A patients. Expression levels were related to the expression of the RNU6B gene and normalized to mean values corresponding to HD samples.
Because a reduced expression of hsa-miR-181c had been associated with the methylation of CpG sites located upstream of this miRNA in gastric carcinomas,30 we conducted these analyses in LCLs from 2 HDs and 2 FA-A patients, and down-regulated expression of hsa-miR-181c was confirmed. However, MSP analyses revealed that the CpG sites upstream of the hsa-miR-181c were strongly methylated in LCLs not only from FA patients but also from HDs (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Pyrosequencing of the 4 CpG dinucleotides located in the genomic sequence that results in mature miRNA hsa-miR-181c showed a similar methylation of the 4 CpG dinucleotides in all samples (supplemental Figure 1B).
hsa-miR-181c regulates the expression of TNFα by interacting with its 3′UTR region
To identify potential targets of the miRNAs that were down-regulated in FA samples, in silico analyses were performed with MIRANDA software (2008 and 2010). These analyses showed that hsa-miR-181c had 3 potential target genes, TNFα, IL-1β, and RAD54B, that could be relevant in the disease. Strikingly, 2 of these genes, TNFα and IL-1β, have been reported to be overexpressed in FA cells.28,31,32
To test the effect of hsa-miR-181c on mRNA expression of TNFα, IL-1β, and RAD54B, LCLs from different FA-A patients were transfected with a control pre-miRNA and Pre-miR-181c. As shown in Figure 2, the ectopic expression of Pre-miR-181c in FA-A LCLs significantly decreased the expression of TNFα, whereas no change in the mRNA expression of IL-1β and RAD54B was observed. Because several studies have shown TNFα overexpression in FA-C cells, the relevance of hsa-miR-181c in the down-regulation of TNFα was also confirmed in FA-C LCLs (supplemental Figure 2). Because miRNAs can also interfere the expression of their target genes by translational repression, our studies of RNA expression do not exclude the possibility that miR-181c may also reduce the IL-1β and/or RAD54B protein levels.
Relative expression of IL-1β, TNFα, and RAD54B in LCLs from FA-A patients transfected with a Pre-miR control and Pre-miR-181c. In all cases, mRNA expression levels were analyzed by qPCR 2 days after transfection with a Pre-miR control (white bars) or Pre-miR-181c (black bars). Expression levels were related to the expression of the human β-actin gene and normalized to mean values corresponding to LCLs transfected with the Pre-miR control. Bars corresponding to IL-1β and TNFα expression represent mean values ± SEM of data deduced from the analysis of 3 FA patients. In the case of RAD54B, data from 2 patients are shown. For each determination, 3 independent experiments were conducted.
Relative expression of IL-1β, TNFα, and RAD54B in LCLs from FA-A patients transfected with a Pre-miR control and Pre-miR-181c. In all cases, mRNA expression levels were analyzed by qPCR 2 days after transfection with a Pre-miR control (white bars) or Pre-miR-181c (black bars). Expression levels were related to the expression of the human β-actin gene and normalized to mean values corresponding to LCLs transfected with the Pre-miR control. Bars corresponding to IL-1β and TNFα expression represent mean values ± SEM of data deduced from the analysis of 3 FA patients. In the case of RAD54B, data from 2 patients are shown. For each determination, 3 independent experiments were conducted.
To investigate whether hsa-miR-181c interacts with the TNFα mRNA, we cloned the 3′UTR region that is hypothetically recognized by the miRNA in a luciferase reporter vector (siCHECK2-TNFα). In addition, a similar vector carrying the 3′UTR from the IL-1β was generated (siCHECK2-IL1β). As a negative control, a vector carrying a mutated 3′UTR of TNFα was constructed (siCHECK2-mutTNFα). As a positive control, a vector with the sequence that exactly resembles the region recognized by the hsa-miR-181c (siCHECK2-TargetmiR181c) was also generated.
In preliminary experiments, we confirmed that the endogenous expression of hsa-miR-181c in RKO cells was similar to that observed in LCLs from HDs (data not shown). To investigate whether the endogenous expression of hsa-miR-181c in RKO cells could be sufficient to inhibit the expression of the Renilla luciferase fused to TNFα 3′UTR, the luciferase activity was first determined in RKO cells transfected with siCHECK2-TNFα and also with the negative and positive controls (siCHECK2-mutTNFα and siCHECK2-TargetmiR181c, respectively). As shown in Figure 3A, a significant decrease in the Renilla luciferase activity (normalized to firefly luciferase) was observed in RKO cells transfected with the positive control (siCHECK2-TargetmiR181c) and with the siCHECK2-TNFα compared with cells transfected with the negative control (siCHECK2-TNFα mutated).
Pre-miR-181c regulates the expression of TNF-α by interacting with its 3′UTR sequence. Shown is the normalized Renilla luciferase activity in RKO cells transfected with the different constructs. (A) RKO cells were transfected with the different siCHECK2 plasmids. (B) RKO cells were co-transfected with Pre-miR control (white bars) or Pre-miR-181c (black bars) together with the different siCHECK2 plasmids. (C) LCLs from HDs were transfected with siCHECK2-mutTNFα or siCHECK2-TNFα and Pre-miR-181c. (D) RKO cells were co-transfected with anti-miR control (white bars) or with an anti–miR-181c (black bars) together with the different siCHECK2 plasmids. In all cases, Renilla luciferase activity was normalized to firefly luciferase activity. Bars show mean values ± SE corresponding to 3-5 independent experiments.
Pre-miR-181c regulates the expression of TNF-α by interacting with its 3′UTR sequence. Shown is the normalized Renilla luciferase activity in RKO cells transfected with the different constructs. (A) RKO cells were transfected with the different siCHECK2 plasmids. (B) RKO cells were co-transfected with Pre-miR control (white bars) or Pre-miR-181c (black bars) together with the different siCHECK2 plasmids. (C) LCLs from HDs were transfected with siCHECK2-mutTNFα or siCHECK2-TNFα and Pre-miR-181c. (D) RKO cells were co-transfected with anti-miR control (white bars) or with an anti–miR-181c (black bars) together with the different siCHECK2 plasmids. In all cases, Renilla luciferase activity was normalized to firefly luciferase activity. Bars show mean values ± SE corresponding to 3-5 independent experiments.
Because other miRNAs apart from hsa-miR-181c that are potentially expressed in RKO cells could account for the down-regulated Renilla luciferase activity observed in siCHECK2-TNFα–transfected cells, in the next set of experiments, RKO cells were co-transfected with different siCHECK2 vectors: siCHECK-TNFα, siCHECK-IL1β, or siCHECK-mutTNFα and also with a Pre-miR control (Figure 3B white bars) or with the Pre-miR-181c (Figure 3B black bars). Three days after co-transfection, the relative Renilla luciferase activity was determined. When cells were transfected with siCHECK-TNFα, a significant decrease of Renilla luciferase activity was induced by the co-transfection with Pre-miR-181c compared with the Pre-miR control (Figure 3B first 2 bars). In samples transfected with siCHECK-IL1β, no significant inhibition was observed between groups co-transfected with Pre-miR-181c and the control Pre-miR (Figure 3B), which agrees well with the data shown in Figure 2. As expected, when siCHECK-mutTNFα was used, no inhibition was induced by Pre-miR-181c compared with the control Pre-miR (Figure 3B).
To verify the interaction of miR-181c with the TNFα 3′UTR region in hematopoietic cells, experiments similar to those shown in RKO cells (Figure 3B) were conducted with LCLs co-transfected with Pre-miR-181c together with siCHECK-TNFα or siCHECK-mutTNFα. As was observed in RKO cells, LCLs transfected with the siCHECK-TNFα showed a reduced luciferase activity compared with cells transfected with siCHECK-mutTNFα (Figure 3C). These results demonstrate that hsa-miR-181c down-regulates the expression of TNFα by the interaction with its 3′UTR.
Finally, to confirm that the TNFα 3′UTR region is a target of hsa-miR-181c, in the next set of experiments, RKO cells were co-transfected with siCHECK-TNFα, siCHECK2-IL1β, or siCHECK-mutTNFα, and also with an anti-miR control (Figure 3D white bars) or with an hsa-miR-181c anti-miR (Figure 3D black bars). Cells transfected with the siCHECK-TNFα vector showed an increased expression of Renilla luciferase activity after co-transfection with the hsa-miR-181c anti-miR compared with the control anti-miR (Figure 3D). Consistent with the data shown in Figure 2, no changes in expression were observed when the siCHECK-IL1β- or the siCHECK-TNFα–mutated vectors were used (Figure 3D). These results confirm that hsa-miR-181c targets the 3′UTR of TNFα, mediating a down-regulated expression of the gene.
Regulation of TNFα by hsa-miR-181c in BM cells from HDs and FA patients
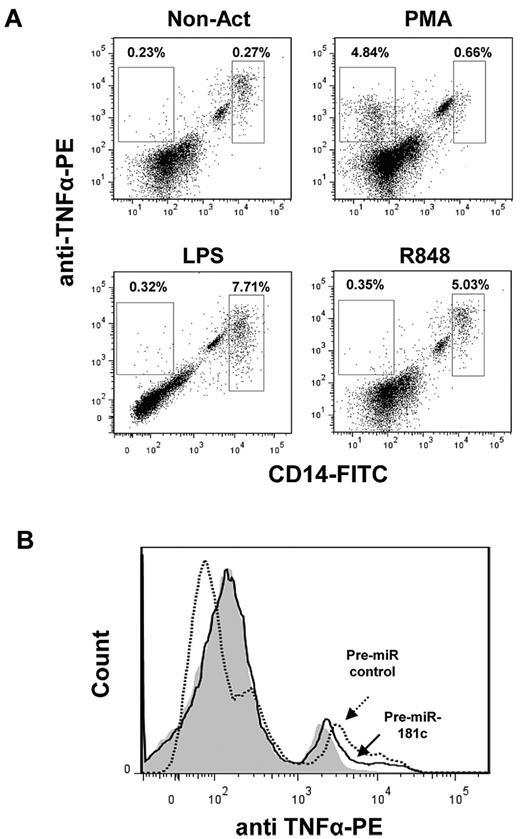
In a next set of experiments, we investigated the production of TNFα in BM cells from HDs and FA patients 48 hours after transfection with the control Pre-miR or with Pre-miR-181c. When TNFα was analyzed in untransfected BM samples from either HDs or FA patients, a very small number of cells expressed detectable levels of intracellular TNFα. Therefore, samples were activated with PMA, LPS, or R848. Activation with LPS or R848 markedly increased the proportion of TNFα-expressing cells, mainly within the CD14+ population, whereas the generation of TNFα-expressing CD14+ cells was much lower in PMA-stimulated cells (for representative analyses, see Figure 4A). When hsa-miR-181c was transfected into LPS- and R848-activated BM cells from a HD, moderate decreases in the intracellular levels of TNFα were observed (Figure 4B). When similar experiments were conducted with BM samples from FA-A patients, less consistent results were obtained. Whereas either LPS or R848 induced a significant TNFα overexpression in cells from 2 FA patients (FA-110 and FA-536), no TNFα overexpression was induced by these molecules on cells from a third FA patient (FA-13). In addition, whereas hsa-miR-181c down-regulated the expression of TNFα in LPS-activated cells, but not in R848-activated cells, from patient FA-110, only R848-activated cells from patient FA-536 showed a down-regulated expression of TNFα (not shown). The use of lentiviral vectors carrying the Pre-miR-181c and the EGFP marker gene clarified that FA cells with a down-regulated expression of TNFα corresponded to hsa-miR-181c–expressing cells (supplemental Figure 3).
Down-regulated expression of TNFα in LPS-activated BM cells transfected with Pre-miR-181c. (A) Representative flow cytometric analysis showing intracellular expression of TNFα in CD14+ cells in fresh BM and BM cells activated with PMA, LPS, or R848. (B) Representative analysis of intracellular TNFα in LPS-activated BM cells transfected with Pre-miR control or with Pre-miR-181c. Gray histogram corresponds to nonactivated cells; empty histograms correspond to LPS-activated cells transduced with the Pre-miR control (dotted line) and with Pre-miR-181c (continuous line). Data correspond to a BM sample from an HD.
Down-regulated expression of TNFα in LPS-activated BM cells transfected with Pre-miR-181c. (A) Representative flow cytometric analysis showing intracellular expression of TNFα in CD14+ cells in fresh BM and BM cells activated with PMA, LPS, or R848. (B) Representative analysis of intracellular TNFα in LPS-activated BM cells transfected with Pre-miR control or with Pre-miR-181c. Gray histogram corresponds to nonactivated cells; empty histograms correspond to LPS-activated cells transduced with the Pre-miR control (dotted line) and with Pre-miR-181c (continuous line). Data correspond to a BM sample from an HD.
hsa-miR-181c transfection in BM cells from FA patients reproducibly improves the growth of FA hematopoietic progenitors in vitro
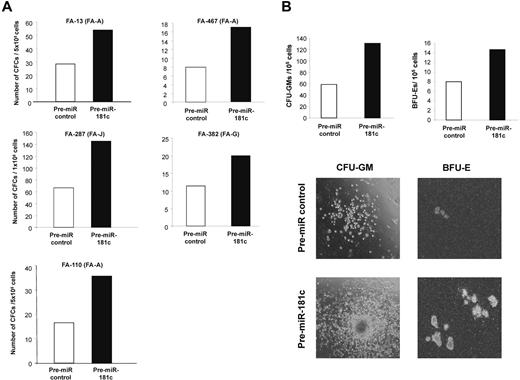
Finally, to investigate the functional consequences mediated by hsa-miR-181c on the growth of FA hematopoietic progenitors, BM cells from 5 FA patients without evidence of myelodysplasia or leukemia deduced from morphological and karyotypic assessment in BM aspirates were transfected with either the control Pre-miR or with Pre-miR-181c and then cultured in semisolid medium for 14 days. As shown in Figure 5A, hsa-miR-181c mediated a marked increase in the number of hematopoietic colonies in all tested FA BM samples. The effects of Pre-miR-181c were evident not only by the increased number of colonies, both myeloid and erythroid, but also by the larger size of the colonies (see representative analyses in Figure 5B). In all cases, the expression of hsa-miR-181c was confirmed by qPCR performed in pooled hematopoietic colonies from each patient (not shown).
Pre-miR-181c improves the clonogenicity of hematopoietic progenitors from FA patients. (A) Analysis of the number of hematopoietic progenitors from FA patients cultured in methylcellulose 48 hours after transfection with a Pre-miR control (white bars) or with Pre-miR-181c (black bars). (B) Representative analysis of CFU-GM and BFU-E colonies corresponding to patient FA-287. Photographs are from patient FA-13.
Pre-miR-181c improves the clonogenicity of hematopoietic progenitors from FA patients. (A) Analysis of the number of hematopoietic progenitors from FA patients cultured in methylcellulose 48 hours after transfection with a Pre-miR control (white bars) or with Pre-miR-181c (black bars). (B) Representative analysis of CFU-GM and BFU-E colonies corresponding to patient FA-287. Photographs are from patient FA-13.
Discussion
In the present study, we sought to determine the potential role of miRNAs in the hematologic manifestations of FA, a severe inherited disease associated with several cellular dysfunctions affecting the proliferation and differentiation of hematopoietic cells. Previous results from Gruber et al demonstrated that mice deficient in Ars2, a protein involved in proper miRNA processing, developed BM failure,33 reinforcing our hypothesis that several miRNAs might be altered in FA. Consistent with this hypothesis, our results showed the down-regulation of 8 miRNAs in LCLs from FA patients, 3 of which (hsa-miR-133a, hsa-miR-135b, and hsa-miR-181c) were also down-regulated in fresh MNCs from the PB of FA patients. Interestingly, one of the miRNAs that was systematically down-regulated in FA cells, hsa-miRNA-181c, had TNFα, IL-1β, and RAD54B as possible target genes, although only TNFα and IL-1β have been reported to be up-regulated in FA cells.28,31,32
Whereas the inhibitory studies with the siCHECK2 vector showed that hsa-miR-181c could target the TNFα 3′UTR, this effect was not observed with the IL-1β 3′UTR (Figure 3). The levels of TNFα down-regulation obtained in our luciferase experiments were similar to those described with hsa-miR-125b, another miRNA also capable of down-regulating TNFα.34 In that study, the investigators observed that miR-125b also targets the 3′UTR of TNFα transcripts and proposed that its down-regulation in response to LPS may be required for proper TNFα production.
Because of the relevance of TNFα in the etiology of the BM failure and cancer progression in FA, we focused our next studies on the effects of hsa-miR-181c on the expression of TNFα in FA BM cells and also on the growth of hematopoietic progenitors from FA patients. Early studies showed that altered responses to this cytokine had a critical role in the pathogenesis of acquired aplastic anemia because of the apoptotic loss of hematopoietic stem and progenitor cells (for review, see Young and Maciejewski35 ). Increased levels of this cytokine were found in the serum of FA patients36,37 and also in supernatants from LCLs37,38 and BM cells from these patients.28 In addition, the neutralization of the interaction of this cytokine with its receptor significantly increased the growth of BFU-E and CFU-GM progenitors from FA patients,28,39 which is consistent with mechanistic studies showing a deregulated apoptosis and hypersensitivity of FA BM progenitor cells exposed to TNFα.38,40-44 In addition to studies showing the relevance of TNFα on the etiology of FA BMF, studies in Fancc−/− mice have shown that this cytokine provides a selective pressure for apoptosis-resistant FA cells, thus facilitating the malignant progression of FA clones with accumulated mutations.45,46
Our data showing the interaction of hsa-miR-181c with the TNFα 3′UTR (Figure 3) are consistent with analyses indicating that this miRNA down-regulates the expression of TNFα in LCLs and primary cells from HDs and FA patients (Figure 2, Figure 4, and supplemental Figure 3). These observations, together with our data showing a reduced expression of hsa-miR-181c in FA cells (Figure 1), indicate that, in addition to other molecules such as MMP-7, this miRNA could be involved in the altered levels of expression/secretion of TNFα in FA cells.31
The functional relevance of hsa-miR-181c on the hematopoiesis of FA patients is apparent from our in vitro culture experiments conducted with BM cells from FA patients (Figure 5). As was the case in cultures of FA BM cells treated with TNFα fusion protein inhibitors,28,39 our data clearly show (Figure 5) that the expression of hsa-miR-181c on the BM of a FA patient improved the growth of myeloid and erythroid progenitors significantly, probably because of the down-regulated expression of TNFα in FA progenitor and/or mature cells (ie, CD14+ cells present in transfected BM or generated during the colony growth).
The fact that BM samples from FA patients corresponding to 3 different complementation groups (FA-A, FA-G, and FA-J) showed similar improvements in colony growth after the ectopic expression of hsa-miR-181c suggests that the down-regulated expression of this miRNA in FA patients should be involved, either directly or indirectly, in the impaired growth of their hematopoietic progenitors independent of the FA complementation group. In addition, the observation of an improved colony growth in cultures of BM cells from patient FA-13, whose TNFα levels were low even after LPS or R848 activation, suggest that this miRNA might also target other negative regulators involved in the growth of hematopoietic progenitors. Our results, together with the fact that miRNAs can be modified to improve their stability after in vivo infusion,47-49 open the possibility of using miRNAs, in particular hsa-miR-181c, for the treatment of the BM failure in FA patients, as has already been done with TNFα inhibitors.50
Interestingly, when BM cells from a FA patient with acute myeloid leukemia were transfected with hsa-miR-181c, no increase in the number of colonies was observed (data not shown). Although further studies are necessary to confirm this observation, it is significant that studies conducted in Fancc−/− mice showed that malignant clones were not sensitive to TNFα,45,46 indicating that these malignant clones may have developed mechanisms of TNFα resistance and/or degradation.
The mechanisms accounting for the decreased expression of different miRNAs, including hsa-miR-181c, in FA cells still remains unclear. Epigenetic modifications of genes encoding for these miRNAs, such as promoter hypermethylation or histone modifications,51-53 may have a critical role in this effect. Moreover, previous results in studies of gastric carcinoma have shown that hsa-miR-181c can be down-regulated by methylation.30 However, methylation analyses conducted in LCLs from HDs and FA-A patients showed a similar heavy methylation of CpG sites located near the hsa-miR-181c and of the CpG dinucleotides located in the mature hsa-miR-181c. These results strongly suggest that the methylation of these CpG sites is not the mechanism that accounts for the down-regulation of this miRNA in FA cells.
The study from O'Connell et al5 showing that hsa-miR-181c is enriched in mouse HSCs and human CB CD34+ cells is of particular interest because of the reduced expression of this miRNA in FA hematopoietic cells and its role in both TNFα down-regulation and hematopoietic progenitor cell growth. In this context, it would be of interest to determine hsa-miR-181c levels in the very rare population of FA CD34+ and HSCs. Similarly, understanding the involvement of this miRNA in the homing and repopulating properties of FA-HSCs would be highly relevant, considering the data by O'Connell et al showing that the ectopic expression of this miRNA impaired the competitive repopulating ability of BM cells from healthy mice.
In the present study, we have shown for the first time that FA hematopoietic cells are characterized by a down-regulated signature of several miRNAs, and have demonstrated that one of these down-regulated miRNAs, hsa-miR-181c, interacts with the 3′UTR of TNFα, inhibiting its expression and toxic effects in hematopoietic progenitors from FA patients. These observations offer new clues to understand the biologic basis of BM failure in FA patients and may help in the development of new therapeutic strategies for the treatment of this severe disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Aurora de La Cal and Sergio Losada (Centro de Investigaciones Energéticas, Medioambientales y Tecnológicas and Centro de Investigación Biomédica en Red de Enfermedades Raras) for technical assistance and the Fundación Marcelino Botín for promoting translational research at the División de Hematopoyesis y Terapia Génica at the Centro de Investigaciones Energéticas, Medioambientales y Tecnológicas. Centro de Investigación Biomédica en Red de Enfermedades Raras is an initiative of the Instituto de Salud Carlos III.
This study was supported by grants from the European program 7FWP, Health (PERSIST; grant 222878); the Ministry of Science and Innovation Programa de Fomento de Cooperación Científica Internacional (110-90.1) and Plan Nacional de Salud y Farmacia (SAF 2009-07164); and Fondo de Investigaciones Sanitarias, ISCIII (Programa RETICS-RD06/0010/0015).
Authorship
Contribution: P.R. and X.A. designed the research, performed the experiments, and wrote the manuscript; L.G., R.B., L.A., E.S.J.-E., and M.G. performed the experiments; I.B. and J.A.C. contributed vital new reagents; and F.P. and J.A.B designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Juan A. Bueren, Hematopoiesis and Gene Therapy Division, CIEMAT/CIBERER, Avenida Complutense 22, 28040 Madrid, Spain; e-mail: juan.bueren@ciemat.es.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal