Abstract
Hypoxia-inducible factor-1α (HIF1α), a master transcriptional regulator of the cellular and systemic hypoxia response, is essential for the maintenance of self-renewal capacity of normal HSCs. It is still unknown whether HIF1α has a role in survival regulation of leukemia stem cells (LSCs) in chronic myeloid leukemia (CML). Using a mouse model of CML, here we report that HIF1α plays a crucial role in survival maintenance of LSCs. Deletion of HIF1α impairs the propagation of CML through impairing cell-cycle progression and inducing apoptosis of LSCs. Deletion of HIF1α results in elevated expression of p16Ink4a and p19Arf in LSCs, and knockdown of p16Ink4a and p19Arf rescues the defective colony-forming ability of HIF1α−/− LSCs. Compared with normal HSCs, LSCs appear to be more dependent on the HIF1α pathway. Together, these results demonstrate that HIF1α represents a critical pathway in LSCs and inhibition of the HIF1α pathway provides a therapeutic strategy for eradicating LSCs in CML.
Introduction
Chronic myeloid leukemia (CML) is a clonal HSC disorder associated with the Philadelphia chromosome resulting from a reciprocal translocation between chromosomes 9 and 22. The molecular basis of this chromosomal translocation is the formation of the chimeric BCR-ABL protein that functions as a constitutively activated tyrosine kinase.1-3 Although BCR-ABL kinase inhibitors are highly effective in treating chronic-phase CML patients,4 they do not efficiently kill leukemia stem cells (LSCs).5 It is generally accepted that eradication of LSCs is required for curing CML. On the one hand, LSCs share similar mechanisms for self-renewal and survival with normal HSCs. For example, some developmental genes essential for normal HSCs, including Wnt and Hedgehog signaling pathways, Bmi-1, and p53, are involved in the regulation of both LSCs and HSCs.6-9 On the other hand, LSCs use pathways relatively specific for LSCs,10 providing a novel strategy for targeting LSCs while sparing normal HSCs. The challenge lies in the identification of genes that play a critical role in survival regulation of LSCs.
Mammalian BM provides a relatively hypoxic niche essential for maintaining the self-renewal and survival of primitive HSCs.11,12 It is widely accepted that gradients of O2 from below 1% in the hypoxic niche to 6% in the sinusoidal cavity exist in human BM.11 The hypoxic niche is crucial for BM function because in vitro culture of hematopoietic progenitors under hypoxic conditions displays a decrease in cellular proliferation accompanied by accumulation of the cells at the G0 phase of the cell cycle.11,13 Although the molecular mechanisms by which hypoxia regulates HSCs remain largely unknown, accumulating evidence suggests that hypoxia-inducible factor 1 (HIF1) plays a critical role in mediating the effect of hypoxia on HSCs.13-15 In addition, it has been shown that cyclin-dependent kinase inhibitors that are regulated by HIF1α are associated with the function of hypoxia in HSCs.15 A recent study on HSCs in an animal model showed that regulation of the HIF1α level is essential for the survival of HSCs.16 In this study, it was found that normal HSCs maintain intracellular hypoxia and stabilize HIF1α protein, and HIF1α deficiency resulted in the loss of cell-cycle quiescence in a p16Ink4a/p19Arf-dependent manner, and subsequently impaired the reconstitution ability of HSCs during various stress settings including serial transplantation, myelosuppression, and aging.
HIF1 belongs to the family of basic helix-loop-helix (bHLH) transcription factors, and is a heterodimer that consists of a constitutively expressed HIF1β subunit and a HIF1α subunit.17 The expression of HIF1α is regulated in multiple ways. Under normoxic conditions, HIF1α is degraded rapidly, which is triggered by the oxygen-hydroxylation of proline residues 402 and 564 by a proline hydroxylase (PHD). The hydroxylated HIF1α is recognized by the Von Hippel-Lindau protein (pVHL), which is the recognition component of an E3 ubiquitin-protein ligase.18,19 HIF1α expression is also regulated by growth factors, cytokines, and other signaling pathways such as PI3K and MAPK pathways.20
Recent studies demonstrate that HIF1α plays a role in cancer progression by activating transcriptional programs for maintaining the ability of self-renewal and multipotency of cancer stem cells in a hypoxic environment,21-23 and that HIF1α was required for stem cell functions of mouse lymphoma and human acute myeloid leukemia.24 However, it is still unknown whether HIF1α plays a role in LSCs in CML. In this study we show that HIF1α is up-regulated in BCR-ABL–expressing LSCs and is required for survival maintenance of LSCs in CML.
Methods
Mice
HIF1αflox/flox C57BL/6J, Vav1-Cre-C57BL/6J, C57BL/6J-CD45.1, and C57BL/6J-CD45.2 mice were obtained from The Jackson Laboratory. All mice were bred and maintained in a temperature- and humidity-controlled environment and given unrestricted access to a 6% chow diet and acidified water.
Generation of retroviral stocks
The retroviral constructs MSCV-IRES-GFP, MSCV-BCR-ABL-IRES-GFP were used to generate high-titer, helper-free, replication-defective ecotropic viral stocks by transient transfection of 293T cells as previously described.25 The MSCV-BCR-ABL-iCre-GFP construct was made by cloning the iCre ORF into the MSCV-BCR-ABL vector, and cloning the IRES-GFP fragment after the iCre ORF, as described previously.26
BM transduction/transplantation
Eight- to 12-week-old C57BL/6 mice were used for BM transduction/transplantation. Retroviral transduction and transplantation of mouse BM cells for inducing CML by BCR-ABL had been described previously.25,27 For secondary transplantation, an equal number of BM cells from primary CML mice was transplanted into lethally irradiated recipient mice.
Flow cytometric analysis
For stem cell analysis, BM cells were incubated with a biotin-labeled lineage Ab cocktail containing a mixture of Abs. After washing, the fluorochrome-labeled secondary Ab (allophycocyanin-Cy7–conjugated streptavidin) for recognizing biotin and PE-conjugated c-Kit and allophycocyanin-conjugated Sca-1 Abs were added to the cells. Long-term and short-term stem cells were distinguished by CD34 and CD135 or SLAM markers CD41, CD48, and CD150. All Abs were purchased from eBioscience.
Cell-cycle analysis of LSCs was performed by staining cells with Abs in combination with Hoechst 33342, followed by flow cytometric analysis. For Ki67 staining, cells were fixed and permeabilized before staining with PerCP-Cy5.5–conjugated anti-Ki67 Ab (from BD Biosciences). To analyze apoptosis of BM cells, the cells were stained with annexin V (from eBioscience), and 7AAD was added before flow cytometric analysis. For active caspase 3 staining, cells were first surface stained, fixed, and permeabilized (Fixation and Permeabilization kit; eBioscience), and stained with PE-conjugated anti–active caspase 3 Ab (from BD Biosciences).
In vitro methylcellulose colony formation assay
Normal stem cells or LSCs (GFP+Lin−Sca-1+c-Kit+) from BM of control and CML mice receiving empty vector or BCR-ABL–transduced WT or HIF1α−/− BM cells were sorted by FACS, and were cultured in methylcellulose medium (Methocult GF M3434; StemCell Technologies) at 37°C in humidified air for 7 days. Colonies were counted under microscope. For the colony forming assay after shRNA-mediated knockdown of CDKN2A, CML BM cells were cultured in methylcellulose media containing 2.5 μg/mL puromycin for selection.
Senescence-associated β-galactosidase staining
LSCs (GFP+Lin−Sca-1+c-Kit+) from BM of CML mice were sorted by FACS, and cytospun on a poly-lysine–coated slide and stained using the senescence-associated β-gal staining kit (Cell Signaling Technology). Micrograph was taken under a microscope (Olympus BX51), and all images were acquired using NIS-Elements BR3.10 software (NIS-Elements) and then constructed in Adobe Photoshop 7.0 (Adobe).
Lentiviral transduction and shRNA-mediated knockdown of p16Ink4a and p19Arf
Lentiviral shRNA vector pLKO.1 were purchased from OpenBiosystems. Sequences of CDKN2a (p16Ink4a and p19Arf) shRNA were as follows: shRNA no. 1: sense 5′-gtgatgatgatgggcaacgtt-3′: shRNA no. 1 antisense 5′-aacgttgcccatcatcatcac-3′; shRNA no. 2 sense 5′-gctcggctggatgtgcgcgat-3′ no. 2 antisense 5′-atcgcgcacatccagccgagc-3′. For transduction of the CDKN2A shRNAs, p53−/−MEF or CML BM cells were transfected with lentirvirus-shRNA, and 24 hours later, were selected under puromycin (2.5 μg/mL) for 48 hours before the next steps.
Abs and Western blot analysis
Abs against p16Ink4a, p19Arf, and actin were purchased from Santa Cruz Biotechnology. Protein lysates were prepared by lysing cells in immunoprecipitation buffer, and Western blotting was carried out as shown previously.28
Real-time PCR
Total RNA was isolated from GFP+Lin−Sca-1+c-Kit+ BM cells from mice using the RNeasy Mini kit (QIAGEN). cDNA was synthesized using the Ovation-Pico cDNA synthesis method. The primer sequences are shown as following: HIF1a sense: tgagcttgctcatcagttgc, HIF1a antisense: ccatctgtgccttcatctca; VEGF sense: caggctgctgtaacgatgaa, VEGF antisense: tttcttgcgctttcgttttt; GLUT1 sense: gctgtgcttatgggcttctc, GLUT1 antisense: agaggccacaagtctgcatt; TGFa sense: agcatgtgtctgccactctg, antisense: tggatcagcacacaggtgat; PGK1 sense: caaggctttggagagtccag, PGK1 antisense: tgtgccaatctccatgttgt; p16Ink4a sense: cgaactctttcggtcgtaccc, p16Ink4a antisense: cgaatctgcaccgtagttgagc; p19Arf sense: gttcttggtcactgtgaggattcag, p19Arf antisense: ccatcatcatcacctggtccag; p53 sense: agagaccgccgaacagaaga, p53 antisense: ctgtagcatgggcatcctt; p57 sense: ctgacctcagacccaattc, p57 antisense: ctcagagaccggctcagttc.
DNA microarray and data analysis
Normal and BCR-ABL–expressing LSK cells (GFP+Lin−Sca-1+c-Kit+; 99% purity) were stored by FACS directly into RNAlater (Ambion) for DNA microarray assay (Affymetrix). To collectively assess HIF1α targets, the GSEA algorithm was used to evaluate the statistical significance of the expression of HIF1α targets.17,29,30 All microarray data are available on the Gene Expression Omnibus (GEO) under accession number GSE35183.
Statistical analysis
Results are given as mean ± SD. Statistical analysis was performed by the Student t test. For survival curves, P values were obtained using a log-rank test.
Results
Hypoxia-responsive genes are up-regulated by BCR-ABL in LSCs
We have shown that BCR-ABL–expressing HSCs (Lin−Sca-1+c-Kit+ cells, LSK cells) function as LSCs in CML mice because this cell population isolated from primary CML mice effectively transfers the disease into lethally irradiated secondary recipient mice.31 This provides us with an assay system to examine whether BCR-ABL alters expression of hypoxia-responsive genes. We conducted a DNA microarray analysis to compare the gene expression profiles between BCR-ABL–expressing LSK cells and normal LSK cells in our BM transplantation (BMT) mouse model of CML (GEO submission: GSE10912). We retrovirally transduced BM cells from C57BL/6J (B6) mice with BCR-ABL-GFP or GFP alone (as a normal HSC control) and transplanted the transduced cells into lethally irradiated B6 recipient mice to induce CML. Two weeks after BMT, we sorted GFP+LSK cells from BM of the mice, and total RNA was extracted from the sorted cells for the Affymetrix microarray analysis. We intended to determine the global status of HIF1α activity in BCR-ABL–expressing LSK cells, and found that the HIF1α and its targets were up-regulated by BCR-ABL in BCR-ABL–expressing LSK cells compared with the LSK cells transduced with GFP alone (Figure 1A-B), which is enriched for normal HSCs. We next examined expression of genes known to be specifically regulated by HIF1α, and found that expression of VEGF, GLUT1, and TGFα, except for PGK1, were significantly higher in BCR-ABL–expressing LSK cells than in normal LSK cells (Figure 1C). Real-time RT-PCR assay confirmed the up-regulation of HIF1α and hypoxia-responsive genes by BCR-ABL in BCR-ABL–expressing LSK cells (Figure 1D). PGK1 was also significantly up-regulated in BCR-ABL–expressing LSK cells (Figure 1D). Together, these results suggest that HIF1α is involved in functional regulation of LSCs and CML development.
Hypoxia-responsive genes are up-regulated in LSCs. BCR-ABL–expressing LSK cells and control LSK cells were sorted from mice receiving BCR-ABL or empty vector-transduced BM cells for DNA microarray analysis. (A) The heatmap compares the activation of the HIF1α signaling pathway in BCR-ABL–expressing LSK cells and control LSK cells. (B) Gene set enrichment analysis (GSEA) displays the expression profiling of HIF1α targets in BCR-ABL–expressing LSK cells (P < .001). (C) Microarray data showed higher expression of HIF1α, vascular endothelial growth factor (VEGF), glucose transporter type 1 (GLUT1), and transforming growth factor α (TGFα) in BCR-ABL–expressing LSK cells comparing to normal LSK cells. (D) Real-time RT-PCR was performed with primers specific for HIF1α, VEGF, GLUT1, TGFα, and PGK1. The results were normalized using actin as a control, and shown as mean ± SD. *P < .05; **P < .01.
Hypoxia-responsive genes are up-regulated in LSCs. BCR-ABL–expressing LSK cells and control LSK cells were sorted from mice receiving BCR-ABL or empty vector-transduced BM cells for DNA microarray analysis. (A) The heatmap compares the activation of the HIF1α signaling pathway in BCR-ABL–expressing LSK cells and control LSK cells. (B) Gene set enrichment analysis (GSEA) displays the expression profiling of HIF1α targets in BCR-ABL–expressing LSK cells (P < .001). (C) Microarray data showed higher expression of HIF1α, vascular endothelial growth factor (VEGF), glucose transporter type 1 (GLUT1), and transforming growth factor α (TGFα) in BCR-ABL–expressing LSK cells comparing to normal LSK cells. (D) Real-time RT-PCR was performed with primers specific for HIF1α, VEGF, GLUT1, TGFα, and PGK1. The results were normalized using actin as a control, and shown as mean ± SD. *P < .05; **P < .01.
HIF1α is essential for CML development
To determine the role of HIF1α in BCR-ABL leukemiogenesis, we asked whether HIF1α is required for CML development. Because deletion of the HIF1α gene is embryonic lethal,32 we crossed mice carrying a loxP-flanked HIF1α allele with Cre transgenic mice in which expression of Cre is driven by the Vav regulatory element (Figure 2A). Because Vav is an adaptor protein expressed predominantly in hematopoietic cells including the HSC population,33 the use of the Vav regulatory element causes the deletion of the HIF1α gene mainly in the hematopoietic system, allowing Vav-Cre-HIF1αflox/flox mice to survive after HIF1α deletion. To examine whether HIF1α was efficiently deleted in Vav-Cre-HIF1αflox/flox mice, we sorted LSK cells from BM and analyzed HIF1 expression by RT-PCR. We did not detect HIF1α expression in LSK cells of Vav-Cre-HIF1αflox/flox mice compared with HIF1αflox/flox mice (Figure 2A). For simplicity, Vav-Cre-HIF1αflox/flox mice will be referred to as HIF1α−/− mice and HIF1αflox/flox mice as wild-type (WT) mice.
HIF1α is essential for CML development. (A) Deletion of HIF1α in hematopoietic cells. Mice carrying the floxed HIF1α allele were crossed with Vav-Cre transgenic mice in which the Vav promoter drives the expression of the Cre recombinase. RT-PCR analysis showed that HIF1α was undetectable in sorted HSCs (Lin−Sca-1+c-Kit+) from HIF1αflox/flox-Vav-Cre mice. (B) Kaplan-Meier survival curves for primary and secondary recipients of empty vector or BCR-ABL–transduced BM cells from WT or HIF1α−/− donor mice. For secondary BM transplantation, BM cells from primary control and CML recipient mice which received empty vector or BCR-ABL–transduced WT and HIF1α−/− BM cells were analyzed by FACS, and BM cells containing equal number of WT or HIF1α−/− GFP+Lin−Sca-1+c-Kit+ cells along with 2 × 105 WT BM cells (CD45.1) were transplanted into each lethally irradiated secondary recipient mouse. (C) FACS analysis of GFP+Gr-1+ cells in PB of primary and secondary recipients of empty vector or BCR-ABL–transduced BM cells from WT or HIF1α−/− donor mice (n = 5). Mean values (± SD) are shown. (D) The total numbers of GFP+Gr-1+ cells in PB of primary and secondary recipients of empty vector or BCR-ABL–transduced BM cells from WT or HIF1α−/− donor mice (n = 5). Mean values (± SD) are shown. *P < .05; **P < .01; ***P < .001. (E) The spleen weight of primary and secondary recipients of empty vector or BCR-ABL–transduced BM cells from WT or HIF1α−/− donor mice (n = 12). Mean values (± SD) are shown; ***P < .001. (F) Gross appearance of the lungs and spleens showed severe lung hemorrhages and splenomegaly in secondary recipients of WT LSCs but not HIF1α−/− LSCs at day 14 after BMT, compared with those from recipients of WT and HIF1α−/− HSCs. (G) H&E staining of tissue sections from lung and spleen of secondary recipients. The scale bar represents 50 μm. (H) Kaplan-Meier survival curves for the secondary BM transplantation. BM cells from WT or HIF1αflox/flox mice were transduced with BCR-ABL-iCre-GFP retrovirus, and transplanted into lethally irradiated recipient mice. CML cells without HIF1α failed to induce CML disease in the secondary BMT. (I) The percentages of leukemia cells in PB at day 14 in WT and HIF1α−/− secondary CML mice.
HIF1α is essential for CML development. (A) Deletion of HIF1α in hematopoietic cells. Mice carrying the floxed HIF1α allele were crossed with Vav-Cre transgenic mice in which the Vav promoter drives the expression of the Cre recombinase. RT-PCR analysis showed that HIF1α was undetectable in sorted HSCs (Lin−Sca-1+c-Kit+) from HIF1αflox/flox-Vav-Cre mice. (B) Kaplan-Meier survival curves for primary and secondary recipients of empty vector or BCR-ABL–transduced BM cells from WT or HIF1α−/− donor mice. For secondary BM transplantation, BM cells from primary control and CML recipient mice which received empty vector or BCR-ABL–transduced WT and HIF1α−/− BM cells were analyzed by FACS, and BM cells containing equal number of WT or HIF1α−/− GFP+Lin−Sca-1+c-Kit+ cells along with 2 × 105 WT BM cells (CD45.1) were transplanted into each lethally irradiated secondary recipient mouse. (C) FACS analysis of GFP+Gr-1+ cells in PB of primary and secondary recipients of empty vector or BCR-ABL–transduced BM cells from WT or HIF1α−/− donor mice (n = 5). Mean values (± SD) are shown. (D) The total numbers of GFP+Gr-1+ cells in PB of primary and secondary recipients of empty vector or BCR-ABL–transduced BM cells from WT or HIF1α−/− donor mice (n = 5). Mean values (± SD) are shown. *P < .05; **P < .01; ***P < .001. (E) The spleen weight of primary and secondary recipients of empty vector or BCR-ABL–transduced BM cells from WT or HIF1α−/− donor mice (n = 12). Mean values (± SD) are shown; ***P < .001. (F) Gross appearance of the lungs and spleens showed severe lung hemorrhages and splenomegaly in secondary recipients of WT LSCs but not HIF1α−/− LSCs at day 14 after BMT, compared with those from recipients of WT and HIF1α−/− HSCs. (G) H&E staining of tissue sections from lung and spleen of secondary recipients. The scale bar represents 50 μm. (H) Kaplan-Meier survival curves for the secondary BM transplantation. BM cells from WT or HIF1αflox/flox mice were transduced with BCR-ABL-iCre-GFP retrovirus, and transplanted into lethally irradiated recipient mice. CML cells without HIF1α failed to induce CML disease in the secondary BMT. (I) The percentages of leukemia cells in PB at day 14 in WT and HIF1α−/− secondary CML mice.
We first studied the role of HIF1α in CML development using HIF1α−/− mice in our BMT model. BM cells from 5-FU–treated WT or HIF1α−/− mice were transduced with empty vector (GFP alone) or BCR-ABL-GFP retrovirus, and then transplanted into lethally irradiated recipient mice. Although recipients of empty vector-transduced WT or HIF1α−/− BM cells did not develop CML disease, recipients of BCR-ABL–transduced WT or HIF1α−/− BM cells developed and succumbed to CML within 4 weeks after BMT; however, no significant difference in survival was observed in WT and HIF1α−/− CML mice (Figure 2B), which correlated with similar percentages at day 14 after BMT (Figure 2C) and total numbers of myeloid leukemia cells in peripheral blood (PB) of CML mice as analyzed at day 8 and 11 after BMT (Figure 2D). However, we noticed that at day 14 after BMT, the number of PB leukemia cells in mice receiving the BCR-ABL–transduced HIF1α−/− BM cells was significantly less than that in mice receiving the BCR-ABL–transduced WT marrow cells (Figure 2D). This observation suggests a possibility that HIF1α plays a role in functional regulation of LSCs in CML. We also examined the spleens and BM of CML mice, and found both WT and HIF1α−/− CML mice displayed similar percentages of leukemia cells in the spleens and BM at 2 weeks after BMT (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In addition, there was no significant difference in the degree of splenomegaly between the 2 groups of the CML mice (Figure 2E).
Therefore, we decided to do a secondary transplantation assay to biologically assess the effect of HIF1α on LSCs. We induced primary CML by transducing BM cells from WT or HIF1α−/− mice, followed by transplantation of the transduced cells into WT recipient mice. We then conducted a secondary BMT by transferring BM cells from primary CML mice at day 14 to secondary recipient mice. Before the transplantation, we analyzed and normalized LSCs (GFP+LSK) from BM of primary CML mice receiving BCR-ABL–transduced WT or HIF1α−/− donor BM cells to ensure that equal numbers of LSCs were injected for the 2 transplantation groups. For control, we also transplanted equal numbers of normal GFP+LSK cells from primary control mice receiving empty vector-transduced WT or HIF1α−/− donor BM cells into secondary recipient mice. To match up with the time frames for CML transplantation, we did secondary transplantation on day 14 after primary transplantation and analyzed the secondary recipient mice within a month. HIF1α−/− LSCs failed to induce CML in the secondary recipient mice, whereas WT LSCs efficiently induced CML and all secondary recipient mice died within 4 weeks post-BMT (Figure 2B). The defective CML phenotype in the absence of HIF1α was consistent with a gradual decrease of the percentages and total numbers of leukemia cells in PB (Figure 2C-D). Unlike CML secondary transplantation (Figure 2C), we observed that the percentages and total numbers of GFP+ cells in PB of non-BCR-ABL mice gradually decreased to low levels in secondary recipients of WT or HIF1α−/− BM cells (Figure 2C-D), presumably because serial BMT within a short time period significantly increases the replicative stress, affecting the cycling ability of control HSCs.34 In addition, recipients of HIF1α−/− LSCs displayed much less severe splenomegaly than did recipients of WT LSCs (Figure 2E-F). Consistently, histologic analysis showed no or modest infiltration of leukemia cells in the lung and spleen of the secondary recipient mice receiving HIF1α−/− LSCs from primary CML mice (Figure 2G). These results indicate that HIF1α is required for CML development.
Constitutive expression of Cre may be toxic.35 It is possible that Vav-driven expression of Cre causes damage on HSCs before BCR-ABL transduction, resulting in the defective CML development. In addition, 5-FU is more detrimental to HIF1α−/− HSCs,16 potentially causing a defect in CML development. To examine these possibilities, we constructed BCR-ABL-iCre-GFP retroviral construct to allow coexpression of BCR-ABL and iCre in the same cell.26 We transduced BM cells from both WT and HIF1αflow/flow mice with BCR-ABL-iCre-GFP retrovirus, and transplanted into recipient mice. If Cre and 5-FU are toxic to BM cells, both WT and HIF1αflow/flow cells should be equally affected, allowing us to assess the additional effect of HIF1α deletion on CML development. Recipients of BCR-ABL-iCre–transduced WT and HIF1αflow/flow BM cells similarly developed CML, consistent with the findings for primary CML using WT and HIF1α−/− mice (Figure 2B). The secondary BMT confirmed the failure of CML development after deletion of HIF1α (Figure 2H-I).
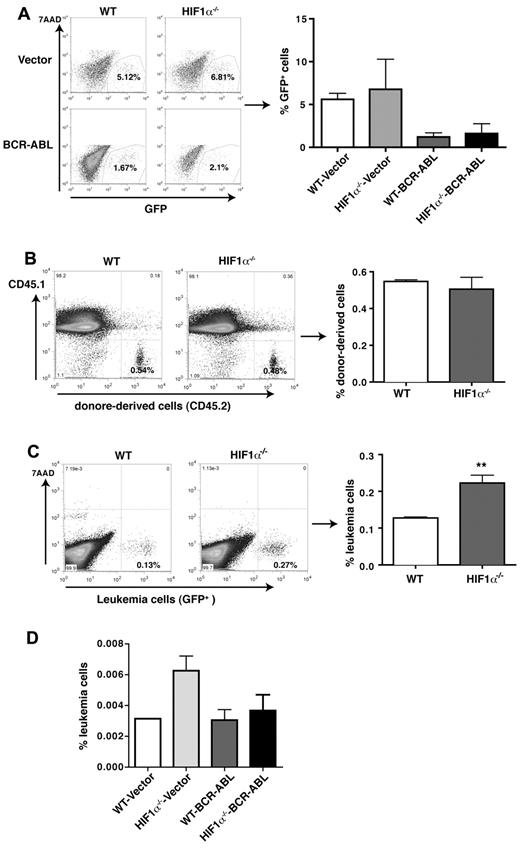
The failure of CML induction in the secondary recipients could also be related to a change in retroviral transduction efficiency caused by the deletion of HIF1α. To rule out this possibility, we compared GFP expression between empty GFP vector and BCR-ABL-GFP–transduced WT or HIF1α−/− BM cells in culture by FACS before BMT, and found that the percentages of GFP+ cells were similar between the transduced WT and HIF1α−/− cells (Figure 3A). These results indicate that HIF1α deletion does not affect retroviral transduction efficiency. In addition, a failure of cell homing potentially resulted from the loss of HIF1α may cause the failure of CML induction. To rule out this possibility, we performed an in vivo homing analysis comparing the ability of WT and HIF1α−/− BM cells. An equal number of WT or HIF1α−/− BM cells was injected into lethally irradiated CD45.1 mice. Three hours after BMT, FACS analysis showed that the percentages of WT and HIF1α−/− donor-derived cells represented by CD45.2 were similar (Figure 3B). To confirm this result in BCR-ABL–expressing leukemia cells, we transplanted an equal number of leukemia cells from BM of primary CML mice receiving BCR-ABL–transduced WT or HIF1α−/− BM cells into recipients. Three hours after BMT, we did not observe a defective homing ability of BCR-ABL–transduced HIF1α−/− cells (Figure 3C). Furthermore, using the same strategy, we compared the homing ability of WT or HIF1α−/− LSCs with that of WT or HIF1α−/− normal stem cells, and did not observe a reduced homing ability of HIF1α−/− LSCs and normal stem cells (Figure 3D).
Effects of HIF1α deletion on retroviral transduction efficiency and the homing ability of normal and BCR-ABL–transduced totalBMcells or LSK cells. (A) HIF1α deletion does not affect retroviral transduction efficiency. WT and HIF1α−/− BM cells were transduced with empty vector or BCR-ABL-GFP retrovirus, and 2 days later, the percentages of GFP+ cells were determined by FACS. (B) FACS analysis showed the similar homing ability of WT and HIF1α−/− BM cells. A total of 3 × 106 BM cells from WT or HIF1α−/− mice (CD45.2) were transplanted into lethally irradiated recipients (CD45.1). The donor-derived BM cells (CD45.2) were detected by FACS in 3 hours after BMT (n = 4). Mean values (± SD) are shown. NS indicates no significance. (C) HIF1α deletion does not cause a reduction of the homing ability of BCR-ABL–transduced BM cells. A total of 2 × 106 leukemia cells (GFP+) from CML mice transplanted with BCR-ABL–transduced WT or HIF1α−/− BM cells were transplanted into lethally irradiated recipients (CD45.1). Three hours later, donor-derived leukemia cells (GFP+) in the BM were detected by FACS (n = 4 for each group). Mean values (± SD) are shown; **P < .01. (D) HIF1α deletion does not cause a reduction of the homing ability of control or BCR-ABL–transduced stem cells. BM cells were transduced with GFP vector or BCR-ABL-GFP, and 2 × 104 sorted normal LSK cells and BCR-ABL–expressing LSK cells (GFP+Lin−Sca-1+c-Kit+) were transplanted into lethally irradiated recipients (CD45.1). Three hours later, donor-derived leukemia cells (GFP+) in the BM were detected by FACS (n = 3 for each group). Mean values (± SD) are shown.
Effects of HIF1α deletion on retroviral transduction efficiency and the homing ability of normal and BCR-ABL–transduced totalBMcells or LSK cells. (A) HIF1α deletion does not affect retroviral transduction efficiency. WT and HIF1α−/− BM cells were transduced with empty vector or BCR-ABL-GFP retrovirus, and 2 days later, the percentages of GFP+ cells were determined by FACS. (B) FACS analysis showed the similar homing ability of WT and HIF1α−/− BM cells. A total of 3 × 106 BM cells from WT or HIF1α−/− mice (CD45.2) were transplanted into lethally irradiated recipients (CD45.1). The donor-derived BM cells (CD45.2) were detected by FACS in 3 hours after BMT (n = 4). Mean values (± SD) are shown. NS indicates no significance. (C) HIF1α deletion does not cause a reduction of the homing ability of BCR-ABL–transduced BM cells. A total of 2 × 106 leukemia cells (GFP+) from CML mice transplanted with BCR-ABL–transduced WT or HIF1α−/− BM cells were transplanted into lethally irradiated recipients (CD45.1). Three hours later, donor-derived leukemia cells (GFP+) in the BM were detected by FACS (n = 4 for each group). Mean values (± SD) are shown; **P < .01. (D) HIF1α deletion does not cause a reduction of the homing ability of control or BCR-ABL–transduced stem cells. BM cells were transduced with GFP vector or BCR-ABL-GFP, and 2 × 104 sorted normal LSK cells and BCR-ABL–expressing LSK cells (GFP+Lin−Sca-1+c-Kit+) were transplanted into lethally irradiated recipients (CD45.1). Three hours later, donor-derived leukemia cells (GFP+) in the BM were detected by FACS (n = 3 for each group). Mean values (± SD) are shown.
HIF1α is essential for survival maintenance of LSCs
Because CML is derived from a stem cell,1 an inhibitory effect of HIF1α deficiency on LSCs is likely a cause for the defect of CML development (Figure 2). To test this idea, we first examined LSCs in primary CML mice, and did not observe significant differences in the percentages and total numbers of LSCs (GFP+LSK and GFP+CD48−CD150+LSK) between WT and HIF1α−/− CML mice (supplemental Figure 1B-C), consistent with the similar survival of WT and HIF1α−/− primary CML mice (Figure 2B). We further analyzed LSCs in the secondary recipient mice receiving WT or HIF1α−/− LSCs from primary CML mice (Figure 4A). Two weeks after the secondary BMT, the percentages and numbers of LSCs (GFP+LSK and GFP+CD48−CD150+LSK) in the BM were dramatically lower in the absence of HIF1α (Figure 4B-C), indicating that HIF1α is required for survival maintenance of LSCs.
HIF1α deletion suppresses LSCs. (A) FACS analysis of LSCs in BM of secondary recipients of WT or HIF1α−/− BM cells from primary CML mice. (B-C) The percentages and total numbers of stem cells (GFP+Lin−Sca-1+c-Kit+ cells and GFP+CD48−CD150+Lin−Sca-1+c-Kit+ cells) in BM of secondary recipient mice receiving BM cells from WT or HIF1α−/− control and CML mice were analyzed by FACS; *P < .05; **P < .01. (D) The loss of HIF1α causes a decrease in the colony-forming ability of BCR-ABL–expressing LSK cells. Sorted normal LSK cells and BCR-ABL–expressing LSK cells (GFP+Lin−Sca-1+c-Kit+ cells) were plated into methycellulose medium, colonies were counted, and cells were serially replated. Data show results from representative experiments. Mean values (± SEM) are shown; *P < .05; ***P < .001.
HIF1α deletion suppresses LSCs. (A) FACS analysis of LSCs in BM of secondary recipients of WT or HIF1α−/− BM cells from primary CML mice. (B-C) The percentages and total numbers of stem cells (GFP+Lin−Sca-1+c-Kit+ cells and GFP+CD48−CD150+Lin−Sca-1+c-Kit+ cells) in BM of secondary recipient mice receiving BM cells from WT or HIF1α−/− control and CML mice were analyzed by FACS; *P < .05; **P < .01. (D) The loss of HIF1α causes a decrease in the colony-forming ability of BCR-ABL–expressing LSK cells. Sorted normal LSK cells and BCR-ABL–expressing LSK cells (GFP+Lin−Sca-1+c-Kit+ cells) were plated into methycellulose medium, colonies were counted, and cells were serially replated. Data show results from representative experiments. Mean values (± SEM) are shown; *P < .05; ***P < .001.
To investigate whether HIF1α affects self-renewal of LSCs, we carried out an in vitro serial colony-forming assay. We sorted normal stem cells and LSCs (GFP+LSK) from control or CML mice receiving empty vector or BCR-ABL–transduced WT or HIF1α−/− BM cells, and plated the cells in vitro. We observed that comparing to WT LSK cells, HIF1α−/− LSK cells generated similar colonies in the first and second plating, and less colonies in the third plating. In contrast, compared with WT BCR-ABL–expressing LSK cells, HIF1α−/− BCR-ABL–expressing LSK cells gave rise to less colonies in all three platings, indicating that deletion of HIF1α results in a more significant reduction of self-renewal ability of LSCs (Figure 4D).
To understand the mechanism by which HIF1α maintains the survival of LSCs, we analyzed the effect of HIF1α on cell-cycle progression and apoptosis of BCR-ABL–expressing LSK cells. We found that deletion of HIF1α resulted in an accumulation of cells in the G0-G1 phase and a concomitant reduction in the S phase of the cell cycle (Figure 5A), suggesting that HIF1α deficiency impaired cell-cycle progression of LSCs. HIF1α is important for maintaining the quiescence of HSCs, and loss of HIF1α promotes cell-cycle entry.15,16 Therefore, we further examined the cell-cycle kinetics of HIF1α−/− LSCs using Ki67. A reduction in the Ki67− G0 fraction and an increase in the Ki67+ G1 fraction were observed in HIF1α−/− BCR-ABL–expressing LSK cells, compared with WT BCR-ABL–expressing LSK cells (Figure 5B). These results indicate that HIF1α regulates cell-cycle progression of LSCs. Furthermore, by staining the cells with annexin V and 7AAD, we found that HIF1α−/− BCR-ABL–expressing LSK cells had a higher apoptotic rate than WT BCR-ABL–expressing LSK cells (Figure 5D). To further demonstrate the induction of apoptosis of LSCs by HIF1α deficiency, we examined active caspase 3 in WT and HIF1α−/− leukemia progenitors. We observed an increase in the number of active caspase 3–positive cells among HIF1α−/− leukemia progenitor cells, compared with WT cells (Figure 5E). We also examined the effect of HIF1α on cell cycle and apoptosis of normal HSCs, and unlike the observed effects on cell-cycle progression and apoptosis of LSCs (Figure 5A,D top panels), we did not observe any significant differences between WT LSK cells and HIF1α−/− LSK cells (Figure 5A,D bottom panels).
HIF1α is required for survival maintenance of LSCs. (A) The cell-cycle analysis of normal LSK cells and BCR-ABL–expressing LSK cells. The percentages of HIF1α−/− BCR-ABL–expressing LSK cells were low in the S phase and higher in the G0-G1 phase, compared with WT BCR-ABL–expressing LSK cells (n = 6). However, normal WT and HIF1α−/− LSK cells showed similar percentages of cells in G0-G1, S, and G2-M phases. (B) The cell-cycle analysis of BCR-ABL–expressing LSK cells using Ki67 and Hoechst 33 342 showed an accumulation of G1 phase of HIF1α−/− BCR-ABL–expressing LSK cells. (C) RT-PCR analysis showed elevated expression of cell-cycle inhibitors p16Ink4a, p19Arf, and p57 in the sorted LSK cells and BCR-ABL–expressing LSK cells from HIF1α−/− control and CML mice compared with the sorted LSK cells and BCR-ABL–expressing LSK cells from WT control and CML mice. BM cells from control and CML mice at day 14 post-BMT were collected, and LSK cells and BCR-ABL–expressing LSK cells were sorted for extracting total RNA. Mean values (± SD) are shown; **P < .01; ***P < .001. (D) The apoptosis of normal LSK cells and BCR-ABL–expressing LSK cells from BM of control and CML mice (n = 6). HIF1α deletion induced the apoptosis of BCR-ABL–expressing LSK cells, but did not affect the apoptosis of normal LSK cells. (E) FACS analysis showed a higher percentage of active caspase positive HIF1α−/− leukemia progenitors. (F) RT-PCR analysis showed elevated expression of p53 in the sorted BCR-ABL–expressing LSK cells from HIF1α−/− CML mice comparing to the sorted BCR-ABL–expressing LSK cells from control CML mice. Mean values (± SD) are shown; **P < .01. (G) Simultaneous knockdown of p16Ink4a and p19Arf rescued the colony-forming ability of HIF1α−/− BCR-ABL–expressing LSK cells. Western blot analysis showed that protein levels of p16Ink4a and p19Arf in p53−/− MEF and CML leukemia cells were dramatically reduced by shRNA no. 1 that targets both p16Ink4a and p19Arf. BM cells from WT and HIF1α−/− normal or CML mice were infected by shRNA no. 1, and plated into methycellulose media after selection by puromycin. Colonies were counted and cells from the colonies were serially replated. Mean values (± SD) are shown; **P < .01; ***P < .001. (H) Loss of HIF1α did not affect the senescence of normal LSK cells and BCR-ABL–expressing LSK cells. Sorted GFP+Lin−Sca-1+c-Kit+ cells from the BM of WT or HIF1α−/− control and CML mice were stained by X-Gal, and the number of β-galactosidase–expressing cells were counted.
HIF1α is required for survival maintenance of LSCs. (A) The cell-cycle analysis of normal LSK cells and BCR-ABL–expressing LSK cells. The percentages of HIF1α−/− BCR-ABL–expressing LSK cells were low in the S phase and higher in the G0-G1 phase, compared with WT BCR-ABL–expressing LSK cells (n = 6). However, normal WT and HIF1α−/− LSK cells showed similar percentages of cells in G0-G1, S, and G2-M phases. (B) The cell-cycle analysis of BCR-ABL–expressing LSK cells using Ki67 and Hoechst 33 342 showed an accumulation of G1 phase of HIF1α−/− BCR-ABL–expressing LSK cells. (C) RT-PCR analysis showed elevated expression of cell-cycle inhibitors p16Ink4a, p19Arf, and p57 in the sorted LSK cells and BCR-ABL–expressing LSK cells from HIF1α−/− control and CML mice compared with the sorted LSK cells and BCR-ABL–expressing LSK cells from WT control and CML mice. BM cells from control and CML mice at day 14 post-BMT were collected, and LSK cells and BCR-ABL–expressing LSK cells were sorted for extracting total RNA. Mean values (± SD) are shown; **P < .01; ***P < .001. (D) The apoptosis of normal LSK cells and BCR-ABL–expressing LSK cells from BM of control and CML mice (n = 6). HIF1α deletion induced the apoptosis of BCR-ABL–expressing LSK cells, but did not affect the apoptosis of normal LSK cells. (E) FACS analysis showed a higher percentage of active caspase positive HIF1α−/− leukemia progenitors. (F) RT-PCR analysis showed elevated expression of p53 in the sorted BCR-ABL–expressing LSK cells from HIF1α−/− CML mice comparing to the sorted BCR-ABL–expressing LSK cells from control CML mice. Mean values (± SD) are shown; **P < .01. (G) Simultaneous knockdown of p16Ink4a and p19Arf rescued the colony-forming ability of HIF1α−/− BCR-ABL–expressing LSK cells. Western blot analysis showed that protein levels of p16Ink4a and p19Arf in p53−/− MEF and CML leukemia cells were dramatically reduced by shRNA no. 1 that targets both p16Ink4a and p19Arf. BM cells from WT and HIF1α−/− normal or CML mice were infected by shRNA no. 1, and plated into methycellulose media after selection by puromycin. Colonies were counted and cells from the colonies were serially replated. Mean values (± SD) are shown; **P < .01; ***P < .001. (H) Loss of HIF1α did not affect the senescence of normal LSK cells and BCR-ABL–expressing LSK cells. Sorted GFP+Lin−Sca-1+c-Kit+ cells from the BM of WT or HIF1α−/− control and CML mice were stained by X-Gal, and the number of β-galactosidase–expressing cells were counted.
To understand the molecular mechanism by which HIF1α regulates the function of LSCs, we compared the gene expression profiling between HIF1α−/− BCR-ABL–expressing LSK cells and WT BCR-ABL–expressing LSK cells and found that the cyclin-dependent kinase inhibitors p16Ink4a and p19Arf were up-regulated in HIF1α−/− BCR-ABL–expressing LSK cells (Tables 1, 2). Real-time PCR confirmed higher levels of expression of p16Ink4a and p19Arf in HIF1α−/− BCR-ABL–expressing LSK cells (Figure 5C). Expression of another cyclin-dependent kinase inhibitor p57 was also higher in HIF1α−/− BCR-ABL–expressing LSK cells than in WT BCR-ABL–expressing LSK cells (Figure 5C). Elevated expression of these inhibitors was also observed in HIF1α−/− normal LSK cells (Figure 5C), which is consistent with previous studies.16 However, expression of p57 was unchanged in HIF1α−/− normal LSK cells (Figure 5C), indicating that loss of HIF1α does not alter an identical set of genes in LSCs and normal HSCs. To test whether p16Ink4a and p19Arf mediate the function of HIF1α in LSCs, we knocked down the expression of p16Ink4a and p19Arf in BCR-ABL–expressing LSK cells using lentivirus-mediated shRNA to evaluate whether repression of p16Ink4a and p19Arf rescues the defective LSC function caused by HIF1α deletion. Because the transcripts of p16Ink4a and p19Arf share exons 2 and 3, we designed a single shRNA to simultaneously knockdown expression of both genes in p53−/−MEF cells which express higher levels of p16Ink4a and p19Arf (Figure 5G). In a colony-forming assay, we infected WT or HIF1α−/− CML BM cells with the p16Ink4a/p19Arf shRNA, and found that HIF1α−/− CML BM cells formed less numbers of colonies than did WT cells, and knockdown of p16Ink4a and p19Arf allowed the formation of more colonies in secondary and third replating. This result indicates that reduced function of HIF1α−/− LSCs was restored by repressing expression of p16Ink4a and p19Arf, demonstrating that the regulatory function of HIF1α in LSCs is at least in part mediated by p16Ink4a and p19Arf.
List of up-regulated genes (HIF1α−/− LSCs vs WT LSCs)
| Symbol . | Gene name . | Relative fold change, Log2 . |
|---|---|---|
| Ces2g | Carboxylesterase 2G | 0.85 |
| Rhag | Rhesus blood group-associated A glycoprotein | 0.83 |
| Cda | cytidine deaminase | 0.81 |
| Cxcl10 | Chemokine (C-X-C motif) ligand 10 | 0.80 |
| Pklr | Pyruvate kinase liver and red blood cell | 0.78 |
| Crip2 | Cysteine-rich protein 2 | 0.71 |
| Epor | Erythropoietin receptor | 0.69 |
| Mmp14 | Matrix metallopeptidase 14 (membrane-inserted) | 0.68 |
| Cd59a | CD59a antigen | 0.63 |
| Ccl19 | Chemokine (C-C motif) ligand 19 | 0.62 |
| Acot2 | Acyl-CoA thioesterase 2 | 0.59 |
| Car1 | Carbonic anhydrase 1 | 0.57 |
| Slc38a5 | Solute carrier family 38, member 5 | 0.57 |
| Bcl2a1a | B-cell leukemia/lymphoma 2–related protein A1a | 0.55 |
| Ott | Ovary testis transcribed | 0.51 |
| Gpr50 | G-protein–coupled receptor 50 | 0.50 |
| Cpox | Coproporphyrinogen oxidase | 0.49 |
| Sphk1 | Sphingosine kinase 1 | 0.47 |
| Glo1 | Glyoxalase 1 | 0.45 |
| Nipa1 | Nonimprinted in Prader-Willi/Angelman syndrome 1 homolog (human) | 0.45 |
| Plek2 | Pleckstrin 2 | 0.44 |
| Camsap1l1 | Calmodulin-regulated spectrin-associated protein 1-like 1 | 0.44 |
| Clcn2 | Chloride channel 2 | 0.43 |
| Slamf1 | Signaling lymphocytic activation molecule family member 1 | 0.43 |
| Pf4 | Platelet factor 4 | 0.42 |
| Mir224 | microRNA 224 | 0.41 |
| Hyal5 | Hyaluronoglucosaminidase 5 | 0.41 |
| Fbxw5 | F-box and WD-40 domain protein 5 | 0.40 |
| Rdh9 | Retinol dehydrogenase 9 | 0.40 |
| Sfrp4 | Secreted frizzled-related protein 4 | 0.38 |
| Ifnb1 | Interferon beta 1, fibroblast | 0.38 |
| Car2 | Carbonic anhydrase 2 | 0.38 |
| Ubxn2a | UBX domain protein 2A | 0.38 |
| Tns1 | Tensin 1 | 0.38 |
| Cited4 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 | 0.37 |
| Mt2 | Metallothionein 2 | 0.37 |
| Trfr2 | Transferrin receptor 2 | 0.37 |
| Bcl11a | B-cell CLL/lymphoma 11A (zinc finger protein) | 0.36 |
| Fat3 | FAT tumor suppressor homolog 3 (Drosophila) | 0.36 |
| Abhd6 | Abhydrolase domain-containing 6 | 0.35 |
| Idi1 | Isopentenyl-diphosphate delta isomerase | 0.35 |
| Ephb1 | Eph receptor B1 | 0.34 |
| Slc19a2 | Solute carrier family 19 (thiamine transporter), member 2 | 0.34 |
| Pou4f1 | POU domain, class 4, transcription factor 1 | 0.34 |
| Ifnab | Interferon alpha B | 0.34 |
| Parm1 | Prostate androgen-regulated mucin-like protein 1 | 0.34 |
| Trim58 | Tripartite motif-containing 58 | 0.34 |
| Phb | Prohibitin | 0.34 |
| Tnfsf4 | Tumor necrosis factor (ligand) superfamily, member 4 | 0.32 |
| Hspe1 | Heat shock protein 1 (chaperonin 10) | 0.32 |
| Ufsp1 | UFM1-specific peptidase 1 | 0.32 |
| Wdr55 | WD repeat domain 55 | 0.32 |
| Cdkn2a | Cyclin-dependent kinase inhibitor 2A | 0.12 |
| Trp53 | Transformation-related protein 53 | 0.1 |
| Symbol . | Gene name . | Relative fold change, Log2 . |
|---|---|---|
| Ces2g | Carboxylesterase 2G | 0.85 |
| Rhag | Rhesus blood group-associated A glycoprotein | 0.83 |
| Cda | cytidine deaminase | 0.81 |
| Cxcl10 | Chemokine (C-X-C motif) ligand 10 | 0.80 |
| Pklr | Pyruvate kinase liver and red blood cell | 0.78 |
| Crip2 | Cysteine-rich protein 2 | 0.71 |
| Epor | Erythropoietin receptor | 0.69 |
| Mmp14 | Matrix metallopeptidase 14 (membrane-inserted) | 0.68 |
| Cd59a | CD59a antigen | 0.63 |
| Ccl19 | Chemokine (C-C motif) ligand 19 | 0.62 |
| Acot2 | Acyl-CoA thioesterase 2 | 0.59 |
| Car1 | Carbonic anhydrase 1 | 0.57 |
| Slc38a5 | Solute carrier family 38, member 5 | 0.57 |
| Bcl2a1a | B-cell leukemia/lymphoma 2–related protein A1a | 0.55 |
| Ott | Ovary testis transcribed | 0.51 |
| Gpr50 | G-protein–coupled receptor 50 | 0.50 |
| Cpox | Coproporphyrinogen oxidase | 0.49 |
| Sphk1 | Sphingosine kinase 1 | 0.47 |
| Glo1 | Glyoxalase 1 | 0.45 |
| Nipa1 | Nonimprinted in Prader-Willi/Angelman syndrome 1 homolog (human) | 0.45 |
| Plek2 | Pleckstrin 2 | 0.44 |
| Camsap1l1 | Calmodulin-regulated spectrin-associated protein 1-like 1 | 0.44 |
| Clcn2 | Chloride channel 2 | 0.43 |
| Slamf1 | Signaling lymphocytic activation molecule family member 1 | 0.43 |
| Pf4 | Platelet factor 4 | 0.42 |
| Mir224 | microRNA 224 | 0.41 |
| Hyal5 | Hyaluronoglucosaminidase 5 | 0.41 |
| Fbxw5 | F-box and WD-40 domain protein 5 | 0.40 |
| Rdh9 | Retinol dehydrogenase 9 | 0.40 |
| Sfrp4 | Secreted frizzled-related protein 4 | 0.38 |
| Ifnb1 | Interferon beta 1, fibroblast | 0.38 |
| Car2 | Carbonic anhydrase 2 | 0.38 |
| Ubxn2a | UBX domain protein 2A | 0.38 |
| Tns1 | Tensin 1 | 0.38 |
| Cited4 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 | 0.37 |
| Mt2 | Metallothionein 2 | 0.37 |
| Trfr2 | Transferrin receptor 2 | 0.37 |
| Bcl11a | B-cell CLL/lymphoma 11A (zinc finger protein) | 0.36 |
| Fat3 | FAT tumor suppressor homolog 3 (Drosophila) | 0.36 |
| Abhd6 | Abhydrolase domain-containing 6 | 0.35 |
| Idi1 | Isopentenyl-diphosphate delta isomerase | 0.35 |
| Ephb1 | Eph receptor B1 | 0.34 |
| Slc19a2 | Solute carrier family 19 (thiamine transporter), member 2 | 0.34 |
| Pou4f1 | POU domain, class 4, transcription factor 1 | 0.34 |
| Ifnab | Interferon alpha B | 0.34 |
| Parm1 | Prostate androgen-regulated mucin-like protein 1 | 0.34 |
| Trim58 | Tripartite motif-containing 58 | 0.34 |
| Phb | Prohibitin | 0.34 |
| Tnfsf4 | Tumor necrosis factor (ligand) superfamily, member 4 | 0.32 |
| Hspe1 | Heat shock protein 1 (chaperonin 10) | 0.32 |
| Ufsp1 | UFM1-specific peptidase 1 | 0.32 |
| Wdr55 | WD repeat domain 55 | 0.32 |
| Cdkn2a | Cyclin-dependent kinase inhibitor 2A | 0.12 |
| Trp53 | Transformation-related protein 53 | 0.1 |
HIF indicates hypoxia-inducible factor; and LSC, leukemia stem cell.
List of down-regulated genes (HIF1α−/− LSCs vs WT LSCs)
| Symbol . | Gene name . | Relative fold change, Log2 . |
|---|---|---|
| Mcpt2 | Mast cell protease 2 | −1.63 |
| Tnfrsf9 | Tumor necrosis factor receptor superfamily, member 9 | −1.61 |
| Mcpt1 | Mast cell protease 1 | −1.57 |
| Rps25 | Ribosomal protein S25 | −1.43 |
| Mcpt4 | Mast cell protease 4 | −1.33 |
| Bnip3 | BCL2/adenovirus E1B-interacting protein 3 | −1.28 |
| Cox7c | Cytochrome c oxidase, subunit viic | −1.20 |
| Tph1 | Tryptophan hydroxylase 1 | −1.16 |
| Gzmb | Granzyme B | −1.01 |
| Bnip3 | BCL2/adenovirus E1B-interacting protein 3 | −0.95 |
| Plod2 | Procollagen lysine, 2-oxoglutarate 5-dioxygenase 2 | −0.94 |
| Ccr2 | Chemokine (C-C motif) receptor 2 | −0.87 |
| Slc16a3 | Solute carrier family 16 (monocarboxylic acid transporters), member 3 | −0.87 |
| Lipo1 | Lipase, member O1 | −0.87 |
| Cma1 | Chymase 1, mast cell | −0.85 |
| Dntt | Deoxynucleotidyltransferase, terminal | −0.85 |
| Xcr1 | Chemokine (C motif) receptor 1 | −0.84 |
| Hmgb1 | High mobility group box 1 | −0.83 |
| Marcks | Myristoylated alanine-rich protein kinase C substrate | −0.82 |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | −0.81 |
| Serpine1 | Serine (or cysteine) peptidase inhibitor, clade E, member 1 | −0.79 |
| Gimap4 | GTPase, IMAP family member 4 | −0.77 |
| Akr1c12 | Aldo-keto reductase family 1, member C12 | −0.74 |
| Itgae | Integrin alpha E, epithelial associated | −0.70 |
| Ebf1 | Early B-cell factor 1 | −0.69 |
| Maob | Monoamine oxidase B | −0.68 |
| Tlr3 | Toll-like receptor 3 | −0.65 |
| Cd36 | CD36 antigen | −0.65 |
| Kmo | Kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) | −0.65 |
| Il13ra1 | Interleukin 13 receptor, alpha 1 | −0.65 |
| Stat4 | Signal transducer and activator of transcription 4 | −0.64 |
| Slamf7 | SLAM family member 7 | −0.63 |
| Il7r | Interleukin 7 receptor | −0.61 |
| Cdkn1b | Cyclin-dependent kinase inhibitor 1B | −0.60 |
| Sucnr1 | Succinate receptor 1 | −0.60 |
| Irf8 | Interferon regulatory factor 8 | −0.59 |
| Cd74 | CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) | −0.58 |
| Nov | Nephroblastoma overexpressed gene | −0.56 |
| Ptprg | Protein tyrosine phosphatase, receptor type, G | −0.55 |
| Socs2 | Suppressor of cytokine signaling 2 | −0.53 |
| Irf5 | Interferon regulatory factor 5 | −0.51 |
| Igfbp5 | Insulin-like growth factor–binding protein 5 | −0.50 |
| Slamf6 | SLAM family member 6 | −0.50 |
| Slc2a1 | Solute carrier family 2 (facilitated glucose transporter), member 1 | −0.50 |
| Lef1 | Lymphoid enhancer-binding factor 1 | −0.49 |
| Cd96 | CD96 antigen | −0.49 |
| Symbol . | Gene name . | Relative fold change, Log2 . |
|---|---|---|
| Mcpt2 | Mast cell protease 2 | −1.63 |
| Tnfrsf9 | Tumor necrosis factor receptor superfamily, member 9 | −1.61 |
| Mcpt1 | Mast cell protease 1 | −1.57 |
| Rps25 | Ribosomal protein S25 | −1.43 |
| Mcpt4 | Mast cell protease 4 | −1.33 |
| Bnip3 | BCL2/adenovirus E1B-interacting protein 3 | −1.28 |
| Cox7c | Cytochrome c oxidase, subunit viic | −1.20 |
| Tph1 | Tryptophan hydroxylase 1 | −1.16 |
| Gzmb | Granzyme B | −1.01 |
| Bnip3 | BCL2/adenovirus E1B-interacting protein 3 | −0.95 |
| Plod2 | Procollagen lysine, 2-oxoglutarate 5-dioxygenase 2 | −0.94 |
| Ccr2 | Chemokine (C-C motif) receptor 2 | −0.87 |
| Slc16a3 | Solute carrier family 16 (monocarboxylic acid transporters), member 3 | −0.87 |
| Lipo1 | Lipase, member O1 | −0.87 |
| Cma1 | Chymase 1, mast cell | −0.85 |
| Dntt | Deoxynucleotidyltransferase, terminal | −0.85 |
| Xcr1 | Chemokine (C motif) receptor 1 | −0.84 |
| Hmgb1 | High mobility group box 1 | −0.83 |
| Marcks | Myristoylated alanine-rich protein kinase C substrate | −0.82 |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | −0.81 |
| Serpine1 | Serine (or cysteine) peptidase inhibitor, clade E, member 1 | −0.79 |
| Gimap4 | GTPase, IMAP family member 4 | −0.77 |
| Akr1c12 | Aldo-keto reductase family 1, member C12 | −0.74 |
| Itgae | Integrin alpha E, epithelial associated | −0.70 |
| Ebf1 | Early B-cell factor 1 | −0.69 |
| Maob | Monoamine oxidase B | −0.68 |
| Tlr3 | Toll-like receptor 3 | −0.65 |
| Cd36 | CD36 antigen | −0.65 |
| Kmo | Kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) | −0.65 |
| Il13ra1 | Interleukin 13 receptor, alpha 1 | −0.65 |
| Stat4 | Signal transducer and activator of transcription 4 | −0.64 |
| Slamf7 | SLAM family member 7 | −0.63 |
| Il7r | Interleukin 7 receptor | −0.61 |
| Cdkn1b | Cyclin-dependent kinase inhibitor 1B | −0.60 |
| Sucnr1 | Succinate receptor 1 | −0.60 |
| Irf8 | Interferon regulatory factor 8 | −0.59 |
| Cd74 | CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) | −0.58 |
| Nov | Nephroblastoma overexpressed gene | −0.56 |
| Ptprg | Protein tyrosine phosphatase, receptor type, G | −0.55 |
| Socs2 | Suppressor of cytokine signaling 2 | −0.53 |
| Irf5 | Interferon regulatory factor 5 | −0.51 |
| Igfbp5 | Insulin-like growth factor–binding protein 5 | −0.50 |
| Slamf6 | SLAM family member 6 | −0.50 |
| Slc2a1 | Solute carrier family 2 (facilitated glucose transporter), member 1 | −0.50 |
| Lef1 | Lymphoid enhancer-binding factor 1 | −0.49 |
| Cd96 | CD96 antigen | −0.49 |
See Table 1 for expansion of abbreviations.
Because elevated p16Ink4a and p19Arf are related to the induction of stem cell senescence,36,37 we examined the senescent status of HIF1α−/− LSCs and normal HSCs. Sorted BCR-ABL–expressing LSK cells and normal LSK cells from WT and HIF1α−/− CML mice and control mice were stained with β-galactosidase. We observed a similar frequency of β-galactosidase–positive BCR-ABL–expressing LSK cells and normal LSK cells (Figure 5H), suggesting that senescence is not responsible for the impaired maintenance of HIF1α−/− LSCs. In addition, we also observed an increased expression of the apoptotic gene p53 in HIF1α−/− BCR-ABL–expressing LSK cells (Figure 5F), which supports our observation of increased apoptosis of HIF1α−/− LSCs (Figure 5D).
Loss of HIF1α has different effects on LSCs and normal HSCs
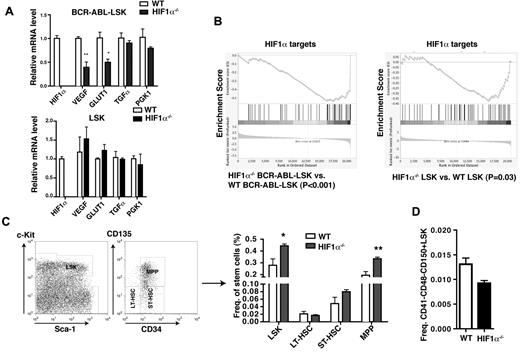
To further examine the involvement of HIF1α in functional regulation of LSCs, we examined whether deletion of HIF1α causes reduced expression of several HIF1α target genes. We found that compared with WT BCR-ABL–expressing LSK cells, expression levels of VEGF and GLUT1 in HIF1α−/− BCR-ABL–expressing LSK cells were reduced by ∼ 50% (Figure 6A); however, there were no significant differences in the expression of TGFα and PGK1 between HIF1α−/− and WT BCR-ABL–expressing LSK cells (Figure 6A). These results suggest that these hypoxia-responsive genes are responsive to HIF1α in LSCs. To determine whether loss of HIF1α also similarly affects normal HSCs, we sorted LSK cells from BM of WT and HIF1α−/− BM cells, and compared expression of these HIF1α target genes by RT-PCR. We found that loss of HIF1α did not cause a significant reduction of expression of VEGF and GLUT1 in normal LSK cells, although expression of these 2 genes was down-regulated in HIF1α−/− BCR-ABL–expressing LSK cells (Figure 6A). Furthermore, we conducted a microarray assay to examine expression of HIF1α target genes in HIF1α−/− BCR-ABL–expressing LSK cells and HIF1α−/− LSK cells and to evaluate the data using the gene set enrichment analysis (GSEA) algorithm.29,30 The enrichment plots showed that HIF1α targets were generally down-regulated in both BCR-ABL–expressing LSK cells and HIF1α−/− LSK cells, but the extent of this down-regulation was much more significant in HIF1α−/− BCR-ABL–expressing LSK cells than HIF1α−/− LSK cells (P < .001 vs P < .03; Figure 6B). Together, these results suggest that relative to normal HSCs, LSCs are more dependent on the HIF1α pathway. From our microarray study, we also identified a group of other HIF1α target genes that were altered in expression in HIF1α−/− BCR-ABL–expressing LSK cells but not in HIF1α−/− normal LSK cells (Table 3).
Effects of HIF1α deficiency on normal HSCs. (A) RT-PCR analysis for the expression of HIF1α, VEGF, GLUT1, TGFα, and PGK1 in WT or HIF1α−/− normal LSK cells and BCR-ABL–expressing LSK cells. (B) Gene set enrichment analysis of DNA microarray data displays gene expression profiling of HIF1α targets in WT or HIF1α−/− normal LSK cells and BCR-ABL–expressing LSK cells. (C) FACS analysis of HSCs in BM cells from WT or HIF1α−/− mice. The percentages of LSK cells (Lin−Sca-1+c-Kit+), LT-HSCs (Lin−Sca-1+c-Kit+CD34−CD135−), and ST-HSCs (Lin−Sca-1+c-Kit+CD34−CD135+) were similar in WT and HIF1α−/− mice; however, HIF1α−/− mice showed a higher percentage of MPP (Lin−Sca-1+c-Kit+CD34+CD135+) (n = 5). (D) Similar percentages of CD41−CD48−CD150+ Lin−Sca-1+c-Kit+ cells in BM cells of WT or HIF1α−/− mice (n = 3).
Effects of HIF1α deficiency on normal HSCs. (A) RT-PCR analysis for the expression of HIF1α, VEGF, GLUT1, TGFα, and PGK1 in WT or HIF1α−/− normal LSK cells and BCR-ABL–expressing LSK cells. (B) Gene set enrichment analysis of DNA microarray data displays gene expression profiling of HIF1α targets in WT or HIF1α−/− normal LSK cells and BCR-ABL–expressing LSK cells. (C) FACS analysis of HSCs in BM cells from WT or HIF1α−/− mice. The percentages of LSK cells (Lin−Sca-1+c-Kit+), LT-HSCs (Lin−Sca-1+c-Kit+CD34−CD135−), and ST-HSCs (Lin−Sca-1+c-Kit+CD34−CD135+) were similar in WT and HIF1α−/− mice; however, HIF1α−/− mice showed a higher percentage of MPP (Lin−Sca-1+c-Kit+CD34+CD135+) (n = 5). (D) Similar percentages of CD41−CD48−CD150+ Lin−Sca-1+c-Kit+ cells in BM cells of WT or HIF1α−/− mice (n = 3).
Differential expression of HIF1α target genes in HIF1α−/− LSCs comparing to HIF1α−/− normal HSCs
| Symbol . | Gene name . | Relative fold change, Log2 . |
|---|---|---|
| c-MET | Met proto-oncogene | −0.2 |
| KRT18 | Keratin 18 | −0.26 |
| Hmox1 | Heme oxygenase-1 | 0.67 |
| MMP2 | Matrix metallopeptidase 2 | 0.74 |
| UPAR | Urokinase-type plasminogen activator receptor | 3.1 |
| ETS1 | E26 avian leukemia oncogene 1 | −1.8 |
| Nr4a1 | Nuclear receptor subfamily 4, group A, member 1 | −0.27 |
| VEGF | Vascular endothelial growth factor A | 1.3 |
| Glut1 | Glucose transporter 1 | 3.4 |
| TGFα | Transforming growth factor alpha | 0.52 |
| Trf | Transferrin | −5.4 |
| P4ha1 | Prolyl 4-hydroxylase, alpha polypeptide I | −2.8 |
| Cited2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 0.77 |
| Symbol . | Gene name . | Relative fold change, Log2 . |
|---|---|---|
| c-MET | Met proto-oncogene | −0.2 |
| KRT18 | Keratin 18 | −0.26 |
| Hmox1 | Heme oxygenase-1 | 0.67 |
| MMP2 | Matrix metallopeptidase 2 | 0.74 |
| UPAR | Urokinase-type plasminogen activator receptor | 3.1 |
| ETS1 | E26 avian leukemia oncogene 1 | −1.8 |
| Nr4a1 | Nuclear receptor subfamily 4, group A, member 1 | −0.27 |
| VEGF | Vascular endothelial growth factor A | 1.3 |
| Glut1 | Glucose transporter 1 | 3.4 |
| TGFα | Transforming growth factor alpha | 0.52 |
| Trf | Transferrin | −5.4 |
| P4ha1 | Prolyl 4-hydroxylase, alpha polypeptide I | −2.8 |
| Cited2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 0.77 |
See Table 1 for expansion of abbreviations.
The differential expression of the HIF1α target genes in LSCs and HSCs prompted us to further examine the biologic effect of HIF1α on normal HSCs in our Vav-Cre-HIF1αflox/flox mice. It is also necessary to point out that in our Vav-Cre-HIF1αflox/flox mice, HIF1α was deleted by the Vav-Cre in contrast to the deletion of HIF1α by Mx1-Cre.16 We observed that the percentage of LSK cells in HIF1α−/− mice was significantly higher than that in WT mice (Figure 6C). The higher percentage of LSK cells in HIF1α−/− mice was largely because of the effect of HIF1α deletion on multipotent progenitor (MPP) population (Lin−Sca-1+c-Kit+CD34+Flt3+) and ST-HSCs (Lin−Sca-1+c-Kit+CD34+Flt3−), but not on LT-HSCs (Lin−Sca-1+c-Kit+CD34−Flt3−), as the percentage of LT-HSCs was only slightly lower in HIF1α−/− mice than in WT mice (Figure 6C). Furthermore, a lower percentage of CD41−CD48−CD150+LSK was also found in HIF1α−/− mice compared with WT mice (Figure 6D).
Discussion
HIF has been shown to be involved in cancer development.17,23,38 In this study, we used mice with conditional deletion for HIF1α to provide the first evidence for an essential role of HIF1α in CML development and maintenance of LSCs. Because HIF1α deficiency results in elevated expression of cell-cycle inhibitors p16Ink4a, p19Arf, and p57 and apoptotic gene p53 in LSCs, inhibition of the HIF1α pathway is likely to be effective in eradicating LSCs, providing a rationale for developing an anti-HIF1α therapy in treating CML.
We demonstrate that HIF1α is required for survival regulation of LSCs in CML mice, and that HIF1α and its target genes are up-regulated in LSCs by BCR-ABL, which is consistent with BCR-ABL up-regulation of HIF1α mRNA expression in a leukemia cell line.39 The PI3K and mTOR pathways appear to be responsible for BCR-ABL–dependent HIF1α expression in Ba/F3 cells.39 Consistent with a previous report showing a higher level of VEGF expression in the plasma and serum of CML patients,40 we show that expression of VEGF is elevated in LSCs in CML mice. We also show a higher level of GLUT1 expression in LSCs, consistent with a previous observation that BCR-ABL expression alters the distribution of GLUT1 in the cell plasma membrane of TonB210 cells.41 Interestingly, HIF1a deletion causes an increased homing ability of BCR-ABL–expressing leukemia cells to BM, consistent with increased expression of some homing-related genes such as MMP-14, CXCL10, CCL19 (Table 1). The underlying mechanisms need to be further studied in the future.
Our study suggests that HIF1α deletion induces a tumor suppressor response in LSCs through induction of expression of p16Ink4a, p19Arf, and p53. In addition, the elevated expression of p16Ink4a and p19Arf is not specific to LSCs, as HIF1α deficiency also induces their expression in normal HSCs.16 This is consistent with a previous study showing that the enforced expression of p16Ink4a and p19Arf in HSCs resulted in cell-cycle arrest and p53-dependent apoptosis, and caused the exhaustion of HSCs.9 Our data suggest that p16Ink4a and p19Arf contribute to the exhaustion of LSCs. On the other hand, it is known that HIF1α is essential for cell-cycle arrest during hypoxia.42 HSCs cultured in hypoxia in vitro displayed decreased proliferation and increased long-term reconstitution, which is mediated by increased expression of CDK inhibitors p21, p27, and p57 in LSK cells by hypoxia.15 Because HIF1α deletion induces the loss of LSC quiescence and accumulation of the cells in the G1 phase, up-regulated expression of p16Ink4a and p19Arf induced by HIF1α deletion may inhibit the transition of G1/S phase of cell cycle through binding to cyclin D–dependent CDK4/6.43 Indeed, we observed an increase of HIF1α−/− LSCs in G1 phase and a decrease of the cells in S phase. Furthermore, knockdown of p16Ink4a and p19Arf rescues the defect of HIF1α−/− LSCs to form colonies. It has been reported that Ink4a-Arf−/− HSCs retain their self-renewal capacity better than wild-type HSCs in a long-term ex vivo culture,44 and repression of p16Ink4a and p19Arf by overexpressing Bmi1 restores the reconstitution ability of HIF1α−/− HSCs.16 However, other cell-cycle regulators might also play a role in mediating the function of HIF1α in LSCs because up-regulated expression of p21 and p27 in HIF1α-deficient cells is also associated with an increased number of cells in the G1 phase and a decreased number of cells in the S phase of the cell cycle.21 Mechanistically, it is also possible that cellular senescence induced by HIF1α deficiency facilitates the exhaustion of HIF1α−/− LSCs and HSCs, which is likely caused by the up-regulation of p16ink4a and p19Arf. Although a previous work suggests that HIF1α plays a protective role against senescence,16 a solid supporting evidence is still lacking. In fact, we observe that HIF1α deficiency causes decreased proliferation of LSCs, indicating a different mechanism.
HIF1a appears to play a protective role under stress setting, such as in vitro culture, 5-FU treatment, and oncogene transformation. In our study, we use the Vav promoter to drive Cre expression to delete the HIF1α gene predominantly in the hematopoietic system. Another advantage of using Vav-driven Cre expression is to avoid the effect of IFN signaling on LSCs and HSCs in the popularly used Mx1-Cre expression system.45,46 Furthermore, Vav-Cre-HIF1αflox/flox mice are viable and have a basically normal lifespan. Thus, our Vav-Cre-HIF1αflox/flox mice will provide a valuable model for in-depth study of the role of HIF1α in LSCs and BCR-ABL leukemogenesis for developing novel therapeutic strategies in CML treatment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Leukemia & Lymphoma Society and National Institutes of Health grants R01-CA122142 and R01-CA114199 (S.L.).
S.L. is a scholar of the Leukemia & Lymphoma Society.
National Institutes of Health
Authorship
Contribution: H.Z. designed and performed experiments, analyzed data, and wrote the manuscript; H.L. helped perform the experiments; H.S.X. analyzed DNA microarray data; and S.L. designed experiments, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shaoguang Li, Department of Medicine, University of Massachusetts Medical School, 364 Plantation St, LRB315, Worcester, MA 01605; e-mail: shaoguang.li@umassmed.edu.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal