Abstract
Anemia of chronic disease is a complication accompanying many inflammatory diseases. The proinflammatory cytokine IFN-γ has been implicated in this form of anemia, but the underlying mechanism remains unclear. Here we describe a novel mouse model for anemia of chronic disease, in which enhanced CD27-mediated costimulation strongly increases the formation of IFN-γ–producing effector T cells, leading to a progressive anemia. We demonstrate that the anemia in these mice is fully dependent on IFN-γ and that this cytokine reduces both the life span and the formation of red blood cells. Molecular analysis revealed that IFN-γ induces expression of the transcription factors of interferon regulatory factor-1 (IRF-1) and PU.1 in both murine and human erythroid precursors. We found that, on IFN-γ stimulation, IRF-1 binds to the promoter of SPI.1 (PU.1) and induces PU.1 expression, leading to inhibition of erythropoiesis. Notably, down-regulation of either IRF-1 or PU.1 expression is sufficient to overcome IFN-γ–induced inhibition of erythropoiesis. These findings reveal a molecular mechanism by which chronic exposure to IFN-γ induces anemia.
Introduction
Maintenance of the number of circulating red blood cells (RBCs) is based on a tight balance between the production of new RBCs by erythroid progenitors and the removal of effete RBCs by cells of the hemophagocytic system. Several processes can negatively affect this erythroid homeostasis and subsequently lead to the development of anemia. Iron deficiency is the primary cause of anemia in the human population, but patients with diseases involving chronic immune activation, such as persistent infections, cancer, and autoimmune diseases, are also commonly found to be anemic. This condition is termed anemia of chronic disease (ACD) or anemia of inflammation, and it is the second most prevalent form of anemia.1 Although the mechanisms involved in the development of ACD are still a matter of debate, proinflammatory cytokines, such as IL-1, IL-6, TNF-α, and IFN-γ, are thought to be important players in its development.1,2 Particularly IFN-γ has been extensively studied in this respect, and it has been shown to have a direct suppressive effect on the formation of erythroid colonies in vitro.3-6 Exposure to IFN-γ in vitro contributes to early erythroblast death because of the induction of proapoptotic molecules, such as TRAIL, TWEAK, and CD95(L).7,8 Whether this mechanism is also the cause of anemia after chronic IFN-γ exposure in vivo is not yet clear. Another possibility is that IFN-γ negatively affects the life span of RBCs because of its ability to activate macrophages and thereby the hemophagocytic system that removes RBCs from the circulation. Moreover, IFN-γ can also induce iron retention in macrophages, which negatively affects iron homeostasis and thereby the erythroid balance.9
We have investigated the consequences of enhanced IFN-γ production on erythroid homeostasis in vivo using a mouse model for sterile chronic immune activation. In this model, we overexpressed the TNF-superfamily member CD70 on B cells, which induces high numbers of IFN-γ–producing effector CD4 and CD8 T cells because of enhanced costimulation through the receptor CD27 on T cells.10 Consequently, these CD70-transgenic (CD70TG) mice display improved T-cell immunity and efficiently counter a challenge with influenza virus or tumor cells.11 However, the increased pool of effector T cells also seriously disrupts the hematopoietic system at different levels, as CD70TG mice gradually lose their B cells10 and eosinophilic granulocytes12 because of chronic exposure to IFN-γ, lose their NK cells,13 and eventually even exhaust their naive T-cell pool.14 Here we describe that CD70TG mice also become anemic, which is fully dependent on the production of IFN-γ. We used this model to examine the cellular and molecular mechanism by which IFN-γ induces anemia. We found that chronic production of IFN-γ in vivo profoundly shortens the life span of mature RBCs and also negatively affects the differentiation capacity of early erythroid progenitors in bone marrow (BM). Molecular analysis of the latter revealed that IFN-γ induces the expression of interferon regulatory factor-1 (IRF-1) in erythroid precursor cells, which up-regulates the transcription factor PU.1 and thereby inhibits erythroid differentiation. These findings demonstrate the profound impact of this cytokine on the erythroid balance in vivo and reveal the molecular mechanism by which IFN-γ inhibits erythropoiesis.
Methods
Mice
For experiments, wild-type (WT), CD70TG,10 IFN-γ−/−, and CD70TG*IFN-γ−/− mice were used. Mice were maintained on a C57BL/6 background in the animal facilities of the Academic Medical Center (University of Amsterdam, Amsterdam, The Netherlands) in specific pathogen-free conditions. Mice were given standard chow and acidified drinking water ad libitum. All animal experiments were approved by the Experimental Animal Committee of the Academic Medical Center, according to institutional and national guidelines.
Peripheral blood analysis
Full blood cell analysis, measuring RBC counts, hemoglobin content, hematocrit, red cell distribution width, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, and mean corpuscular volume, was performed on heparinized blood with an automated hemocytometer (Vet ABC Counter, SCIL; Coulter Ac•T Diff2, Beckman Coulter).
Flow cytometry and cell sorting
Single-cell suspensions from spleen were obtained by mincing them through 40-μm cell strainers. To obtain splenic macrophages, spleens were first digested with Liberase/DNAse (Roche Diagnostics) for 30 minutes at 37°C. Single-cell suspensions from BM were obtained by crushing femurs and tibiae and subsequently filtering the suspension through a 40-μm cell strainer. Where possible, cells were incubated with anti-CD16/CD32 block (2.4G2; kind gift from Louis Boon, Bioceros). Monoclonal antibodies used (all from eBioscience, unless stated otherwise) were: c-Kit–allophycocyanin (APC; 2B8); CD71-phycoerythrin (PE; R17217), Ter119-PE-Cy5.5 (Ly-76), F4/80-fluorescein isothiocyanate/APC (BM8), CD11b-APC (M1/70), PU.1-Alexa488 (9G7, Cell Signaling), and major histocompatibility complex class II-biotin (M5/114). For identification of common myeloid progenitors (CMPs) and megakaryocyte-erythroid precursors (MEPs), cells were incubated with a lineage cocktail of biotin-conjugated antibodies directed against CD4 (GK1.5), CD8α (53-6.7), B220 (RA3-6B2), CD11b (M1/70), Gr1 (RB6-8C5), and Ter119 (Ly-76). After washing, cells were incubated with streptavidin-PE, CD34-fluorescein isothiocyanate (RAM34), CD127-peridinin chlorophyll protein-Cy5.5 (A7R34), CD16/32-PE-Cy7 (93) and c-Kit-APC (2B8). For sorting, BM cells were enriched with anti-CD117 microbeads (Miltenyi Biotec), stained with the antibodies described in the preceding sentence, and CMPs and MEPs were subsequently sorted on a FACSAria (BD Biosciences). Because of the up-regulation of Sca-1 by IFN-γ on all BM cells in CD70TG mice,15 this marker was not included in the analysis or sorting strategy for CMPs and MEPs, which did not compromise our findings (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). For FACS analysis of reticulocytes, 3-5 μL of heparinized blood was washed in FACS buffer and surface stained for CD71. Cells were washed and resuspended in FACS buffer, after which Thiazole Orange (Sigma-Aldrich) was added to a final concentration of 1 ng/mL and immediately analyzed. Data acquisition was done with a FACSCalibur (BD Biosciences) or FACSCanto II (BD Biosciences), and data were analyzed using FlowJo Version 9.2 software (TreeStar).
EPO-ELISA
Serum erythropoietin (EPO) levels of mice were quantified using the Quantikine Mouse/Rat EPO Immunoassay (R&D Systems) as described by the manufacturer.
Biotinylation of RBCs
In vivo biotinylation was achieved by intravenous injection of 100 μL of 30 mg/mL sulfo-N-hydroxysuccinimide-long chain-biotin (sulfo-NHS-LC-biotin, Pierce Chemical). At regular time intervals, a few microliters of blood were isolated via vena saphena puncture, stained with PE-conjugated–streptavidin and erythroid specific antibodies and analyzed using FACS.
For adoptive transfer of biotin-labeled-RBCs, blood was drawn from WT donor mice, washed with PBS supplemented with 0.1% glucose PBS-G), and subsequently incubated with 0.1 mg/mL sulfo-NHS-LC-biotin in PBS-G for 15 minutes at room temperature. After washing, 200 μL of biotin-labeled RBCs was injected intravenously. RBC turnover was analyzed as described in the preceding paragraph.
Erythrophagocytosis assay
WT whole blood was washed with PBS, resuspended to a concentration of 2 × 108 cells/mL, and subsequently labeled with a final concentration of 25μM CFSE (Invitrogen), according to the manufacturer's protocol. Single-cell suspensions from Liberase-digested spleens were cultured for 2 hours in 12-well plates in phenol-red free DMEM (Lonza)/10% FCS for 2 hours to allow macrophage adherence. Unbound cells were removed and CFSE-labeled RBCs were added in a concentration of 2 × 107 RBCs/mL and cocultured for 2 hours. Cells were subsequently washed, nonphagocytosed RBCs were lysed with ammonium chloride, and the macrophages were isolated for FACS analysis.
Semisolid colony assays
For burst-forming unit-erythroid (BFU-e) and colony-forming unit-erythroid (CFU-e) assays, 2 × 105 nucleated total BM cells were plated in methylcellulose medium (MethoCult M3234; StemCell Technologies). Medium was supplemented with 4 U/mL human recombinant EPO (Janssen-Cilag), 100 ng/mL murine recombinant stem cell factor, 20 μg/mL human holo-transferrin (SCIPAC, T101-5), 2 × 10−4M hemin (Sigma-Aldrich, H9039), and 1% penicillin/streptomycin/l-glutamine solution (Invitrogen). CFU-e colonies were counted on day 3, and BFU-e colonies on day 8. When appropriate, mouse recombinant IFN-γ (PeproTech) was added. FACS-sorted BM CMPs and MEPs were cultured in complete methylcellulose medium (Methocult M3434; StemCell Technologies) at 250 cells per 35-mm culture dish and analyzed on day 8.
Microarray
CD71+ cells were MACS-enriched by labeling with biotin-conjugated CD71 antibodies (eBioscience) and streptavidin-conjugated microbeads (Miltenyi Biotec). Total RNA was extracted using Trizol (Invitrogen). Initial RNA yield and subsequent quality of the labeled fragmented cRNA was determined using the 2100 Bioanalyzer (Agilent Technologies). A total of 100-300 ng of total RNA was hybridized to Mouse Gene 1.0 ST Array GeneChips, according to the manufacturer's protocols (Affymetrix). Single array expression analysis was performed using Genespring GX v7.3.1 software (Agilent Technologies). This platform generates a list of differentially expressed genes after filtering absent, marginal, or AFFY control probe sets and applying a log2 transformation. A 1.5-fold change threshold and test statistic of P < .05 were used as cut-off. These data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE22656. Further analysis was performed using Ingenuity Pathways Analysis (Ingenuity Systems). Potential PU.1 and Gata1 target gene classification was obtained by comparing our list of IFN-γ–dependent differentially expressed genes with unpublished (L.G.), published PU.1 (GDS9011)16 and Gata1 ChIP-seq data (ERA000161)17 using Ingenuity Pathways Analysis.
Quantitative PCR
Total RNA was isolated using Trizol (Invitrogen) and reverse-transcribed to cDNA using random hexamers and Superscript II reverse transcriptase (Roche Diagnostics). Quantitative PCR was performed using the StepOnePlus RT-PCR system (Applied Biosystems) using Express SYBR GreenER (Invitrogen). Primer sequences are available on request.
Western blot
Total protein extracts were made according to standard methods. Proteins were separated on a 12.5% SDS-PAGE gels and subsequently transferred onto polyvinylidene difluoride membranes (Sigma-Aldrich) by electroblotting. Membranes were blocked with PBS containing 1% BSA or 3% milk and 0.25% Tween. Blots were incubated overnight at 4°C with primary antibodies, thoroughly washed with PBS containing 0.25% Tween, incubated with appropriate HRP-conjugated secondary antibodies, and developed in enhanced chemiluminescence reagents (GE Healthcare). Antibodies used were: PU.1 (T21, sc-352), Gata1 (N6, 265 and H200, sc-13053), IRF-1 (H8, sc-74530), GAPDH (MAB374 Chemicon Millipore), beta-actin (ab-6276), and Nucleophosmin (FC8229, ab-10530).
Cell culture of human erythroid progenitors and shRNA-mediated knockdown
In accordance with institutional guidelines provided by the Erasmus MC Medical Ethical Committee, human erythroid progenitor cells were cultured as described,18 in the presence of recombinant human EPO (1 unit/mL, kind gift of Ortho-Biotech), recombinant human stem cell factor (50 ng/mL, kind gift of Amgen), and dexamethasone (5 × 10−7M; Sigma-Aldrich).
Lentivirus was produced by transient transfection of 293T cells.19 Supernatant was harvested over 3 consecutive days after transfection, kept at 4°C and pooled. Pooled supernatant was filtered and concentrated by centrifugation at 72 000g for 2 hours at 4°C. Human erythroid progenitor cells were transduced in 6-well plates at 2-3 × 106 cells per well and sufficient amounts of virus to transduce approximately 80% of the cells. When appropriate, puromycin (1 μg/mL final concentration) was added after 1 day, and selection was performed overnight.
For knockdown experiments, clones from the RNAi Consortium (TRC15; Sigma-Aldrich) were used. The nontarget SHC002 vector was used as a control. (SHC002: 5′-CAACAAGATGAAGAGCACCAA-3′). We tested 5 shRNA clones directed against PU.1 and IRF-1 mRNA, and we selected the ones with stronger down-regulation for further experiments: PU.1 (TRCN0000020538: 5′-GAAGAAGCTCACCTACCAGTT-3′), and IRF-1 (TRCN0000014671: 5′-AGATGCTAAGAGCAAGGCCAA-3′).
At days 1 to 3 after selection, cells were counted and plated in triplicate in methylcellulose medium (MethoCult H4434, StemCell Technologies). We used 2 × 105 cells for CFU-e counts and 2 × 106 cells for BFU-e counts. Medium was supplemented with 1% penicillin/streptomycin solution (Invitrogen) and, when appropriate, with 200 ng/mL of human recombinant IFN-γ (PeproTech). CFU-e colonies were counted on day 8 and BFU-e colonies on day 16.
ChIP
Human erythroid progenitor cells were cultured and grown in sufficient numbers and pulsed or mock pulsed overnight with 200 ng/mL human recombinant IFN-γ. Cells were collected, and ChIP was performed as described20 with IRF-1 (H8, sc-74530) and CD71 antibody (347510, BD Biosciences) as a negative control. Quantitative PCR was performed on the input and immunoprecipitated samples using primers for the IRF-1–binding site at the TAP121 and SPI.1 (PU.1) promoter. The relative fold enrichment was calculated as RFE = 2−[(CT ChIP sample − CT input sample). Primers used are as follows: (1) TAP1 promoter, amplicon size 94 bp: 5′-GGCGAGAAGCTCAGCATT-3 and 5′-TAGTCTGGGCAGGCCACTTT-3′; and (2) SPI.1 (PU.1) promoter, amplicon size 121 bp: 5′-CTGGTCTGAAGTGCCTTTCTTTG-3 and 5′-AAGAAGGAGTTGAGGAGCCAC-3′.
Statistical analysis
Results are expressed as mean ± SD. Statistical analysis between groups was performed with Graphpad Prism 5, using either a paired or nonpaired 2-tailed Student t test comparing 2 groups or a 1-way or 2-way ANOVA test with Bonferroni correction comparing > 2 groups. Area under the curve analysis was performed using SPSS Version 15.0.1 software. P values < .05 were considered to be statistically significant.
Results
CD70TG mice develop an IFN-γ–dependent anemia
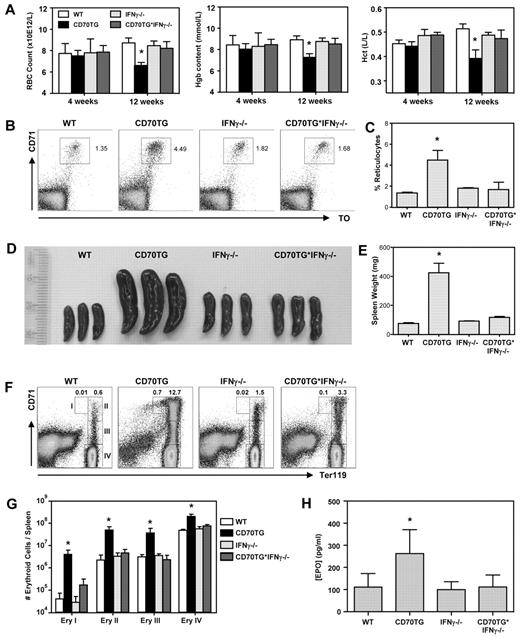
As CD70 overexpression induces formation of high numbers of IFN-γ–producing effector T cells, we used this model of chronic immune activation to examine the impact of IFN-γ on RBC homeostasis in vivo.10 Although CD70TG mice have normal RBC numbers, hemoglobin levels, and hematocrit at 4 weeks of age, they develop severe anemia over time, as is evident from the significant decrease of these parameters in 12-week-old mice (Figure 1A). Cytometric analysis indicated that CD70TG mice develop a normochromic (normal mean corpuscular hemoglobin concentration) and normocytic (normal mean corpuscular volume) anemia (Table 1). Importantly, this anemia is fully dependent on IFN-γ, as it does not occur in IFN-γ–deficient CD70TG (CD70TG*IFN-γ−/−) mice. Mean corpuscular hemoglobin is slightly increased in CD70TG mice, but not in CD70TG*IFN-γ−/− mice (Table 1), which correlates with an IFN-γ–dependent increase in reticulocytes in peripheral blood (Figure 1B-C).
CD70TG mice develop IFN-γ–dependent anemia. Analysis of WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. (A) Hemocytometric analysis at 4 and 12 weeks of age: Hgb indicates hemoglobin; and Hct, hematocrit. Data are mean ± SD bar graphs for 3-5 mice per group. *Significant difference (P < .05) between CD70TG mice and all other groups (2-way ANOVA with Bonferroni correction). (B-C) Representative dot plots and bar graph depicting the percentage of blood reticulocytes (ie, CD71+ and thiazole orange-positive cells). (D) Spleens. (E) Spleen weight. (F) Representative dot plots of the splenic erythroid compartment. Numbers I, II, III, and IV refer to the respective erythroblast subsets defined by Socolovsky et al.22 (G) Absolute numbers of indicated erythroblast subsets per spleen. Data are mean ± SD in bar graphs for 3 mice per group. Results are representative from 3 independently performed experiments. *Significant difference (P < .05) between CD70TG mice and all other groups (1-way ANOVA). (H) Plasma EPO levels. Data are mean ± SD in bar graphs for 5-8 mice per group. *Significant difference (P < .05) between CD70TG mice and all other groups (1-way ANOVA with Bonferroni correction). (B-H) Mice were used in the age range of 10-16 weeks old.
CD70TG mice develop IFN-γ–dependent anemia. Analysis of WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. (A) Hemocytometric analysis at 4 and 12 weeks of age: Hgb indicates hemoglobin; and Hct, hematocrit. Data are mean ± SD bar graphs for 3-5 mice per group. *Significant difference (P < .05) between CD70TG mice and all other groups (2-way ANOVA with Bonferroni correction). (B-C) Representative dot plots and bar graph depicting the percentage of blood reticulocytes (ie, CD71+ and thiazole orange-positive cells). (D) Spleens. (E) Spleen weight. (F) Representative dot plots of the splenic erythroid compartment. Numbers I, II, III, and IV refer to the respective erythroblast subsets defined by Socolovsky et al.22 (G) Absolute numbers of indicated erythroblast subsets per spleen. Data are mean ± SD in bar graphs for 3 mice per group. Results are representative from 3 independently performed experiments. *Significant difference (P < .05) between CD70TG mice and all other groups (1-way ANOVA). (H) Plasma EPO levels. Data are mean ± SD in bar graphs for 5-8 mice per group. *Significant difference (P < .05) between CD70TG mice and all other groups (1-way ANOVA with Bonferroni correction). (B-H) Mice were used in the age range of 10-16 weeks old.
IFN-γ–dependent normochromic and normocytic anemia in CD70TG mice
| . | WT . | CD70TG . | IFN-γ−/− . | CD70TG* IFN-γ−/− . |
|---|---|---|---|---|
| RBCs × 1012/L | 9.13 (0.84) | 6.58 (1.08)* | 8.59 (0.41) | 8.30 (0.57) |
| Hemoglobin, mM | 9.30 (0.75) | 7.18 (1.01)* | 8.89 (0.32) | 8.56 (0.59) |
| Hematocrit, L/L | 0.49 (0.04) | 0.39 (0.07)* | 0.49 (0.03) | 0.47 (0.02) |
| RDW, % | 18.86 (1.57) | 18.04 (1.69) | 19.31 (2.24) | 23.32 (3.61) |
| MCH, fmol | 1.03 (0.02) | 1.10 (0.03)* | 1.04 (0.02) | 1.04 (0.02) |
| MCV, fL | 54.17 (3.66) | 53.37 (6.22) | 51.27 (3.42) | 51.27 (5.35) |
| MCHC, mM | 19.17 (1.54) | 20.26 (1.63) | 19.30 (1.09) | 19.85 (1.60) |
| . | WT . | CD70TG . | IFN-γ−/− . | CD70TG* IFN-γ−/− . |
|---|---|---|---|---|
| RBCs × 1012/L | 9.13 (0.84) | 6.58 (1.08)* | 8.59 (0.41) | 8.30 (0.57) |
| Hemoglobin, mM | 9.30 (0.75) | 7.18 (1.01)* | 8.89 (0.32) | 8.56 (0.59) |
| Hematocrit, L/L | 0.49 (0.04) | 0.39 (0.07)* | 0.49 (0.03) | 0.47 (0.02) |
| RDW, % | 18.86 (1.57) | 18.04 (1.69) | 19.31 (2.24) | 23.32 (3.61) |
| MCH, fmol | 1.03 (0.02) | 1.10 (0.03)* | 1.04 (0.02) | 1.04 (0.02) |
| MCV, fL | 54.17 (3.66) | 53.37 (6.22) | 51.27 (3.42) | 51.27 (5.35) |
| MCHC, mM | 19.17 (1.54) | 20.26 (1.63) | 19.30 (1.09) | 19.85 (1.60) |
Hemocytometric analysis of 10- to 16-week-old WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. Values indicate the mean (SD) from 10-18 mice.
RBC indicates red blood cell; RDW, red cell distribution width; MCH, mean corpuscular hemoglobin; MCV, mean corpuscular volume; and MCHC; mean corpuscular hemoglobin concentration.
Significant difference (P < .05) between CD70TG mice and all other groups (1-way ANOVA with Bonferroni correction).
As mice typically respond to anemia and hypoxic stress by inducing extramedullary stress erythropoiesis in the spleen, we examined spleens of CD70TG mice in more detail. CD70TG mice develop a severe IFN-γ–dependent splenomegaly (Figure 1 D-E). This increase in spleen size cannot be attributed to a leukocyte expansion, as CD70TG mice have reduced splenocyte numbers because of progressive depletion of B cells.10 Therefore, we examined the splenic erythroid compartment, based on the differential expression of TER119 and CD71.22 We found that CD70TG mice have a dramatic increase of all erythroblast subsets, whereas CD70TG*IFN-γ−/− mice do not (Figure 1F-G). Finally, CD70TG mice also displayed a significant, IFN-γ–dependent increase in plasma EPO levels (Figure 1H), consistent with the notion that these mice have hypoxia. These data demonstrate that CD70TG mice develop an IFN-γ–dependent anemia and concomitant stress erythropoiesis in the spleen. We used this model to investigate the mechanism by which IFN-γ contributes to ACD.
IFN-γ enhances RBC turnover and increases macrophage erythrophagocytosis
As normocytic anemia in combination with reticulocytosis is indicative of accelerated RBC destruction, we examined whether IFN-γ affected the life span of RBCs in CD70TG mice. Therefore, mice were injected intravenously with biotin, which stably labels all RBCs in the circulation23 and allowed us to determine RBC turnover (Figure 2A). Whereas WT mice lost 50% of their biotinylated RBCs in approximately 23 days, CD70TG mice reached this point already after 12 days (Figure 2B). This increased loss of biotinylated RBCs was fully IFN-γ–dependent, as RBC removal was normal or even slightly delayed in CD70TG*IFN-γ−/− mice (Figure 2B; supplemental Figure 2a).
Constitutive IFN-γ exposure in vivo enhances RBC turnover and increases macrophage erythrophagocytosis. (A) Flow cytometric analysis of in vivo biotinylated peripheral blood using fluorescently labeled streptavidin and Ter119. Representative dot plots of day 17 after transfer are shown. (B) Turnover of in vivo biotinylated RBCs from WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice as measured by flow cytometry. (C) Turnover of adoptively transferred ex vivo biotinylated WT RBCs in WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice as measured by flow cytometry. (D) Major histocompatibility complex class II surface expression of splenic red pulp macrophages (F4/80+ CD11blow) expressed as geometric MFI. (E) Uptake of CFSE-labeled erythrocytes by splenic red pulp macrophages (F4/80+) expressed as the ratio of CFSE geometric MFI compared with background. (E) Data are representative for 2 experiments. Data are mean ± SD for 4 mice (B-C), 3 mice (D), or duplicate analysis (E) per group. *Significant difference (P < .05) between CD70TG or CD70TG*IFN-γ−/− mice and all other groups (1-way ANOVA with Bonferroni correction).
Constitutive IFN-γ exposure in vivo enhances RBC turnover and increases macrophage erythrophagocytosis. (A) Flow cytometric analysis of in vivo biotinylated peripheral blood using fluorescently labeled streptavidin and Ter119. Representative dot plots of day 17 after transfer are shown. (B) Turnover of in vivo biotinylated RBCs from WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice as measured by flow cytometry. (C) Turnover of adoptively transferred ex vivo biotinylated WT RBCs in WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice as measured by flow cytometry. (D) Major histocompatibility complex class II surface expression of splenic red pulp macrophages (F4/80+ CD11blow) expressed as geometric MFI. (E) Uptake of CFSE-labeled erythrocytes by splenic red pulp macrophages (F4/80+) expressed as the ratio of CFSE geometric MFI compared with background. (E) Data are representative for 2 experiments. Data are mean ± SD for 4 mice (B-C), 3 mice (D), or duplicate analysis (E) per group. *Significant difference (P < .05) between CD70TG or CD70TG*IFN-γ−/− mice and all other groups (1-way ANOVA with Bonferroni correction).
Because IFN-γ is a potent activator of macrophages and the hemophagocytic system, we determined whether IFN-γ accelerated RBC turnover in CD70TG mice in an extrinsic manner. Therefore, we transferred ex vivo–biotinylated WT RBCs to the 4 different groups of mice and followed the fate of these cells over time. This approach revealed that WT RBCs were also removed more rapidly from the circulation of CD70TG mice, which was not seen in CD70TG*IFN-γ−/− mice (Figure 2C; supplemental Figure 2b). This increase in hemophagocytic capacity correlated well with the finding that splenic red pulp macrophages (F4/80hi, CD11blo) were activated in CD70TG mice, but not in CD70TG*IFN-γ−/− mice, based on the increased expression of major histocompatibility complex class II (Figure 2D). To establish the erythrophagocytic capacity of these cells, splenic red pulp macrophages were isolated, cocultured for 2 hours with CFSE-labeled WT RBCs, followed by lysis of the remaining nonphagocytosed RBCs. These experiments indicate that, compared with those from WT mice, red pulp macrophages from CD70TG mice phagocytose more erythrocytes, whereas red pulp macrophages from CD70TG*IFN-γ−/− mice phagocytose fewer (Figure 2E). This correlates well with the degree of RBC turnover observed in vivo (Figure 2B). Taken together, these data indicate that IFN-γ produced during chronic inflammation activates the splenic hemophagocytic compartment, thereby enhancing the turnover of RBCs.
IFN-γ inhibits erythroid output from BM
As IFN-γ can also negatively regulate erythropoiesis of BM progenitor cells,4,6,24 we analyzed the erythroid compartment in BM from CD70TG mice and found significantly fewer erythroblasts (Ery II, III, and IV) than in WT BM (Figure 3A). For the early subsets, this was to a certain extent also seen in CD70TG*IFN-γ−/− mice, but the final stage of orthochromatophilic erythroblasts (Ery IV) was not affected in these mice (Figure 3A). This correlated well with the observations that BM of CD70TG mice was paler compared with WT, IFN-γ−/−, and CD70TG*IFN-γ−/− mice (data not shown), which is also a clear sign of a reduction in hemoglobinized cells. Furthermore, we found a relative accumulation of pro-erythroblasts in CD70TG mice (Figure 3B), which, together with the decrease in absolute number of mature erythroid cells (Figure 3A), indicates that erythropoiesis in CD70TG BM is hampered. We found no evidence for increased apoptosis of erythroid precursor populations in CD70TG mice, based on annexin V stainings of BM (supplemental Figure 3). Yet, reduced erythropoiesis was confirmed when the erythroid-forming capacity of the BM was functionally tested using colony-forming assays, as BFU-e numbers were strongly reduced in CD70TG BM compared with WT BM. This reduction was dependent on IFN-γ (Figure 3C). Formation of more mature CFU-e was not affected, but the negative impact of IFN-γ on erythroid formation was evident from the fact that both IFN-γ−/− and CD70TG*IFN-γ−/− mice had more CFU-e than WT and CD70TG mice (Figure 3D). Moreover, purified CMPs from CD70TG mice formed fewer erythroid colonies compared with WT mice, but more myeloid colonies (Figure 3E), indicating a decreased commitment of hematopoietic progenitors to the erythroid lineage. Subsequently, we examined BM of CD70TG mice for the presence of IFN-γ–producing T cells, which revealed that CD70TG mice have 7-fold more IFN-γ–producing CD4+ T cells and 4-fold more IFN-γ–producing CD8+ T cells than WT mice (Figure 3F-G). Serum IFN-γ levels were undetectable12 (and data not shown), strongly suggesting that a local increase in IFN-γ production is responsible for the decreased BFU-e capacity in CD70TG mice. Moreover, we demonstrate that IFN-γ is sufficient to inhibit the outgrowth of BFU-e from WT BM (Figure 3H), whereas CFU-e are less sensitive to this cytokine (Figure 3I), which is consistent with previous findings.5 In conclusion, these data imply that prolonged exposure to high IFN-γ levels in vivo causes ACD, not only by increased turnover of RBCs, but also by a reduction in the erythroid-forming capacity of hematopoietic progenitor cells in BM.
IFN-γ production in CD70TG mice inhibits erythroid BM output. (A) Absolute numbers of various erythroblast subsets or (B) relative contribution of pro-erythroblasts (percentage Ery I [Ter119medCD71high] from all erythroid [Ter119med/high cells]) in BM (isolated from 2 femurs and 2 tibiae) of WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. (C-D) BFU-e and CFU-e numbers of unfractionated BM from WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. (E) Colony assay of FACS-sorted CMPs from WT and CD70TG mice. (F-G) Absolute numbers of IFN-γ–producing CD4 and CD8 T cells in BM of WT and CD70TG mice, measured after phorbol myristate acetate/ionomycin stimulation. (H-I) Effect of IFN-γ on the in vitro BFU-e and CFU-e colony-forming capacity of BM cells from WT mice. Data are mean ± SD for 3 mice per group (A-D,F-G) or triplicate analysis (E,H-I). Results are representative from 3 (A-D) or 2 (E-I) independently performed experiments. *P < .01 (A) or P < .05 (B-H) using 1-way ANOVA with Bonferroni correction (A-D) or nonpaired Student t test (E-I).
IFN-γ production in CD70TG mice inhibits erythroid BM output. (A) Absolute numbers of various erythroblast subsets or (B) relative contribution of pro-erythroblasts (percentage Ery I [Ter119medCD71high] from all erythroid [Ter119med/high cells]) in BM (isolated from 2 femurs and 2 tibiae) of WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. (C-D) BFU-e and CFU-e numbers of unfractionated BM from WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. (E) Colony assay of FACS-sorted CMPs from WT and CD70TG mice. (F-G) Absolute numbers of IFN-γ–producing CD4 and CD8 T cells in BM of WT and CD70TG mice, measured after phorbol myristate acetate/ionomycin stimulation. (H-I) Effect of IFN-γ on the in vitro BFU-e and CFU-e colony-forming capacity of BM cells from WT mice. Data are mean ± SD for 3 mice per group (A-D,F-G) or triplicate analysis (E,H-I). Results are representative from 3 (A-D) or 2 (E-I) independently performed experiments. *P < .01 (A) or P < .05 (B-H) using 1-way ANOVA with Bonferroni correction (A-D) or nonpaired Student t test (E-I).
IFN-γ induces expression of PU.1
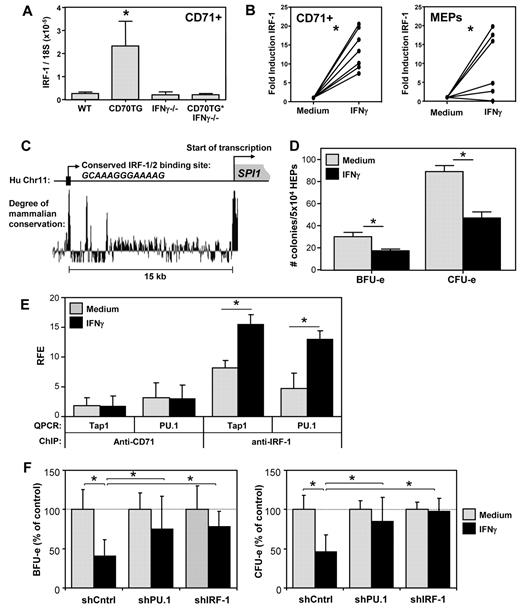
To investigate the molecular mechanism by which IFN-γ affects erythropoiesis, we performed microarray analysis on CD71+ erythroblasts from BM of WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. Unsupervised cluster analysis showed that erythroblasts from CD70TG mice clustered separately from the other mice, independently of their ability to produce IFN-γ (Figure 4A). However, we also found a set of 191 transcripts that was differentially expressed in CD70TG mice compared with CD70TG*IFN-γ−/− as well as WT and IFN-γ−/− mice, and we decided to further focus on these genes, as they were directly or indirectly regulated by IFN-γ. Comparison with other datasets16,17 revealed that 128 of these 191 genes (67%) are potential targets of the transcription factors PU.1 or GATA-1 (Figure 4B; supplemental Figure 4), which are key regulators of myeloid and erythroid differentiation, respectively. During normal erythroid differentiation, PU.1 is down-regulated, whereas GATA-1 up-regulation mediates differentiation of erythroid precursors.25 Expression of these factors has to be tightly controlled during hematopoiesis, as PU.1 and GATA-1 physically interact and thereby block each other's function.26-28 Protein expression analysis of these transcription factors revealed that PU.1 was highly up-regulated in CD71+ cells of CD70TG mice, but not in CD70TG*IFN-γ−/−, and even decreased in IFN-γ−/− mice; expression of GATA-1 was not altered between the different groups of mice (Figure 4C). Flow cytometric analysis of earlier progenitors revealed that PU.1 was also up-regulated in megakaryocyte/erythrocyte lineage-restricted progenitors (MEPs) of CD70TG, but not of CD70TG*IFN-γ−/− mice (Figure 4D). Finally, we found that IFN-γ treatment of purified CD71+ cells (Figure 4E) or MEPs (Figure 4F) was sufficient to up-regulate PU.1 expression, whereas it did not affect GATA-1 (Figure 4F). Because PU.1 is a known inhibitor of erythroid differentiation,27-29 these data strongly suggest that IFN-γ blocks erythropoiesis through the induction of PU.1.
IFN-γ induces expression of PU.1 in erythroblasts. (A) Microarray heat map of unsupervised cluster analysis for genes expressed in CD71+ erythroblasts from BM of WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. Red represents high expression; and green, low expression. Three mice were analyzed per group. (B) Venn diagram displaying potential PU.1 and GATA-1 targets among the IFN-γ–regulated genes in CD70TG mice. (C) Western blot analysis of PU.1 and GATA-1 expression on purified CD71+ BM cells from 2 mice per experimental group. (D) Flow cytometric analysis of PU.1 expression in MEPs of WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. Horizontal line represents the value of the isotype control. Data are mean ± SD for 3 mice per group. Results are representative from 2 independently performed experiments. *Significant difference (P < .05) between CD70TG mice and all other groups (1-way ANOVA with Bonferroni correction). (E) Quantitative PCR analysis of PU.1 mRNA on WT CD71+ erythroblasts cultured overnight with or without IFN-γ. (F) Quantitative PCR analysis of PU.1 and GATA-1 mRNA on WT MEPs cultured overnight with or without IFN-γ. (E-F) Data are presented as the fold induction of gene expression with IFN-γ compared with the medium control for 7 (E), or 9 and 6 (F) mice per group, pooled from at least 2 independently performed experiments. *P < .01 (E) or P < .05 (F) using paired Student t test. n.s. indicates not significant.
IFN-γ induces expression of PU.1 in erythroblasts. (A) Microarray heat map of unsupervised cluster analysis for genes expressed in CD71+ erythroblasts from BM of WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. Red represents high expression; and green, low expression. Three mice were analyzed per group. (B) Venn diagram displaying potential PU.1 and GATA-1 targets among the IFN-γ–regulated genes in CD70TG mice. (C) Western blot analysis of PU.1 and GATA-1 expression on purified CD71+ BM cells from 2 mice per experimental group. (D) Flow cytometric analysis of PU.1 expression in MEPs of WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. Horizontal line represents the value of the isotype control. Data are mean ± SD for 3 mice per group. Results are representative from 2 independently performed experiments. *Significant difference (P < .05) between CD70TG mice and all other groups (1-way ANOVA with Bonferroni correction). (E) Quantitative PCR analysis of PU.1 mRNA on WT CD71+ erythroblasts cultured overnight with or without IFN-γ. (F) Quantitative PCR analysis of PU.1 and GATA-1 mRNA on WT MEPs cultured overnight with or without IFN-γ. (E-F) Data are presented as the fold induction of gene expression with IFN-γ compared with the medium control for 7 (E), or 9 and 6 (F) mice per group, pooled from at least 2 independently performed experiments. *P < .01 (E) or P < .05 (F) using paired Student t test. n.s. indicates not significant.
IFN-γ up-regulates PU.1 expression through induction of IRF-1
Next, we set out to determine the molecular mechanism by which IFN-γ signaling induces PU.1 expression. The microarray analysis revealed that IFN-γ also induced expression of the transcription factor IRF-1 and its downstream target genes (Table 2; supplemental Figure 4). We validated by quantitative PCR that IRF-1 was up-regulated in CD71+ BM cells of CD70TG mice in an IFN-γ–dependent manner (Figure 5A). Moreover, incubation with IFN-γ was sufficient to induce IRF-1 in CD71+ cells and MEPs (Figure 5B). Examination of the SPI.1 (PU.1) promoter revealed a potential IRF-1/2 binding site approximately 15 kb upstream of the transcription start site, which was highly conserved among mammals (Figure 5C). To determine whether IRF-1 protein indeed interacted with this domain, we performed ChIP experiments, using expanded primary human erythroid progenitor cells; these cells can be expanded to sufficient numbers required for these experiments,18 and they enabled us to validate the effect of IFN-γ also on human erythroid precursors. We found that IFN-γ inhibited the outgrowth of BFU-e and CFU-e of human erythroid progenitor cells in vitro (Figure 5D), which corroborates comparable experiments with progenitor cells from human BM.3,4,6 Moreover, IFN-γ induced expression of both IRF-1 and SPI.1 (PU.1) also in these primary human cells (supplemental Figure 5a). Subsequent ChIP analysis revealed that IFN-γ treatment increased IRF-1 binding to the highly conserved IRF-1/2 motifs in the PU.1 locus (Figure 5E). IFN-γ treatment also increased binding of IRF-1 to an IRF-1 binding site in the Tap1 promoter,21 which served as a positive control. No significant enrichment was observed when a negative control antibody (anti-CD71) was used for the immunoprecipitation.
Relative expression of IFN-γ–inducible genes in erythroblasts of CD70TG mice and their relationship with IRF-1
| IRF-1–related gene . | Fold difference in expression between . | Relationship with IRF-1 . | ||||
|---|---|---|---|---|---|---|
| CD70TG vs WT . | CD70TG vs CD70TG*IFN-γ−/− . | WT vs IFN-γ−/− . | WT vs CD70TG*IFN-γ−/− . | IFN-γ−/− vs CD70TG*IFN-γ−/− . | ||
| IRF-1 | 3.0 | 5.0 | 1.1 | 1.6 | 1.5 | IRF-1 itself |
| CD274 (PD-L1) | 2.1 | 2.1 | −1.0 | 1.0 | 1.0 | IRF-1–dependent IFN-γ–inducible gene30 |
| GBP1 | 1.9 | 1.9 | 1.0 | 1.0 | 1.0 | IRF-1–dependent IFN-γ–inducible gene31 |
| GBP2 | 12.2 | 21.5 | 1.4 | 1.8 | 1.3 | IRF-1–dependent IFN-γ–inducible gene32 |
| GBP4 | 4.0 | 5.9 | 1.1 | 1.5 | 1.3 | IRF-1–dependent IFN-γ–inducible gene32 |
| PSMB9 (LMP-2) | 3.2 | 4.3 | 1.1 | 1.3 | 1.2 | IRF-1–dependent IFN-γ–inducible gene33 |
| TAP1 | 3.3 | 4.6 | 1.1 | 1.4 | 1.2 | IRF-1–dependent IFN-γ–inducible gene33 |
| IFI47 (IRG47) | 3.0 | 3.6 | −1.0 | 1.2 | 1.2 | IRF-1–independent IFN-γ–inducible gene32 |
| IGTP | 4.1 | 6.5 | −1.1 | 1.6 | 1.7 | IRF-1–independent IFN-γ–inducible gene32 |
| IIGP1 | 12.7 | 14.7 | 1.2 | 1.2 | 1.0 | IRF-1–independent IFN-γ–inducible gene32 |
| IRGM (LRG47) | 1.7 | 2.3 | −1.1 | 1.3 | 1.5 | IRF-1–independent IFN-γ–inducible gene32 |
| TGTP | 4.9 | 8.8 | 1.1 | 1.9 | 1.7 | IRF-1–independent IFN-γ–inducible gene32 |
| IRF-1–related gene . | Fold difference in expression between . | Relationship with IRF-1 . | ||||
|---|---|---|---|---|---|---|
| CD70TG vs WT . | CD70TG vs CD70TG*IFN-γ−/− . | WT vs IFN-γ−/− . | WT vs CD70TG*IFN-γ−/− . | IFN-γ−/− vs CD70TG*IFN-γ−/− . | ||
| IRF-1 | 3.0 | 5.0 | 1.1 | 1.6 | 1.5 | IRF-1 itself |
| CD274 (PD-L1) | 2.1 | 2.1 | −1.0 | 1.0 | 1.0 | IRF-1–dependent IFN-γ–inducible gene30 |
| GBP1 | 1.9 | 1.9 | 1.0 | 1.0 | 1.0 | IRF-1–dependent IFN-γ–inducible gene31 |
| GBP2 | 12.2 | 21.5 | 1.4 | 1.8 | 1.3 | IRF-1–dependent IFN-γ–inducible gene32 |
| GBP4 | 4.0 | 5.9 | 1.1 | 1.5 | 1.3 | IRF-1–dependent IFN-γ–inducible gene32 |
| PSMB9 (LMP-2) | 3.2 | 4.3 | 1.1 | 1.3 | 1.2 | IRF-1–dependent IFN-γ–inducible gene33 |
| TAP1 | 3.3 | 4.6 | 1.1 | 1.4 | 1.2 | IRF-1–dependent IFN-γ–inducible gene33 |
| IFI47 (IRG47) | 3.0 | 3.6 | −1.0 | 1.2 | 1.2 | IRF-1–independent IFN-γ–inducible gene32 |
| IGTP | 4.1 | 6.5 | −1.1 | 1.6 | 1.7 | IRF-1–independent IFN-γ–inducible gene32 |
| IIGP1 | 12.7 | 14.7 | 1.2 | 1.2 | 1.0 | IRF-1–independent IFN-γ–inducible gene32 |
| IRGM (LRG47) | 1.7 | 2.3 | −1.1 | 1.3 | 1.5 | IRF-1–independent IFN-γ–inducible gene32 |
| TGTP | 4.9 | 8.8 | 1.1 | 1.9 | 1.7 | IRF-1–independent IFN-γ–inducible gene32 |
Selection of IRF-1–related genes (based on the microarray data described in Figure 4 and supplemental Figure 3) that were significantly up-regulated in CD70TG mice compared with WT as well as CD70TG*IFN-γ−/− mice. These IFN-γ–inducible genes were categorized as being either IRF-1–dependent (top half) or –independent (bottom half), based on indicated literature references (right column).
IFN-γ up-regulates PU.1 expression in erythroblasts through IRF-1. (A) Quantitative PCR analysis of the expression of IRF-1 mRNA in CD71+ erythroblasts from WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. Data are mean ± SD for 3 mice per group. *Significant difference (P < .05) between CD70TG mice and all other groups (1-way ANOVA with Bonferroni correction). (B) Quantitative PCR analysis of IRF-1 mRNA in CD71+ erythroblasts or MEPs from WT mice cultured overnight with or without IFN-γ. Data are the fold induction of IRF-1 expression with IFN-γ compared with the medium control for 7 (CD71+) or 6 (MEPs) mice per group, pooled from 2 independently performed experiments. *P < .001 (CD71+) or P < .05 (MEPs; paired Student t test). (C) Sequence analysis of the promoter region of the human SPI1 (PU.1) gene, displaying a conserved putative IRF-1/2 binding site approximately 15 kb upstream of the transcription start site (based on the UCSC Genome Browser). (D) Effect of IFN-γ on the in vitro BFU-e and CFU-e colony-forming potential of human erythroid precursor cells cultured overnight with or without IFN-γ. Data are mean ± SD from triplicate analysis. Results are representative from 2 independently performed experiments. *P < .05 (nonpaired Student t test). (E) ChIP analysis of the binding of IRF-1 to the putative IRF-1/2 binding site in the SPI1 (PU.1) promoter of human erythroid precursor cells cultured overnight with or without IFN-γ. RFE indicates relative fold enrichment. Data are mean ± SD from 3 independent experiments. *P < .05 (nonpaired Student t test). (F) Effect of IFN-γ on the BFU-e and CFU-e potential of human erythroid precursor cells transduced with shRNA directed against IRF-1 or PU.1. Data are mean ± SD from 3 independent experiments. *P < .05 (1-way ANOVA with Bonferroni correction).
IFN-γ up-regulates PU.1 expression in erythroblasts through IRF-1. (A) Quantitative PCR analysis of the expression of IRF-1 mRNA in CD71+ erythroblasts from WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. Data are mean ± SD for 3 mice per group. *Significant difference (P < .05) between CD70TG mice and all other groups (1-way ANOVA with Bonferroni correction). (B) Quantitative PCR analysis of IRF-1 mRNA in CD71+ erythroblasts or MEPs from WT mice cultured overnight with or without IFN-γ. Data are the fold induction of IRF-1 expression with IFN-γ compared with the medium control for 7 (CD71+) or 6 (MEPs) mice per group, pooled from 2 independently performed experiments. *P < .001 (CD71+) or P < .05 (MEPs; paired Student t test). (C) Sequence analysis of the promoter region of the human SPI1 (PU.1) gene, displaying a conserved putative IRF-1/2 binding site approximately 15 kb upstream of the transcription start site (based on the UCSC Genome Browser). (D) Effect of IFN-γ on the in vitro BFU-e and CFU-e colony-forming potential of human erythroid precursor cells cultured overnight with or without IFN-γ. Data are mean ± SD from triplicate analysis. Results are representative from 2 independently performed experiments. *P < .05 (nonpaired Student t test). (E) ChIP analysis of the binding of IRF-1 to the putative IRF-1/2 binding site in the SPI1 (PU.1) promoter of human erythroid precursor cells cultured overnight with or without IFN-γ. RFE indicates relative fold enrichment. Data are mean ± SD from 3 independent experiments. *P < .05 (nonpaired Student t test). (F) Effect of IFN-γ on the BFU-e and CFU-e potential of human erythroid precursor cells transduced with shRNA directed against IRF-1 or PU.1. Data are mean ± SD from 3 independent experiments. *P < .05 (1-way ANOVA with Bonferroni correction).
Finally, to demonstrate that IRF-1 and PU.1 were responsible for the IFN-γ–induced inhibition of erythroid colony formation, we transduced human erythroid progenitor cells with lentiviruses expressing an shRNA to either IRF-1 or PU.1, stimulated these cells with IFN-γ, and tested their ability to form erythroid colonies. These constructs strongly inhibited the up-regulation of their cognate target mRNAs on IFN-γ stimulation (supplemental Figure 4b). In addition, inhibition of IRF-1 expression efficiently prevented the IFN-γ–mediated up-regulation of PU.1 and vice versa (supplemental Figure 5b), indicating that IRF-1 and PU.1 reciprocally enhance each other's expression. Importantly, subsequent colony assays revealed that inhibition of either IRF-1 or PU.1 expression was sufficient to overcome the IFN-γ–induced reduction in CFU-e and BFU-e numbers (Figure 5F). These data demonstrate that IFN-γ blocks erythroid differentiation by inducing the expression of PU.1 in an IRF-1–dependent manner.
Discussion
Next to its well-established proinflammatory and antimicrobial role, IFN-γ can also influence the hematopoietic process in the BM, as it affects HSC maintenance34 and inhibits the formation of B cells10,35 and eosinophilic granulocytes.12 IFN-γ also has a strong inhibitory effect on erythroid colony formation in vitro,3-6 and it is therefore assumed that IFN-γ plays a major role in the development of ACD in patients with cancer or chronic inflammatory conditions.1,2 However, the impact of chronic IFN-γ production on erythroid homeostasis in vivo and the molecular mechanism by which IFN-γ inhibits erythroid differentiation have been poorly investigated. This is because most methods used to induce chronic inflammation not only elicit the production of IFN-γ, but a whole plethora of proinflammatory cytokines, which can also act synergistically with IFN-γ on erythroid differentiation, as has been shown for TNF-α and type I IFNs.4,5 Of interest in this respect is the recent finding that the anemia accompanying the acute phase of Toxoplasma gondii infection is dependent on the production of IL-15, which is regulated by IFN-γ.24 Yet, the underlying molecular mechanism linking IFN-γ to anemia in either this model or in ACD remains unclear. Using a sterile form of chronic inflammation elicited by enhanced and sustained T-cell activation, we describe here that the ensuing increase in IFN-γ production induces anemia because of the concomitant inhibition of BM erythropoiesis and the enhanced turnover of circulating RBCs. Although T cells in CD70TG mice do not have increased production of IL-2, TNF-α,10 or type I IFNs (data not shown), we cannot exclude that other proinflammatory mediators are induced in CD70TG mice that contribute to the severity of the anemia. Importantly, we could not detect any significant changes in the serum levels of IL-10, IL-1α, IL-6, or TNF-α in these mice (data not shown) Yet, the phenotype of CD70TG*IFN-γ−/− mice demonstrates that, even if the (local) production of such factors would be increased, they are either not sufficient to induce anemia and/or stress erythropoiesis or they are induced by IFN-γ itself.
Although we demonstrated that IFN-γ negatively affects both formation and life span of RBCs, it has been described that IFN-γ induced by in vivo administration of CpG-oligodeoxynucleotides does mediate erythroid suppression, but not reduction of RBC survival.36 However, this study addressed the role of IFN-γ in acute inflammatory responses, whereas we have used a valid model for chronic inflammation. We hypothesized that a chronic response could result in macrophage activation and thereby affect RBC uptake. Concordantly, we found an IFN-γ–dependent increase of RBC uptake by activated splenic macrophages in vitro (Figure 2D-E), as well as increased iron storage in the spleens of CD70TG mice (data not shown). The latter observation suggests potential defects in iron release, which also impedes the course of erythropoiesis.37 However, we did not find altered expression of the iron-regulating hormone hepcidin38 in the liver of CD70TG mice (supplemental Figure 2a-b), indicating that hepcidin is not causally involved in the anemic phenotype of these mice.
Next to the clear impact on RBC life span, we also found that IFN-γ impaired BM erythropoiesis, as was evident from strongly reduced BFU-e numbers (Figure 3C) and fewer mature erythroblasts (Figure 3A) in CD70TG mice, but not CD70TG*IFN-γ−/− mice. It could be that the observed decrease is an underestimation and that this defect was already partly compensated in vivo by the increased EPO levels in these mice because EPO is sufficient to increase erythropoiesis not only in the spleen, but also in BM.39 Regarding the spleen, CD70TG mice had splenomegaly because of the induction of stress erythropoiesis, which is a physiologic response to anemia in rodents. Yet, the fact that CD70TG mice still become progressively anemic (Figure 1A) at least demonstrates that this form of “emergency” erythropoiesis is not sufficient to prevent anemia in these mice. It could be that stress erythropoiesis is negatively affected by the combination of increased local production of IFN-γ and enhanced RBC destruction in the spleen. This issue is currently under investigation.
At a molecular level, we demonstrate that IFN-γ inhibits erythropoiesis by activation of an IRF-1-PU.1 axis. Because type I IFNs can also induce expression of IRF-1 and efficiently suppress erythroid colony formation in vitro,3,4 we expect that chronic production of IFN-α or IFN-β can elicit a comparable degree of inhibition of BM erythropoiesis. This has been indirectly achieved in mice by knocking out Irf2, a suppressor of type I IFN signaling.40 These mice become anemic, which is rescued in the absence of Ifnar1, a member of the type I IFN receptor complex. However, type I IFNs are less potent activators of macrophages, which suggests that chronic production of type I IFNs has a less significant impact on RBC turnover and does not induce anemia as efficiently as IFN-γ. In steady-state conditions, EPO levels in IRF2−/− mice are not increased to the same extent as in our mouse model of sterile chronic inflammation.
PU.1 is a well-known transcriptional regulator of myelopoiesis.25 In the erythroid lineage, PU.1 activity is repressed by GATA-1, which is essential for normal erythropoiesis.26,41 The interplay of PU.1 and GATA-1 is relevant for lineage commitment, and high expression of PU.1 is known to repress GATA-1 activity and thereby erythroid differentiation.27-29 We identified a highly conserved IRF-1 binding site in the distal promoter of the SPI1 (PU.1) gene locus and showed that IFN-γ induces PU.1 expression via IRF-1. IFN-γ can induce apoptosis of erythroid progenitors through TRAIL, TWEAK, and CD95,8 and indeed we found that TRAIL (TNFSF10), a target of PU.1, is up-regulated by IFN-γ in CD71+ cells of CD70TG mice (supplemental Figure 4). We found that IRF-1 and PU.1 are interdependent because shRNA-mediated down-regulation of each factor resulted in down-regulation of the expression levels of the other. These findings are supported by the fact that IRF-1−/− mice have defective myelopoiesis, which is partly caused by down-regulation of PU.1,42 and that infection of neutrophil precursors with Anaplasma phagocytophilum results in down-regulation of both IRF-1 and PU.1.43 A cooperative function between IRF-1 and PU.1 has been reported during all-trans retinoic acid-mediated granulopoiesis44 and in IFN-γ–stimulated myeloid cells.45 Furthermore, IRF-1 is naturally down-regulated toward the final stages of erythroid maturation,46 which overall sustains the notion that suppression of the IRF-1–PU.1 axis is a prerequisite for normal erythropoiesis. Our findings provide the molecular mechanism by which IFN-γ affects erythropoiesis, which implicates transcriptional regulation of hematopoietic-lineage differentiation in response to inflammation.
Regarding the physiologic rationale behind the impact of IFN-γ on erythropoiesis, we postulate that during immune activation, IFN-γ temporarily shifts the balance of hematopoietic differentiation toward myeloid cells to combat an infection. Activated T cells might play an important role in this process, as they can migrate to the BM and modulate hematopoiesis.10,12,47 A temporary increase of myelopoiesis at the cost of erythropoiesis will not be detrimental to the host and will benefit the ongoing immune response, as the life span of RBCs is 10- to 50-fold longer than that of neutrophils or monocytes.48,49 However, during chronic immune activation, the prolongation of such a shift can lead to anemia, as observed in CD70TG mice, which also have increased monocyte formation.12 Our findings not only emphasize the importance of IFN-γ as a potent regulator of hematopoiesis but also reveal the potential molecular mechanism of inflammation-induced anemia that occurs frequently in patients with chronic inflammatory diseases, such as HIV infection and rheumatoid arthritis. Interestingly, BFU-e colony formation is strongly reduced in BM of anemic patients with chronic idiopathic neutropenia, which is significantly increased when cells are cultured with IFN-γ–neutralizing antibodies.50 It will be important to determine the contribution of this cytokine to the development of anemia in several chronic inflammatory diseases and whether neutralization of IFN-γ could be an effective treatment for different forms of ACD.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ernie de Boer (Erasmus MC) for sharing Gata1 ChIP-seq data, Berend Hooibrink (Academic Medical Center) for cell sorting, the staff of the animal facility of the Academic Medical Center for excellent animal care, Dr Reuben Tooze (Leeds) for discovering the IRF-1/2 binding site in the PU.1 promoter and for critical reading of the manuscript, and Prof Dr Rene van Lier, Dr Marieke von Lindern, and Dr Esther Nolte-‘t Hoen for critical reading of the manuscript.
This work was supported by The Netherlands Organization of Scientific Research (VENI grant 863.09.012, L.G.; and VIDI grant 917.76.310, M.A.N.), an EMBO Fellowship (ASTF 15-2010; P.P.), and the Landsteiner Foundation for Bloodtransfusion Research (project grant 0607; M.A.N.).
Authorship
Contribution: S.F.L. and L.G. designed and performed experiments, analyzed the data, and wrote the paper; A.M.d.B. and F.M.W. performed experiments and analyzed the data; P.P. cultured human erythroid progenitor cells; W.v.I.J. and Z.Ö. collected and analyzed microarray data; S.P. supervised the experimental design and contributed to writing of the manuscript; and M.A.N. designed and performed experiments, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for L.G. is Department of Blood Cell Research, Sanquin Research and Landsteiner Laboratory, Amsterdam, The Netherlands. The current affiliation for P.P. is Department of Molecular Cell Biology and Immunology, Vrije Universiteit Medical Center, Amsterdam, The Netherlands. The current affiliation for S.F.L. and M.A.N. is Department of Hematopoiesis, Sanquin Research and Landsteiner Laboratory, Amsterdam, The Netherlands.
Correspondence: Martijn A. Nolte, Department of Hematopoiesis, Sanquin Research and Landsteiner Laboratory Academic Medical Center/UvA, Plesmanlaan 125, 1066 CX Amsterdam, The Netherlands; e-mail: m.nolte@sanquin.nl.
References
Author notes
S.F.L. and L.G. contributed equally to this study.


![Figure 3. IFN-γ production in CD70TG mice inhibits erythroid BM output. (A) Absolute numbers of various erythroblast subsets or (B) relative contribution of pro-erythroblasts (percentage Ery I [Ter119medCD71high] from all erythroid [Ter119med/high cells]) in BM (isolated from 2 femurs and 2 tibiae) of WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. (C-D) BFU-e and CFU-e numbers of unfractionated BM from WT, CD70TG, IFN-γ−/−, and CD70TG*IFN-γ−/− mice. (E) Colony assay of FACS-sorted CMPs from WT and CD70TG mice. (F-G) Absolute numbers of IFN-γ–producing CD4 and CD8 T cells in BM of WT and CD70TG mice, measured after phorbol myristate acetate/ionomycin stimulation. (H-I) Effect of IFN-γ on the in vitro BFU-e and CFU-e colony-forming capacity of BM cells from WT mice. Data are mean ± SD for 3 mice per group (A-D,F-G) or triplicate analysis (E,H-I). Results are representative from 3 (A-D) or 2 (E-I) independently performed experiments. *P < .01 (A) or P < .05 (B-H) using 1-way ANOVA with Bonferroni correction (A-D) or nonpaired Student t test (E-I).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/9/10.1182_blood-2010-10-315218/4/m_zh89991177000003.jpeg?Expires=1765918404&Signature=iQjAI6vM8soF3EbBHOgbNGaiThWxRxD5pdYJsSzno9YtyHLR0YuaZTcxWAj-gmYgHOysKUVU8MUn1yvqJz4D1XSqHS-i9v6lN3ZZfgeB9gOtW9iYENUXCnryaZpeYrgk12Mg7DWQZRj5vS6s7176doCGhC51kNvU14c2jF2ZxNnkFR~gMVq-WBKBjf8fk~C4EKwi6x5MzrnLm2Kt2jaX34-G9fydbZafsx5pZ7NZAi6A1RhMnZ3MwWxrfGEwo7fWE0k4ASonr1gJH8hekdZvCnimfowYfFZoTSXGcnOg8Zn0BOvqwz2CgZVaAMj6TnFPQRFtn9-ITlXlPGv9iE3gUg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal