Abstract
To identify cooperating lesions in core-binding factor acute myeloid leukemia, we performed single-nucleotide polymorphism-array analysis on 300 diagnostic and 41 relapse adult and pediatric leukemia samples. We identified a mean of 1.28 copy number alterations per case at diagnosis in both patient populations. Recurrent minimally deleted regions (MDRs) were identified at 7q36.1 (7.7%), 9q21.32 (5%), 11p13 (2.3%), and 17q11.2 (2%). Approximately one-half of the 7q deletions were detectable only by single-nucleotide polymorphism-array analysis because of their limited size. Sequence analysis of MLL3, contained within the 7q36.1 MDR, in 46 diagnostic samples revealed one truncating mutation in a leukemia lacking a 7q deletion. Recurrent focal gains were identified at 8q24.21 (4.7%) and 11q25 (1.7%), both containing a single noncoding RNA. Recurrent regions of copy-neutral loss-of-heterozygosity were identified at 1p (1%), 4q (0.7%), and 19p (0.7%), with known mutated cancer genes present in the minimally altered region of 1p (NRAS) and 4q (TET2). Analysis of relapse samples identified recurrent MDRs at 3q13.31 (12.2%), 5q (4.9%), and 17p (4.9%), with the 3q13.31 region containing only LSAMP, a putative tumor suppressor. Determining the role of these lesions in leukemogenesis and drug resistance should provide important insights into core-binding factor acute myeloid leukemia.
Introduction
Acute myeloid leukemia (AML) with t(8;21)(q22;q22) [RUNX1-RUNX1T1] and inv(16)(p13.1q22) or t(16;16)(p13.1;q22) [CBFB-MYH11], commonly referred to as core-binding factor (CBF)–AML, is a clinicopathologic-genetic entity recognized in the World Health Organization category “AML with recurrent genetic abnormalities”1 and accounts for approximately 15% of AMLs.2 Patients with CBF-AML have a relatively favorable prognosis, particularly when they are treated with repetitive cycles of high-dose cytarabine.3-7 Nevertheless, only 40% to 50% of adult and 80% to 90% of pediatric patients achieve long-term remission.3-5,8,9 Therefore, it is important to identify the genetic basis underlying the clinical heterogeneity of the disease so that alternative treatment strategies can be developed.
At the molecular level, t(8;21) and inv(16) result in the formation of the fusion genes RUNX1-RUNX1T1 and CBFB-MYH11, respectively, that alter the individual components of the heterodimeric CBF transcription complex, a master regulator of definitive hematopoiesis. Both fusion proteins impair the normal transcriptional activity of RUNX1/CBFB, leading to enhanced self-renewal of hematopoietic progenitors and predisposing the cells to leukemic transformation. Importantly, the expression of RUNX1-RUNX1T1 and CBFB-MYH11 in HSCs and progenitors is insufficient on its own to induce leukemia.10 Moreover, low levels of the fusion transcript have been detected in myeloid and B cells in patients who have been in long-term remission, supporting the observation that additional genetic lesions are required for the occurrence of leukemia.11
The analysis of primary clinical material has identified several cooperating events. Common secondary chromosome abnormalities identified in CBF-AMLs include loss of sex chromosomes (-Y or -X) and del(9q) in t(8;21) and trisomy 22 and 8 in inv(16).3,5 The presence of trisomy 22 in AML with inv(16) has been associated with a significantly better rate of relapse-free survival in adult patients.3,4 Molecular analyses have provided further insight into the range of cooperating mutations, including activating mutations of KIT, NRAS, and FLT3.6 The presence of a KIT mutation has been shown to have an unfavorable influence on outcome.6 Furthermore, using global gene expression profiling, we recently identified hierarchical cluster-based subclasses of CBF-AML of prognostic significance and a gene signature associated with secondary KIT mutations.12,13
During recent years, the development of high-resolution genome-wide scanning technologies has enabled the detection of subtle copy number alterations (CNAs) and regions of copy-neutral loss-of-heterozygosity (CN-LOH). By the use of these tools, a high frequency of genetic alterations of key regulators of B-lymphoid development and cell cycle in B-progenitor pediatric acute lymphoblastic leukemia (ALL) has recently been identified, and similar results have independently been reported for adult ALL.14-17 In AML and other myeloid neoplasms, the application of increasingly greater-resolution single-nucleotide polymorphism (SNP)–array platforms has enabled the detection of submicroscopic CNAs and regions of CN-LOH.18-23
Here, we report the results from high-resolution SNP-array profiling of 300 diagnostic leukemia samples from pediatric and adult CBF-AML patients and of 41 matched relapse samples. Although the number of CNAs was low, our analysis identified novel recurrent genetic alterations harboring known and potentially new cancer genes.
Methods
Patients and samples
The study included 300 diagnostic samples from CBF-AML patients: t(8;21), n = 157 (adult n = 114; pediatric, n = 43); inv(16), n = 143 (adult, n = 104; pediatric, n = 39); CBF-AML was identified by conventional cytogenetics and/or reverse-transcriptase PCR. Patient characteristics are summarized in supplemental Table 1 (see the Supplemental Materials link at the top of the article). Intraindividual germline DNA from remission BM or peripheral blood was available for paired analysis in 175 cases [t(8;21), n = 95; inv(16), n = 80], and relapse samples were available in 41 cases [t(8;21), n = 25; inv(16), n = 16]. Samples originated from the biobanks of the German-Austrian AML Study Group (AMLSG; n = 207), St Jude Children's Research Hospital (n = 82), and University Hospital Ibn Rochd, Morocco (n = 11). Written informed consent and University of Ulm ethics committee approval was obtained in accordance with the Declaration of Helsinki for all patients.
Gene mutation analyses
Samples were analyzed for AML-associated mutations in FLT3, NRAS (exons 1 and 2), KIT (exons 8, 10, 11, and 17; n = 243; Table 2), JAK2 (n = 221), WT1 (n = 201), KRAS (n = 192), NPM1 (n = 183), MLL-partial tandem duplications (MLL-PTD, n = 144), CEBPA (n = 43), and CBL (n = 160). The specific mutation assays are described elsewhere.13,24-26
On the basis of the SNP-array data, additional sequence analysis was performed for NF1 [3 cases with del(17q)] as previously reported,25 and for MLL3 [14 cases with del(7q), 31 cases without del(7q), and 1 case with CN-LOH(7q)] as described in supplemental Methods.
Copy number and CN-LOH analysis
DNA extracted from frozen blood or BM samples enriched for leukemic blasts and obtained at diagnosis, during remission, or at relapse was genotyped with Affymetrix GeneChip Human Mapping 6.0 (Affymetrix) according to manufacturer's protocol, as previously described.15 DNA copy number and paired LOH analyses were performed by the use of reference alignment,27 dChipSNP,28 and circular binary segmentation29 as previously reported and as described in detail in supplemental Methods. Only segments with a window of 5 consecutive markers and segment copy number means of > 0.2 or < 0.2 were treated as true CNAs. If CN-LOH was detected, defects > 20 Mb in size and/or with > 20 consecutive markers containing < 10% intervening or conflicting calls were considered true lesions according to previous experiences.21 LOH from unpaired analysis was only counted as true lesion for regions recurrently affected by LOH in paired analysis. Size, position, and location of genes were identified with UCSC Genome Browser (http://genome.ucsc.edu/). The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE32462 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32462).
Fluorescence in situ hybridization
On the basis of tissue availability, a subset of identified recurrent CNAs was validated by the use of FISH as previously reported.30 DNA clones were ordered at www.imagenes-bio.de, and the following chromosomal locations were analyzed (respective clones in parentheses): 3q13 (RP11-490B, RP11-10I2, RP11-263A24, RP11-220G20); 7q36 (RP5-1070G24, RP4-563H24, RP11-796I2, RP5-981O7), 11p13 (RP11-710L2), and 17q11.2 (RP11-142O6).
Statistical analysis
The Genomic Identification of Significant Targets in Cancer (GISTIC) algorithm31 was applied to the curated segmentation results as previously described.21 The q value cut-off was set at 0.25, as shown by the green vertical line in Figure 1B.
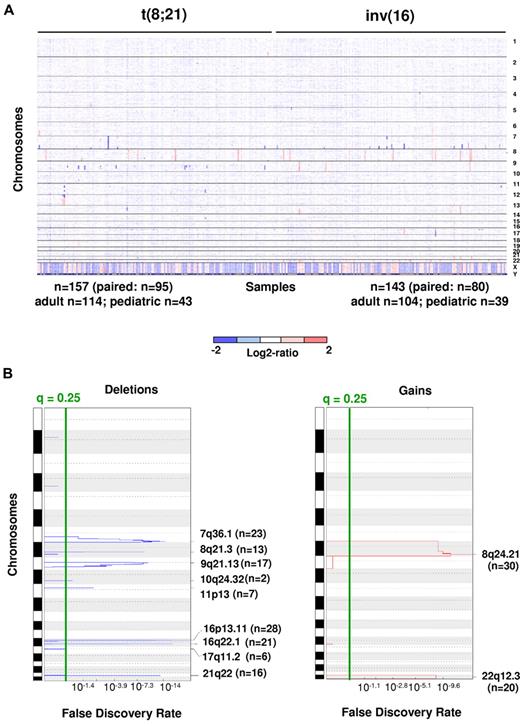
DNA CNAs in 300 newly diagnosed adult and pediatric patients with CBF-AML. (A) Log2 ratio SNP copy number data of diagnostic leukemia cells (median-smoothed with a window of 5 markers; blue indicates deletion and red gain). Each column represents a case, and the SNPs are arranged in rows according to chromosomal location. Cases are arranged by subgroup. Gross changes can be observed for example in chromosome 8 (17 cases with trisomy 8). (B) Analysis via GISTIC of copy number losses (left) and gains (right). False-discovery rate q values are plotted along the x-axis with chromosomal position along the y-axis. Altered regions with significance levels exceeding 0.25 (marked by vertical green line) were deemed significant. Nine significant regions of deletion, and 2 significant regions of amplification were identified. Chromosomal positions are shown for each significant region on the right side of the plots.
DNA CNAs in 300 newly diagnosed adult and pediatric patients with CBF-AML. (A) Log2 ratio SNP copy number data of diagnostic leukemia cells (median-smoothed with a window of 5 markers; blue indicates deletion and red gain). Each column represents a case, and the SNPs are arranged in rows according to chromosomal location. Cases are arranged by subgroup. Gross changes can be observed for example in chromosome 8 (17 cases with trisomy 8). (B) Analysis via GISTIC of copy number losses (left) and gains (right). False-discovery rate q values are plotted along the x-axis with chromosomal position along the y-axis. Altered regions with significance levels exceeding 0.25 (marked by vertical green line) were deemed significant. Nine significant regions of deletion, and 2 significant regions of amplification were identified. Chromosomal positions are shown for each significant region on the right side of the plots.
To investigate the association between genetic variables and patient survival, estimates of event-free survival (EFS) and overall survival (OS) were computed with the Kaplan-Meier method, and comparisons were performed with a Monte Carlo approximation to the exact log-rank test on the basis of 10 000 permutations of the assignment of mutation status to survival data. OS was defined as the time elapsed from initiation of therapy to death, with living patients' survival times censored at last follow-up. EFS was defined as the time from the initiation of therapy to induction failure, relapse, development of a second malignancy, or death from any cause. No adjustments for multiple-testing were performed in this exploratory analysis. A binomial test was used to test equality of the proportion of patients with more gains than losses and the proportion of patients with more losses than gains. These analyses were performed with R statistical software (www.r-project.org) Windows Version 2.13.9.
Results
Identification of CNAs
High-resolution genome-wide copy number analysis was performed on leukemic blasts from diagnostic BM aspirate or peripheral blood samples from 300 CBF-AML patients via use of Affymetrix 6.0 SNP microarrays. Matched intraindividual control DNA from remission BM aspirates or peripheral blood samples were available for paired analysis in 175 cases. The CBF-AML cohort included 218 adult [114 t(8;21) and 104 inv(16)] and 82 pediatric [43 t(8;21) and 39 inv(16)] patients. Paired analysis revealed a mean of 1.28 somatically acquired CNAs per case, with deletions more common than gains [t(8;21): 0.94 losses/case vs 0.2 gains/case; 51 cases had more losses than gains and 6 cases had more gains than losses, P < 10−9; inv(16): 0.95 vs 0.5, 32 cases had more losses than gains and 14 cases had more gains than losses, P = .01; Figure 1A). No significant difference was noted in the number of CNAs between t(8;21) [1.14; range, 0-5] and inv(16) [1.45; range, 0-9; P = .37] or between adult [1.31; range, 0-9] and pediatric cases [1.25; range, 0-5; P = .65].
Despite the limited number of CNAs per case, several recurrent lesions were identified (Table 1; all lesions from paired analysis are provided in supplemental Table 2). Focal deletions (6-4511 Kb in size) were a common event at the breakpoints adjacent to the genes involved in the t(8;21) and inv(16) in the respective CBF-AMLs (18.5% of t(8;21) cases, 8q21.3 [n = 13] and 21q22 [n = 16] and in 34.3% of inv(16)/t,(16;16) 16p13.11 [n = 28] and 16q22.11 [n = 21]). Additional recurrent deletions and amplifications involving entire chromosomes, chromosomal arms, or large chromosomal regions are also listed in Table 1. Del(9q) and loss of the sex chromosomes were among the most common CNAs detected and were restricted to t(8;21)–containing cases, which is consistent with published findings.3-6 Del(17q), although a relatively rare event, was a recurrent CNA that was restricted to inv(16)/t(16,16)–containing cases. Trisomy 22 or gain of 22q was the most common region of gain and was restricted to inv(16)/t(16,16)–containing cases. Trisomy 8/+8q and gains of 13q and 11q were also recurrent CNAs, with the latter 2 being more prevalent in inv(16)/t(16;16)–containing cases.
Recurrent CNAs in 300 newly diagnosed adult and pediatric CBF-AMLs
| Lesion type and cytoband . | Loc. start† . | Loc. end† . | MDR size, Kb . | No. total lesions at locus . | No. t(8;21) lesions at locus . | No. inv(16) lesions at locus . | Gene and miRNA symbols (if < 15 or number) . |
|---|---|---|---|---|---|---|---|
| del(7q36.1)* | 151 119 304 | 151 766 515 | 647 | 23 | 10 | 13 | PRKAG2, GALNTL5, GALNT11, MLL3 |
| del(7q36.1)* | 148 970 700 | 150 326 128 | 1355 | 23 | 9 | 14 | 22 genes |
| del(7q33-34)* | 136 930 471 | 139 330 291 | 2400 | 19 | 7 | 12 | 18 genes |
| gain(8q24.21) | 130 655 287 | 130 793 607 | 138 | 30 | 16 | 14 | CCDC26 |
| del(9q21.32) | 84 807 339 | 85 927 065 | 1128 | 17 | 17 | 0 | RASEF, FRMD3, C9orf103, UBQLN1, KIF27, C9orf64, HNRNPK, RMI1;hsa-mir-7-1 |
| del(10q24.32-q24.33) | 104 388 971 | 104 991 787 | 602 | 2 | 1 | 1 | TRIM8, ARL3, SFXN2, C10orf26, CYP17A1, C10orf32, AS3MT, CNNM2, NT5C2 |
| del(11p13) | 32 406 375 | 32 535 970 | 130 | 7 | 2 | 5 | WT1, WIT1 |
| gain(11q25) | 133 855 317 | 134 223 724 | 368 | 5 | 0 | 5 | LOC238177 |
| gain(13q14.1-q23.2) | 82 686 052 | 114 126 487 | 31 440 | 5 | 0 | 5 | 473 genes; 9 miRNA |
| del(16p11.2) | 29 748 299 | 30 390 041 | 642 | 3 | 0 | 3 | 33 genes |
| del(17q11.2) | 26 543 169 | 27 444 641 | 901 | 6 | 0 | 6 | NF1, OMG, EVI2B, EVI2A, RAB11FIP4, C17orf79, UTP6, SUZ12, LRRC37B, SH3GLIP1;hsa-mir-193a, hsa-mir-365-2 |
| gain(22q11.21-q13.33) | 17 410 722 | 49 581 309 | 32 171 | 20 | 0 | 20 | 795 genes; 15 miRNA |
| -X | Whole chromosome | 11 | 11 | 0 | |||
| -Y | Whole chromosome | 44 | 42 | 2 | |||
| t(8;21) and inv(16) breakpoint-associated losses | |||||||
| del(8q21.3) | 93 152,074 | 93 180 996 | 29 | 13 | 13 | 0 | RUNX1T1 |
| del(21q22.12) | 35 124,310 | 35 130 211 | 6 | 16 | 16 | 0 | RUNX1 |
| del(16p13.11) | 15 725,642 | 15 738 236 | 13 | 28 | 0 | 28 | NDE1,MYH11 |
| del(16q22.1) | 65 676,333 | 65 712 933 | 36 | 21 | 0 | 21 | CBFB, C16orf70 |
| Lesion type and cytoband . | Loc. start† . | Loc. end† . | MDR size, Kb . | No. total lesions at locus . | No. t(8;21) lesions at locus . | No. inv(16) lesions at locus . | Gene and miRNA symbols (if < 15 or number) . |
|---|---|---|---|---|---|---|---|
| del(7q36.1)* | 151 119 304 | 151 766 515 | 647 | 23 | 10 | 13 | PRKAG2, GALNTL5, GALNT11, MLL3 |
| del(7q36.1)* | 148 970 700 | 150 326 128 | 1355 | 23 | 9 | 14 | 22 genes |
| del(7q33-34)* | 136 930 471 | 139 330 291 | 2400 | 19 | 7 | 12 | 18 genes |
| gain(8q24.21) | 130 655 287 | 130 793 607 | 138 | 30 | 16 | 14 | CCDC26 |
| del(9q21.32) | 84 807 339 | 85 927 065 | 1128 | 17 | 17 | 0 | RASEF, FRMD3, C9orf103, UBQLN1, KIF27, C9orf64, HNRNPK, RMI1;hsa-mir-7-1 |
| del(10q24.32-q24.33) | 104 388 971 | 104 991 787 | 602 | 2 | 1 | 1 | TRIM8, ARL3, SFXN2, C10orf26, CYP17A1, C10orf32, AS3MT, CNNM2, NT5C2 |
| del(11p13) | 32 406 375 | 32 535 970 | 130 | 7 | 2 | 5 | WT1, WIT1 |
| gain(11q25) | 133 855 317 | 134 223 724 | 368 | 5 | 0 | 5 | LOC238177 |
| gain(13q14.1-q23.2) | 82 686 052 | 114 126 487 | 31 440 | 5 | 0 | 5 | 473 genes; 9 miRNA |
| del(16p11.2) | 29 748 299 | 30 390 041 | 642 | 3 | 0 | 3 | 33 genes |
| del(17q11.2) | 26 543 169 | 27 444 641 | 901 | 6 | 0 | 6 | NF1, OMG, EVI2B, EVI2A, RAB11FIP4, C17orf79, UTP6, SUZ12, LRRC37B, SH3GLIP1;hsa-mir-193a, hsa-mir-365-2 |
| gain(22q11.21-q13.33) | 17 410 722 | 49 581 309 | 32 171 | 20 | 0 | 20 | 795 genes; 15 miRNA |
| -X | Whole chromosome | 11 | 11 | 0 | |||
| -Y | Whole chromosome | 44 | 42 | 2 | |||
| t(8;21) and inv(16) breakpoint-associated losses | |||||||
| del(8q21.3) | 93 152,074 | 93 180 996 | 29 | 13 | 13 | 0 | RUNX1T1 |
| del(21q22.12) | 35 124,310 | 35 130 211 | 6 | 16 | 16 | 0 | RUNX1 |
| del(16p13.11) | 15 725,642 | 15 738 236 | 13 | 28 | 0 | 28 | NDE1,MYH11 |
| del(16q22.1) | 65 676,333 | 65 712 933 | 36 | 21 | 0 | 21 | CBFB, C16orf70 |
CBF-AML indicates core-binding factor acute myeloid leukemia; CNA, copy number alterations; Loc., location on chromosome; and MDR, minimally detected regions.
In total, 25 cases with del(7q) were identified, with large overlapping deletions in most of the cases. Three distinct critical regions were defined by cases with focal deletions.
hg18.
By applying the GISTIC algorithm, we were not only able to identify the CNAs listed previously but also the focal regions of gains within 8q and 22q and focal regions of loss within 7q36.1, 9q21.13, 10q24.32, 11p13, and 17q11.2 (Figure 1B; supplemental Table 4a). By performing GISTIC analyses separately on t(8;21) and inv(16) cases, we identified additional focal deletions at 4q28.3 [t(8;21) cases] and 16p11.2 [inv(16)/t(16;16) cases; supplemental Table 4b-c)]. FISH validation was performed with the use of probes from the minimally deleted regions (MDRs) at 7q36 (n = 5), 11p13 (n = 2), and 17q11.2 (n = 3), and each lesion was confirmed in the examined cases.
Delineation of critical regions of CNAs
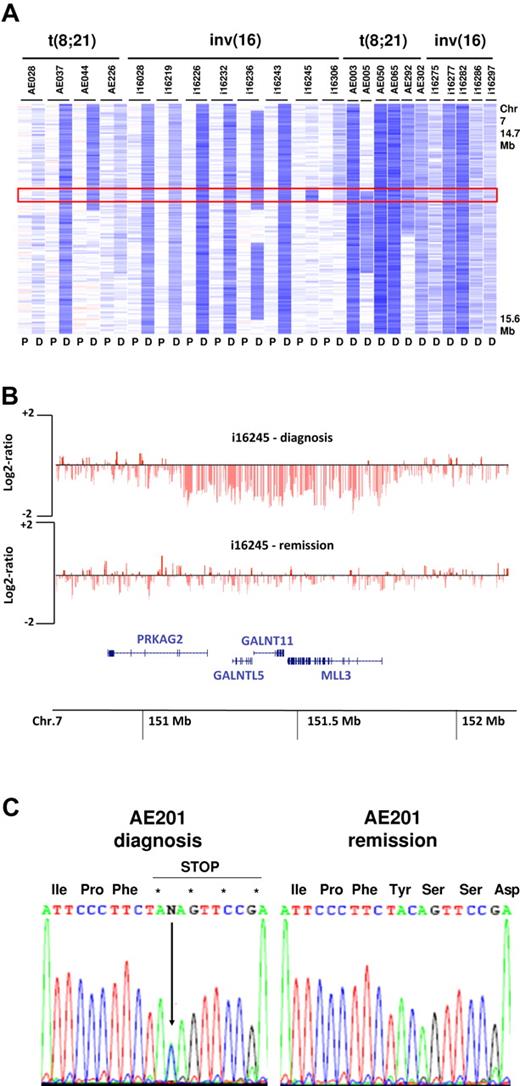
Excluding the recurrent lesions associated with t(8;21) and inv(16)/t(16;16), we found that several of the identified MDRs were < 1.5 Mb in size. In the 25 cases with deletions involving 7q, a MDR was identified at 7q36.1, affecting 23 of the 25 cases. This MDR was 647 Kb in size and contained 4 genes, PRKAG2, GALNTL5, GALNT11, and MLL3 (Figure 2A-B). MLL3 is a member of the Drosophila trithorax-related (SET) domain and plant homeodomain (PHD) containing family of transcriptional regulators and chromatin remodelers, a gene family in which several members have been implicated in hematopoietic and epithelial cancers.32 The loss of 1 allele of MLL3 in approximately 8% of CBF-AMLs raises the possibility that this gene may function as a haploinsufficient tumor suppressor in leukemogenesis. To further explore this possibility, we sequenced all coding exons and the splice junctions of MLL3 in 14 cases with del(7q) and 32 cases that lacked CNAs involving 7q. This analysis identified a single case lacking a del(7q) that contained a MLL3 single base-pair deletion that resulted in a premature stop codon (Figure 2C).
Deletions of 7q36.1 involving MLL3. (A) Log2 ratio SNP copy number data in 23 cases at diagnosis with focal or more extensive deletions (P indicates paired normal; and D, diagnosis). (B) Minimally deleted region in 7q36.1 (647 Kb in size) defined by case i16245 containing the 4 genes PRKAG2, GALNTL5, GALNTL11, and MLL3. Each vertical red line represents the genomic position and log2 ratio copy number of an individual marker. (C) Sequencing of MLL3 showing a truncating mutation leading to a premature stop codon in a single case (AE201) without del(7q).
Deletions of 7q36.1 involving MLL3. (A) Log2 ratio SNP copy number data in 23 cases at diagnosis with focal or more extensive deletions (P indicates paired normal; and D, diagnosis). (B) Minimally deleted region in 7q36.1 (647 Kb in size) defined by case i16245 containing the 4 genes PRKAG2, GALNTL5, GALNTL11, and MLL3. Each vertical red line represents the genomic position and log2 ratio copy number of an individual marker. (C) Sequencing of MLL3 showing a truncating mutation leading to a premature stop codon in a single case (AE201) without del(7q).
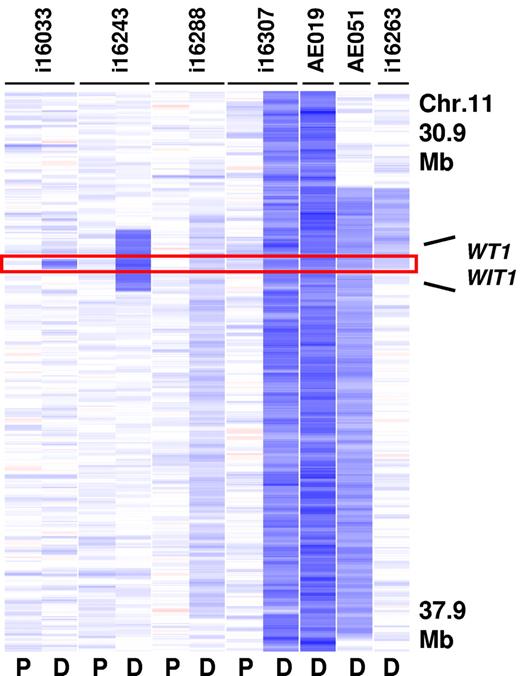
A second MDR at 11p13 was 130 Kb in size and contained only WT1 and the Wilms tumor upstream neighbor 1 (WIT1) gene (Figure 3). WT1 encodes a zinc-finger transcription factor that has been previously found to be mutated in s subset of adult and pediatric AMLs.26,33 Deletion of this gene was detected in 7 cases (2.3% of CBF-AMLs) and was more prevalent in inv(16)/t(16;16)–containing cases (3.5%). Importantly, sequence analysis of WT1 in 4 cases with 11p13 deletions identified a frame shift mutation in exon 7 (p.Val378Leufs*2) in the remaining allele in 1 case. Sequence analysis of WT1 in additional cases lacking a 11p13 deletion revealed mutations in 8.8% (9 of 102) of inv(16)/t(16;16)–containing cases and 0.6% (1 of 95) of t(8;21)–containing cases (Table 2).
Recurrent deletions at 11p13. The MDR (130 Kb in size) is defined by case i16033 and contains the WT1 and the WIT1 gene (P indicates paired normal; and D, diagnosis).
Recurrent deletions at 11p13. The MDR (130 Kb in size) is defined by case i16033 and contains the WT1 and the WIT1 gene (P indicates paired normal; and D, diagnosis).
Mutation frequencies in CBF-AML
| Mutated gene/locus . | t(8;21) cases n, (%) . | inv(16) cases n, (%) . | All cases n, (%) . |
|---|---|---|---|
| NRAS, 1p13.2 | |||
| Sequence mutation | 25/130 (19) | 53/135 (39) | 78/265 (29) |
| CNA | 0 | 0 | 0 |
| LOH | 2 | 1 | 3 |
| KRAS, 12p12.1 | |||
| Sequence mutation | 5/95 (5) | 12/97 (12) | 17/192 (9) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| FLT3-TKD, 13q12.2 | |||
| Sequence mutation | 4/125 (3) | 19/133 (14) | 23/258 (9) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| FLT3-ITD, 13q12.2 | |||
| Sequence mutation | 12/126 (10) | 7/133 (5%) | 19/259 (7) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| KIT, 4q12 | |||
| Sequence mutation | 30/120 (25) | 44/124 (35) | 74/244 (30) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| CBL, 11q23.3 | |||
| Sequence mutation | 0/81 (0) | 4/79 (5) | 4/160 (3) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| JAK2, 9p24.1 | |||
| Sequence mutation | 3/110 (3) | 0/111 (0) | 3/221 (1) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| WT1, 11p13 | |||
| Sequence mutation | 1/95 (1) | 10/106 (9) | 11/201 (5) |
| CNA | 2 | 5 | 7 |
| LOH | 0 | 1 | 1 |
| CEBPA, 19q13.11 | |||
| Sequence mutation | 1/27 (4) | 1/16 (6) | 2/43 (5) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| NPM1, 5q35.1 | |||
| Sequence mutation | 0/90 (0) | 1/93 (1) | 1/183 (1) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| MLL-PTD, 11q23.3 | |||
| Sequence mutation | 1/74 (1) | 0/70 (0) | 1/144 (1) |
| CNA | 0 | 1 | 1 |
| LOH | 0 | 0 | 0 |
| MLL3, 7q36.1 | |||
| Sequence mutation | 1/23 (4) | 0/23 (0) | 1/46 (2) |
| CNA | 10 | 13 | 23 |
| LOH | 1 | 0 | 1 |
| Mutated gene/locus . | t(8;21) cases n, (%) . | inv(16) cases n, (%) . | All cases n, (%) . |
|---|---|---|---|
| NRAS, 1p13.2 | |||
| Sequence mutation | 25/130 (19) | 53/135 (39) | 78/265 (29) |
| CNA | 0 | 0 | 0 |
| LOH | 2 | 1 | 3 |
| KRAS, 12p12.1 | |||
| Sequence mutation | 5/95 (5) | 12/97 (12) | 17/192 (9) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| FLT3-TKD, 13q12.2 | |||
| Sequence mutation | 4/125 (3) | 19/133 (14) | 23/258 (9) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| FLT3-ITD, 13q12.2 | |||
| Sequence mutation | 12/126 (10) | 7/133 (5%) | 19/259 (7) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| KIT, 4q12 | |||
| Sequence mutation | 30/120 (25) | 44/124 (35) | 74/244 (30) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| CBL, 11q23.3 | |||
| Sequence mutation | 0/81 (0) | 4/79 (5) | 4/160 (3) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| JAK2, 9p24.1 | |||
| Sequence mutation | 3/110 (3) | 0/111 (0) | 3/221 (1) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| WT1, 11p13 | |||
| Sequence mutation | 1/95 (1) | 10/106 (9) | 11/201 (5) |
| CNA | 2 | 5 | 7 |
| LOH | 0 | 1 | 1 |
| CEBPA, 19q13.11 | |||
| Sequence mutation | 1/27 (4) | 1/16 (6) | 2/43 (5) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| NPM1, 5q35.1 | |||
| Sequence mutation | 0/90 (0) | 1/93 (1) | 1/183 (1) |
| CNA | 0 | 0 | 0 |
| LOH | 0 | 0 | 0 |
| MLL-PTD, 11q23.3 | |||
| Sequence mutation | 1/74 (1) | 0/70 (0) | 1/144 (1) |
| CNA | 0 | 1 | 1 |
| LOH | 0 | 0 | 0 |
| MLL3, 7q36.1 | |||
| Sequence mutation | 1/23 (4) | 0/23 (0) | 1/46 (2) |
| CNA | 10 | 13 | 23 |
| LOH | 1 | 0 | 1 |
CBF-AML indicates core-binding factor acute myeloid leukemia; CNA, copy-number alterations; ITD, internal tandem duplications; LOH, loss of heterozygosity; PTD, partial tandem duplications; and TKD, tyrosine kinase domain.
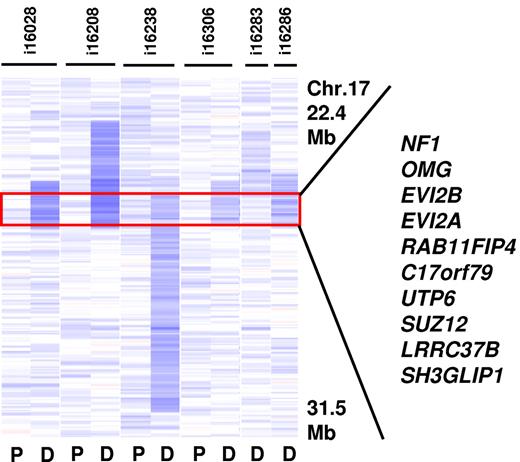
Although relatively rare in the analyzed CBF-AML cohort, a recurrent 17q11.2 deletion was detected in 6 cases and consisted of a MDR of 902 Kb containing 10 genes, including the known tumor suppressor NF1 (Figure 4) and 2 micro-RNAs. Sequence analysis of all coding exons and splice sites of NF1 in 4 cases with 17q11.2 deletion revealed no mutation in the retained allele.
Recurrent deletion at 17q11.2. The MDR (901 Kb in size) is defined by the 2 cases i16238 and i16306 and contains 10 genes including the tumor suppressor NF1 (P indicates paired normal; and D, diagnosis).
Recurrent deletion at 17q11.2. The MDR (901 Kb in size) is defined by the 2 cases i16238 and i16306 and contains 10 genes including the tumor suppressor NF1 (P indicates paired normal; and D, diagnosis).
Additional focal CNAs included a MDR of 1125 Kb at 9q21.32 that was detected in 17 cases [10.8% of t(8;21) cases; no inv(16) cases] and contained 8 genes and 1 micro-RNA (supplemental Figure 1) and 2 regions of focal gain, the first at 8q24.21 (minimally gained region: 138 Kb), and the second at 11q25 (368 Kb). Both of these latter regions contained only a single noncoding RNA gene, CCDC26 and LOC283177, respectively (Table 1).
Identification of CBF-AML associated CN-LOH
On the basis of 175 paired-analyzed cases, 13 regions of CN-LOH were identified, 7 in 95 (7.4%) t(8;21)– and 6 in 80 (7.5%) inv(16)/t(16;16)–containing cases. The frequency was the same in children (4/53, 7.5%) and in adults (9/122, 7.4%). To identify recurrent regions of CN-LOH, we extended our analysis to leukemic samples that lacked a matched germline sample. For these latter cases, we only considered an identified region of CN-LOH to be valid if the same region had been detected in paired analysis. Including these latter cases, we identified 3 regions of CN-LOH that were recurrent in our CBF-AML cohort (supplemental Table 5). These included a region at 1p36.33-p12 (n = 3; paired, n = 2; unpaired, n = 1; minimal size: 119 Mb), 4q22.1-q25 (n = 3; paired, n = 2, unpaired, n = 1; minimal size: 17 Mb), and 19p13.3-p13.2 (n = 2; both paired; minimal size: 9 Mb). Sequence analysis of 2 of the cases with CN-LOH at 1p36.33-p12 revealed NRAS activating mutations in both cases, although the mutation was homozygous in only a single case (homozygous NRAS-G12D activating mutation in AE037 and a heterozygous NRAS-G12D activating mutation in i16236). Similarly, sequence analysis of the known cancer gene TET2 contained within the 4q22.1-q25 region revealed a homozygous TET2 mutation leading to a stop codon (p.Q461X) in only 1 of the 2 cases containing this alteration.
Several nonrecurrent CN-LOH regions colocalized with regions affected by recurrent deletions, including 7q, 11p, and 17q, suggesting the existence of genes within the altered regions that are involved in leukemogenesis. Two nonrecurrent CN-LOH regions at 6p21.1 and 8p23.3-p23.5 also were affected by nonrecurrent deletions, again raising the possibility of cancer genes within the defined region.
Identification of relapse associated CNAs
BM samples obtained at the time of relapse were available from 41 CBF-AML patients. Matched diagnosis samples were available from 39 of these patients, and germline remission BM samples from 23. To map the evolutionary relationship of samples, we carefully analyzed the changes of CNAs between matched remission, diagnostic, and relapse material using the 23 cases with available samples from all these occasions. This analysis demonstrated a significant increase in the mean number of somatic CNAs from diagnosis to relapse (supplemental Table 6, mean of 1.28 to 3.17 CNAs per case; 0.94 deletions at diagnosis vs 1.87 deletions per case at relapse; 0.34 gains at diagnosis vs 1.3 gains per case at relapse; P = .04). There were no significant differences between t(8;21)– and inv(16)/t(16;16)–containing cases (P = .37). Among these 23 relapse samples, 8 (35%) showed no change in CNAs from diagnosis, 1 (4%) only lost a diagnostic CNA, 9 (39%) gained new CNAs, and 5 (22%) both lost and gained CNAs (supplemental Table 6). For 5 relapse cases, there were no CNAs in common between the diagnostic and relapse sample. Nevertheless, in 3 of these cases, the same translocation fusion transcript was detected, suggesting a clonal relationship. Common chromosomal aberrations found at diagnosis that were frequently lost at relapse, include nullisomy Y (4/4 cases), X (5/5 cases), and trisomy 8 (1/1 cases). The loss of some lesions present at the time of diagnosis coupled with the persistence of other diagnostic lesions, with or without the acquisition of new relapse-specific lesions, is consistent with the relapse clone arising from a clone that is ancestral to both the diagnostic and relapse clones.
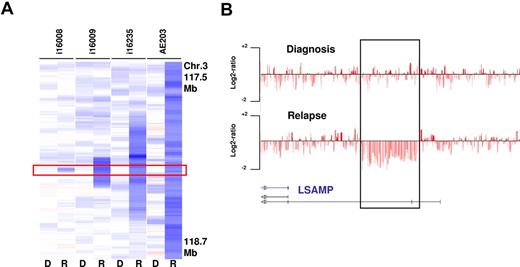
To identify recurrent relapse associated CNAs, we analyzed all 39 cases with available relapse and diagnosis material. The most frequent CNA acquired at relapse was a deletion of 3q13.31, which was identified in 5 cases and confirmed by FISH in 3 cases with available material. The MDR of 46 Kb defined by case i16008 contained a single transcript that contains exons of the annotated gene LSAMP, a putative tumor suppressor (Figure 5). Moreover, LSAMP is contained within each of the other larger relapse-associated deletions. LSAMP has previously been reported to be frequently deleted, down-regulated, or epigenetically silenced in osteosarcoma, brain tumors, and renal cell carcinoma34-36 ; however, to date no association of this lesions with relapsed AML has been reported.
Relapse-associated deletions at 3q13.31 in 4 paired analyzed cases. (A) The MDR is defined by case i16008 that carries a deletion of 46 Kb containing a LSAMP transcript (R indicates relapse; and D, diagnosis). (B) SNP coverage of the locus in case i16008.
Relapse-associated deletions at 3q13.31 in 4 paired analyzed cases. (A) The MDR is defined by case i16008 that carries a deletion of 46 Kb containing a LSAMP transcript (R indicates relapse; and D, diagnosis). (B) SNP coverage of the locus in case i16008.
Other recurrent CNAs acquired at relapse were losses in 2q33.3 (n = 3; 9 Kb, no gene), 5q15-q23.2 (n = 2; 26 678 Kb, 106 genes), 7q21-q31.1 (n = 2; 72 519 Kb, 1779 genes), 17p13 (n = 2; 6130 Kb, 114 genes, not including TP53), and gains of the whole chromosome 8 (n = 3), 2q37.1 (n = 2; 14 Kb; 1 gene: TRPM8), and 17q11.2-q25.3 (n = 2; 54 259 Kb, 740 genes). Several regions of CN-LOH were identified in single-relapse samples, including 2p23.1-p22.1 (9757 Kb), 5q23.2-q33.1 (22 377 Kb), and 16q23.2-q24.2 (6380 Kb; supplemental Table 5).
Correlation with clinical data
For recurrent genomic alterations observed in at least 5 subjects, we performed exploratory analysis for an association with patient outcome. In inv(16)/t(16;16)–containing cases, trisomy 22 was associated with a significantly better 5-year EFS (P = .02) and OS (P = .02) compared with cases without trisomy 22; these data are consistent with our previously published data.3 No significant association with EFS or OS was detected for del(11p13) or del(17q11.2) in inv(16)/t(16;16)–containing cases, or for trisomy 8, gain of 8q24.21, nullisomy Y in male, or loss of X in female patients with t(8;21)–containing leukemia (data not shown).
Discussion
High-resolution SNP-array analysis of 300 pediatric and adult CBF-AMLs revealed remarkably few somatic genetic alterations, with only 1.28 CNAs/case and CN-LOH observed in only 7% of cases. Moreover, no significant differences were noted in the number of CNAs between inv(16) and t(8;21) cases, nor in the number or type of lesions seen in the pediatric and adult patients. The latter observation suggests that differences in prognosis between children and adults likely reflect age-dependent host factors and not leukemia-specific biologic differences. The low burden of CNAs in our study is in line with previous reported studies performed on unselected AML cohorts21-23 and is in sharp contrast to ALL.14-17 The low burden of CNAs suggests that genomic instability is not a prominent mechanism of leukemogenesis in CBF-AML.
The most common recurrent CNAs were adjacent to the breakpoints of the chromosomal rearrangements that define the CBF-leukemias; t(8;21) [RUNX1-RUNX1T1] and inv(16)/t(16;16) [CBFB-MYH11]. In addition, several other recurrent genetic lesions were identified that contain genes whose alterations may cooperate with the translocation-encoded CBF fusion protein to induce overt leukemia. One of the most frequent CNAs was deletion on 7q. Del(7q), including del(7q36.1), is a recurrent alteration frequently seen in therapy-related AML and AML with complex karyotype30,37,38 but is less common in CBF-AML.3,4,8 In our study, a deletion on 7q was found in approximately 10% of cases, with approximately one-half of the deletions detectable only by SNP-array analysis because of their limited size. A MDR at 7q36.1 of 647 Kb was defined within a larger MDR that we previously described at 7q35-q36.30 This smaller MDR contained 4 genes, including MLL3, 1 of 6 members of a myeloid-lymphoid mixed-lineage leukemia gene family that encode H3K4-specific methyltransferases with homology to the Drosophila-trithorax gene. The MLL proteins are responsible for the transcriptional regulation of developmental genes including the homeobox (HOX) gene family, with each MLL family member thought to target different subsets of HOX genes and thus regulated the development of different lineages.
MLL1, the parent of this gene family, is the target of recurrent translocations in ALL and AML, with more than 40 different partner genes identified.39 More recently, other members of the MLL gene family have been implicated in cancer, including MLL2 in renal cell carcinoma,40 diffuse large B-cell lymphoma and follicular lymphoma,41,42 and MLL3 in breast cancer43 and medulloblastoma.44 Moreover, germline mutations of MLL2 are the underlying cause of a subset of patients with Kabuki syndrome, a developmental syndrome characterized by distinctive facial appearance, cardiac anomalies, skeletal abnormalities, and mild-to-moderate intellectual disability.45 Sequence analysis of MLL3 in 46 CBF-AMLs with and without del(7q), identified a single somatic heterozygous frame-shift mutation in a case without del(7q). Taken together, our data suggest that haploinsufficiency of MLL3 may contribute to leukemogenesis in a subset of CBF-AMLs. Whether additional genes contained within the MDR, or within the larger region of deletion on 7q, contribute to leukemogenesis remains to be determined.
Another recurrent lesion identified in the CBF-AMLs was deletion at 11p13. This chromosomal region has previously been shown to be affected by CN-LOH and rare deletions in unselected AMLs18,46 and AMLs with normal karyotype.23 WT1 has been suggested to be the primary target of these alterations, with homozygous WT1 mutations reported in cases with CN-LOH and heterozygous mutations found in a significant percentage of AMLs with normal karyotype.26,33 In our study, we identified 8 cases with alterations at 11p13 by SNP-array analysis [7 with del(11p13) and 1 with CN-LOH], with the MDR containing only WT1 and WIT1. DNA sequence analysis of WT1 in 201 of the CBF-AMLs that lacked CNAs in this region identified somatic WT1 mutations in 9.4% and 1.1% of inv(16) and t(8;21) cases, respectively. Thus, inactivating somatic mutations in the WT1 tumor suppressor are common alterations in CBF-AMLs, particularly in inv(16)/t(16;16)–containing cases.
A rarer recurrent CNA that was limited to inv(16)–containing cases localized to 17q11.2 and included heterozygous deletions in 6 cases and CN-LOH in 1 case. The MDR was 901 Kb in size and contained 14 genes, including NF1, a known tumor suppressor previously shown to be involved in myeloid malignancies.47 Sequence analysis of NF1 in 3 of the cases with heterozygous deletion failed to reveal any evidence of inactivating mutations in the nondeleted allele, suggesting that the genetic alteration leads primarily to haploinsufficiency although homozygous mutations are occasionally seen. Moreover, mutations that activate signaling through the RAS pathway have been previously reported to occur at a high frequency in CBF-AMLs.6,48
Consistent with these studies, we identified activating mutations in KIT, FLT3, JAK2, NRAS, and/or KRAS, in approximately two-thirds of cases analyzed by both CNAs and targeted sequencing of these genes. This pattern of mutation is consistent with the suggestion that leukemogenesis requires at a minimum 2 classes of mutations, one affecting genes encoding transcription factors, such as RUNX1-RUNX1T1 and CBFB-MYH11 (class II mutations), and another involving genes such as KIT, FLT3, NRAS, or KRAS, whose mutations lead to increased cell proliferation, survival, or both (class I mutations).49 Somewhat surprising was the observation that in some cases more than 1 gene within the RAS/Kinase signaling pathway was activated, including examples with mutations in NRAS and KRAS, NRAS and KIT, and FLT3 and NRAS.
9q deletions are recurrent secondary alterations in AML with t(8;21). Recently, Dayyani et al50 characterized a MDR of 2.4 Mb at 9q21.32, and they identified TLE1 and TLE2 as potential tumor suppressor targets in this region. We also found frequent 9q21.32 deletions in t(8;21)–cases and delineated a MDR that mapped into the previously defined segment. However, we were able to narrow down the MDR to a 1.1 Mb-sized genomic fragment containing only 9 genes; TLE1 and TEL4 are localized 1.3 and 3.2 Mb upstream of this MDR making a role of these genes less likely.
Besides known chromosomal gains, such as trisomy 22 in inv(16) cases, and trisomy 8 in both CBF-AML subtypes, we identified focal genomic gains at 8q24.1. The minimally gained region was 138 Kb in size and contained CCDC26, a previously identified transcriptional unit that appears to play a role in retinoic acid–induced myeloid cell differentiation. In our previous analysis of CNAs in pediatric AMLs, we also identified focal gains of this region as a rare event in nonCBF-AMLs.21 Although the biologic function of CCDC26 and/or other noncoding transcripts expressed from this region are yet unknown, our findings suggest that alteration of this region may function as a cooperating lesion in CBF-AML.
We found CN-LOH in only approximately 7% of CBF-AMLs and, thus, were somewhat less frequent than previously reported for AML.18 Recurrent CN-LOH were only detected at 1p, 4q, and 19p. NRAS was mutated in 2 of 2 cases with CN-LOH(1p), and TET2 in 1 of 2 cases with CN-LOH(4q). Several nonrecurrent CN-LOH were detected in regions also affected by recurrent or nonrecurrent CNAs, highlighting the enormous heterogeneous pattern of secondary lesions in CBF-AML.
In contrast to the low number of CNAs at diagnosis, our analysis of relapse CBF-AML samples revealed a significant increase in the mean number of CNAs per case (mean of 1.28 at diagnosis to 3.17 at relapse; P = .04). This finding is similar to the previously reported increase in the mean number of CNAs from diagnosis to relapse in ALL.15 Importantly, an analysis of the evolutionary relationship between diagnostic and relapse samples revealed that approximately two-thirds of the cases either showed no change in CNAs from diagnosis, or gained CNAs consistent with simple clonal evolution. By contrast, one-third of the cases lost diagnostic CNAs, with or without the acquisition of new relapse-specific lesions, consistent with the relapse clone arising from a clone ancestral to the diagnostic clones. Despite the increase in the overall number of CNAs at relapse, very few recurrent lesions were identified. The most common recurrent CNA acquired at relapse was del(3q13.31) found in 5 of 41 (12%) cases. The MDR contained a single transcript that includes exons from the annotated gene LSAMP, a putative tumor suppressor located 404 Kb upstream of the deletion. How these 3q13.31 losses affect LSAMP expression and whether its alteration contributes to relapse of CBF-AML remains to be determined.
In summary, these data demonstrate a low burden of genomic CNAs in CBF-AMLs at the time of diagnosis, with few recurrent lesions identified. Moreover, when CNAs were coupled with sequence analysis of a selected set of candidate cancer genes, a marked degree of heterogeneity in the specific cooperating mutations was observed within individual CBF-AMLs. Although a majority of cases contains activating mutations in RAS/kinase signaling pathway genes, each of the other identified recurrent somatic mutations occurred in only a minority of cases. Although our genome-wide analysis is limited to a single class of mutations/CNAs, these data suggest that alterations in a range of distinct biologic pathways can cooperate with CBF mutations to induce leukemia. Similarly, no common mechanism of relapse is suggested from our analysis. A range of different lesions were identified with only a subset showing low levels of recurrency within the analyzed cohort. Whether this picture of mutational heterogeneity will change to a more uniform involvement of a limited number of pathways after the application of next-generation sequencing technologies remains to be seen.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Frank Rücker, Guangchun Song, Caroline Obert, and Claire Boltz for help with data analysis and Zhongling Cai, Jianling Armstrong, Katja Urlbauer, Karin Lanz, Karina Eiwen, and Marianne Habdank for excellent technical assistance. They also thank all members of the German-Austrian AMLSG for their participation in the clinical trials and for providing leukemia samples.
This study was supported by National Institutes of Health grants P01 CA040046 (to J.R.D.) and P30 CA021765 (Cancer Center Support Grant to St Jude Children's Research Hospital), a Leukemia & Lymphoma Society Specialized Center of Research grant LLS7015 (to J.R.D.), the American Lebanese Syrian Associated Charities of St Jude Children's Research Hospital, and by a grant from the Deutsche Forschungsgemeinschaft DO-704-3-1 (to K.D.).
National Institutes of Health
Authorship
Contribution: M.W.M.K. and I.R. designed/performed research, analyzed/interpreted data, wrote the paper; L.B., S.G., and J.C. performed research and analyzed/interpreted data; J.E. analyzed/interpreted data; J.G. performed research; X.S. contributed analytical tools and analyzed/interpreted data; P.P. performed research; S.P. performed statistical analysis, analyzed/interpreted data; J. Krauter, A.G., A.Q., and R.R. analyzed/interpreted data; V.I.G. performed research; S.S. analyzed/interpreted data; J. Krönke performed research; K.H. performed research and analyzed/interpreted data; J.M. contributed analytical tools and analyzed/interpreted data; R.F.S. and J.E.R. analyzed/interpreted data; and K.D., H.D., and J.R.D. designed research, analyzed/interpreted data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: James R. Downing, MD, Scientific Director, St Jude Children's Research Hospital, 262 Danny Thomas Pl, Memphis, TN 38105; e-mail: james.downing@stjude.org; and Hartmut Döhner, MD, Department of Internal Medicine III, University Hospital of Ulm, Albert-Einstein-Allee 23, 89081 Ulm, Germany; e-mail: hartmut.doehner@uniklinik-ulm.de.
References
Author notes
M.W.M.K., I.R., H.D., and J.R.D. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal