Abstract
G-CSF signals contribute to granulocyte lineage specification. We previously found that G-CSF induces SHP2 tyrosine phosphorylation and that chemical inhibition of SHP1/SHP2 reduces CFU-G and prevents G-CSF but not M-CSF activation of ERK. We now find that SHP2 shRNA knockdown in the 32Dcl3 granulocytic line reduces ERK activation, diminishes CEBPA protein and RNA expression and promoter histone acetylation, and inhibits granulopoiesis. Exogenous, shRNA-resistant SHP2 rescues these effects of SHP2 knockdown, exogenous C/EBPα rescues granulocytic markers, and exogenous RUNX1 rescues C/EBPα. 32Dcl3 lines with knockdown of ERK1 and ERK2 retain normal levels of C/EBPα and differentiate normally in G-CSF despite also having reduced proliferation. SHP2 knockdown reduces CEBPA levels in lineage-negative murine marrow cells cultured in TPO, Flt3 ligand, and SCF, without affecting the rate of cell expansion. On transfer to IL-3, IL-6, and SCF to induce myelopoiesis, levels of granulocytic RNAs are reduced and monocyte-specific RNAs are increased by SHP2 knockdown, and there is a reduction in the percentage of CFU-G that form in methylcellulose and of granulocytes that develop in liquid culture. In summary, SHP2 is required for induction of C/EBPα expression and granulopoiesis in response to G-CSF or other cytokines independent of SHP2-mediated ERK activation.
Introduction
Transcription factors are key mediators of granulocyte versus monocyte lineage specification.1 Homozygous deletion of the gene encoding C/EBPα in neonatal or adult mice reduces formation of granulocyte-monocyte progenitors,2-4 and deletion of the gene encoding PU.1 prevents formation of B lymphoid cells, blocks monopoiesis, and markedly reduces granulopoiesis.5,6 Gfi-1 is required for granulopoiesis,7,8 and AP-1 proteins contribute to monopoiesis via interaction with PU.1 or C/EBPs.9-11 Levels of several of these transcription factors also contribute to myeloid lineage commitment – reduced PU.1 in heterozygous mice or in mice lacking the −14 kb enhancer allows granulopoiesis in preference to monopoiesis,12-14 and reduced C/EBPα in mice lacking NF-κB p50 or RUNX1 favors monopoiesis.15,16
Cytokine signals also contribute to myelopoiesis, likely modifying activity or expression of transcriptional regulators. Observation of individual granulocyte-monocyte progenitors (GMP) exposed to G-CSF or M-CSF demonstrates that these cytokines contribute to granulocyte versus monocyte lineage specification, respectively.17 We compared G-CSF with M-CSF signals induced in lineage-negative murine marrow cells and in Ba/F3 cells expressing their receptors.18 Both cytokines activate MEK to induce ERK phosphorylation, but M-CSF does so with greater potency, likely dependent on the unique ability of the M-CSF receptor to activate phospholipase C-γ (PLCγ) and so protein kinase C. Activated ERK in turn stabilizes c-Fos, providing one potential link between M-CSF signaling and induction of monopoiesis. Consistent with a role for activated ERK in monocyte specification, chemical inhibition of MEK by addition of U0126 to murine marrow cells cultured with IL-3, IL-6, and SCF reduces CFU-M without affecting CFU-G.
In contrast to M-CSF, G-CSF more potently activates STAT3 and induces phosphorylation of Y542 and Y580 of the SHP2 tyrosine phosphatase. STAT3 is required for optimal neutrophil production during emergency but not basal granulopoiesis.19,20 SHP2 tyrosine phosphorylation can increase its intrinsic enzymatic activity but might also alter its substrate specificity.21 Methylcellulose culture of marrow cells in the presence of NSC-87877, a chemical inhibitor that targets both SHP1 and SHP2, reduces CFU-G but not CFU-M. SHP2 activates ERK, potentially dependent on dephosphorylation of the adaptor protein Gab2,22 and NSC-87877 prevents ERK activation mediated by G-CSF but not M-CSF, underscoring its ability to specifically effect G-CSF signaling.
To further evaluate the role of SHP2 during granulopoiesis, we have now targeted SHP2 RNA in the 32Dcl3 granulocyte progenitor cell line and in lineage-negative murine marrow cells. SHP2 knockdown impaired 32Dcl3 differentiation in response to G-CSF and shifted marrow cell differentiation in response to IL-3, IL-6, and SCF from granulopoiesis toward monopoiesis, associated in both instances with reduced C/EBPα RNA and protein expression. In contrast, simultaneous knockdown of both ERK1 and ERK2 in 32Dcl3 cells did not reduce C/EBPα or impair granulopoiesis. These results support the idea that SHP2 tyrosine phosphorylation induced by G-CSF or other cytokines contributes to granulopoiesis via stimulation of C/EBPα gene expression but independent of SHP2-mediated ERK activation.
Methods
Cell line culture and transduction
32Dcl3 cells23 were maintained in IMDM with 10% heat-inactivated FBS (HI-FBS) with 1 ng/mL murine IL-3 (Peprotech). To induce differentiation, cells were washed twice with PBS and transferred to IMDM with 10% HI-FBS and 20 ng/mL human G-CSF (Amgen). Lentiviral vectors were packaged by transient transfection into 293T cells with packaging plasmids at a ratio of 3.75 μg pLKO.1 vector: 5 μg pCMV-ΔR8.91: 1.25 μg pMD.G(VSV.G) per 100 mm dish using 20 μL Lipofectamine 2000 (Invitrogen), and retroviral vectors were packaged by cotransfection of 2 μg pkat2ecopac with 8 μg pBabeNeo vector.24 293T cells were maintained in DMEM with 10% HI-FBS. After transfection, media was changed to IMDM with 1% HI-FBS, which was collected 2 days later and filtered using 0.45 μm filters. 32Dcl3 cells were transduced by addition of viral supernatant in the presence of 4 μg/mL Polybrene (Sigma-Aldrich). After 48 hours, stable transductants were selected in the presence of 2 μg/mL puromycin or 1.2 mg/mL G418, and subclones were obtained by limiting dilution. Viable cell counts were enumerated using Trypan blue dye and a hemocytometer, and cell morphology was assessed by cytospin followed by Wright-Giemsa staining. Photomicrographs were taken using a Zeiss Axiophot microscope (Carl Zeiss), a Kontron Electronik Progress 3012 camera (Kontron), and a 63×/1.40 NA oil objective.
shRNAs and SHP2 or C/EBPα rescue
To target murine SHP2 (Ptpn11), we screened shRNAs TRCN29874-29878 in pLKO.1-puro (Open Biosystems), which we abbreviate as shRNA74-78. To target murine ERK2 we screened 10 shRNAs and used TRCN54729 in pLKO.1-Puro. To subsequently target ERK1 we again screened several shRNAs and used TRCN23184, which we inserted into a modified version of pLKO.1 with the puromycin-resistance gene replaced by the G418-resistance gene (pLKO.1-Neo). To rescue SHP2 expression we modified the human SHP2 cDNA to be resistant to shRNA77 by changing the targeted site CGT-GTT-AGG-AAC-GTC-AAA-GAA to CGC-GTG-CGC-AAT-GTG-AAG-GAA, with changes italicized, using site-directed mutagenesis (Stratagene). The sequence of the entire, modified SHP2 cDNA was then confirmed by DNA sequencing. The SHP2 cDNA was then subcloned into the pBabeNeo retroviral vector, as was the cDNA encoding C/EBPα-ER. C/EBPα-ER–transduced cells were cultured using phenol-red free media, and the transgene was activated using 1μM estradiol.
Marrow culture and transduction
Marrow isolated from 8-16 week old C57BL/6 female mice was subjected to red cell lysis with NH4Cl and the cells were lineage-depleted using biotin-labeled B220, Gr-1, Mac-1, Ter119, and CD3 antibodies (BD Pharmingen), anti-biotin microbeads, and MACS columns (Miltenyi Biotec). The cells were then cultured in IMDM with 10% HI-FBS containing 20 ng/mL murine SCF, 100 ng/mL murine flt3 ligand (FL), and 10 ng/mL murine thrombopoietin (TPO, Peprotech) for 24 hours. The cells were transduced by spinoculation at 2000g for 3 hours after addition of lentiviral supernatant and 8 μg/mL Polybrene. Coculture with viral supernatant and 4 μg/mL Polybrene was then continued for 2 days, followed by addition of 2 μg/mL puromycin for 3 additional days. Viable cells were then collected using Lympholyte-M polysucrose (Cedarlane labs) and transferred to IMDM with 10% HI-FBS with IL-3, IL-6, and SCF or to Methocult M3534 containing methylcellulose with IMDM, FBS, and these same cytokines (StemCell Technologies). CFUs were enumerated 8 days later. Cells in liquid culture were subjected to FACS analysis using PE-anti–Mac-1 and FITC-anti–Gr-1.
Western blotting, RNA analysis and chromatin immunoprecipitation
Cells were washed with PBS, and total cellular proteins were prepared by addition of Laemmli sample buffer followed by incubation at 100°C for 5 minutes. Extracts corresponding to 5 × 105 cells were subjected to polyacrylamide gel electrophoresis using 8%-10% gels and transferred to Hybond-P membrane (Amersham), followed by probing with primary antibodies. Antibodies used were SHP2, P-SHP2 (Tyr542), and P-ERK (197G2; Cell Signaling Technology), ERK (K-23) and C/EBPα (14AA; Santa Cruz Biotechnology), and β-actin (AC-15; Sigma-Aldrich). After addition of HRP-conjugated secondary antibody and subsequent washes, a signal was generated using HyGlo chemiluminescence reagents (Denville Scientific) and detected by autoradiography. Total cellular RNA was prepared from 107 cells using the NucleoSpin RNA II kit, including use of RNase-free DNase (Machery-Nagel). First strand cDNA was prepared using AMV reverse transcriptase (Promega) and oligodT primer at 42°C for 1 hour. Quantitative PCR was carried out using 50 ng of each cDNA using iQ SYBR Green supermix (Bio-Rad). Oligonucleotides used were:
S16-F, 5′-ATATTCGGGTCCGTGTGAAG; S16-R, 5′-CTTGGAGGCTTCATCCACAT
MPO-F, GCTCCGCCCGCATTCCTTGT; MPO-R, TTGAGCTGTGTGGCCAGCCG
LF-F, TATTTCTTGAGGCCCTTGGA; LF-R, TCTCATCTCGTTCTGCCACC
NE-F, CAGGCATCTGCTTCGGGGAC; NE-R, AGGGGCGAAGGCATCTGGGT
PR3-F, AGCTACCCATCCCCCAAG; PR3-R, TCGTGCCCACCTACAATCTT
C/EBPϵ-F, AGAGGGCAACCGAGGCACCA; C/EBPϵ-R, GCTGCCACAGGGGCCTTGAG
GCSFR-F, CTGATCTTCTTGCTACTCCCCA; GCSFR-R, GGTGTAGTTCAAGTGAGGCAG
C/EBPα-F, CGGTGCGCAAGAGCCGAGAT; C/EBPα-R, CCCGCAGCGTGTCCAGTTCA
PU.1-F, CCTTCGTGGGCAGCGATGGA; PU.1-R, TGTAGCTGCGGGGGCTGCAC
CD14-F, ATCTACCGACCATGGAGCGT; CD14-R, TCCACATCTGCCGCCCCCAA
MCSFR-F, TCCGGTGGTGGTGGCCTGTA; MCSFR-R, AGCGCACCTGGTACTTCGGC
CD68-F, TTGCTAGGACCGCTTATAGC; CD68-R, GCAGGAGAGTAACGGCCTTT.
Cells (5 × 106 32Dcl3) were subjected to ChIP using 5 μg anti-acetyl histone H3 antiserum (Millipore), specific for H3 with acetylation of H3K9 and H3K14, or rabbit Ig followed by real time PCR for the CEBPA or β-actin promoters, as described,25 using the following primers:
CEBPA(-991)-F, GAACACTTGACTAGAGTGCTC;
CEBPA(-873)-R, CTCGTCCACTCGCCTAGG;
actin(-1520)-F, GGGAAAGTTCTCTCAGGGTTGG;
actin(-1418)-4, TGCTGTGAACTGGAAACACACC.
The Student t test was used for statistical comparisons. Band intensities were quantified using National Institutes of Health ImageJ 1.38 software.
Results
SHP2 knockdown impairs granulopoiesis and CEBPA expression
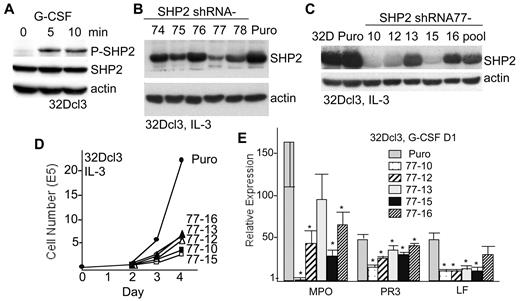
We previously found that G-CSF induces SHP2 tyrosine phosphorylation in Ba/F3 cells expressing exogenous G-CSF receptor or in murine lineage-negative marrow cells.18 G-CSF similarly induces SHP2 Y580 phosphorylation in 32Dcl3 cells (Figure 1A). To reduce SHP2 expression, 32Dcl3 cells were transduced with pLKO.1 lentiviral vectors expressing one of several SHP2 shRNAs or with the empty vector, and stable, pooled-transductants were selected with puromycin. SHP2 shRNA-77 produced effective knockdown, whereas the other shRNAs were less effective (Figure 1B). The pool of 32Dcl3 cells expressing shRNA-77 was then subcloned, identifying 3 lines (77-10, 77-12, and 77-15) with marked and 2 lines (77-13 and 77-16) with modest SHP2 RNA knockdown (Figure 1C), though even lines with modest knockdown expressed approximately 3-fold reduced SHP2 protein. SHP2 knockdown slowed 32Dcl3 cell proliferation in IL-3 compared with vector-transduced cells (Puro), with inhibition of proliferation strong in all cases but most evident in subclones with the greatest degree of SHP2 knockdown (Figure 1D). Vector-transduced cells or the 5 subclones expressing SHP2 shRNA-77 were transferred to G-CSF for 1 day and total cellular RNA was evaluated for expression of myeloperoxidase (MPO), proteinase 3 (PR3), and lactoferrin (LF), markers of granulocytic differentiation (Figure 1E). Induction of these markers was impaired in all 5 subclones with reduced SHP2 expression, with the exception of MPO in 32Dcl3(shRNA77)-13 cells and LF in the 77-16 line. MPO and to a lesser extent PR3 were more affected in lines 77-10, 77-12, and 77-15 that had the greatest degree of SHP2 knockdown. SHP2 knockdown in 32Dcl3 cells also markedly impaired Mac-1 and Gr-1 cell surface expression in IL-3 and prevented their induction by G-CSF (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
SHP2 knockdown suppresses 32Dcl3 cell granulopoiesis. (A) 32Dcl3 cells were removed from IL-3 for 3 hours and than exposed to 20 ng/mL G-CSF for 0, 5, or 10 minutes. Total cellular proteins were collected and then subjected to Western blotting for phosphorylation of SHP2 on Y580 (P-SHP2), total SHP2, and β-actin. (B) 32Dcl3 cells were transduced with a panel of lentiviral vectors expressing SHP2 shRNAs(74-78) or with the empty vector (Puro), and pooled transductants were selected. Total cellular proteins were then subjected to Western blotting for SHP2 or β-actin. (C) Similar Western blot analysis was conducted using total cellular protein samples prepared from parental 32Dcl3 cells (32D), vector-transduced cells, subclones of the 32Dcl3(shRNA-77) pool (10,12,13,15,16), and the shRNA-77 cellular pool. (D) Growth curves show daily viable cell counts for the cells transduced with empty vector and for the indicated shRNA-77 subclones. (E) The vector-transduced cells or the 5 32Dcl3(shRNA-77) subclones were transferred to G-CSF and total cellular RNAs were prepared after 24 hours. These were converted to first-strand cDNA and the relative expression of MPO, PR3, and LF, normalized to ribosomal protein S16 RNA expression, were analyzed by real-time PCR analysis. Shown are means and SE from 3 independent RNA preparations. * indicates P < .05 for each RNA level relative to expression in the vector-transduced cells.
SHP2 knockdown suppresses 32Dcl3 cell granulopoiesis. (A) 32Dcl3 cells were removed from IL-3 for 3 hours and than exposed to 20 ng/mL G-CSF for 0, 5, or 10 minutes. Total cellular proteins were collected and then subjected to Western blotting for phosphorylation of SHP2 on Y580 (P-SHP2), total SHP2, and β-actin. (B) 32Dcl3 cells were transduced with a panel of lentiviral vectors expressing SHP2 shRNAs(74-78) or with the empty vector (Puro), and pooled transductants were selected. Total cellular proteins were then subjected to Western blotting for SHP2 or β-actin. (C) Similar Western blot analysis was conducted using total cellular protein samples prepared from parental 32Dcl3 cells (32D), vector-transduced cells, subclones of the 32Dcl3(shRNA-77) pool (10,12,13,15,16), and the shRNA-77 cellular pool. (D) Growth curves show daily viable cell counts for the cells transduced with empty vector and for the indicated shRNA-77 subclones. (E) The vector-transduced cells or the 5 32Dcl3(shRNA-77) subclones were transferred to G-CSF and total cellular RNAs were prepared after 24 hours. These were converted to first-strand cDNA and the relative expression of MPO, PR3, and LF, normalized to ribosomal protein S16 RNA expression, were analyzed by real-time PCR analysis. Shown are means and SE from 3 independent RNA preparations. * indicates P < .05 for each RNA level relative to expression in the vector-transduced cells.
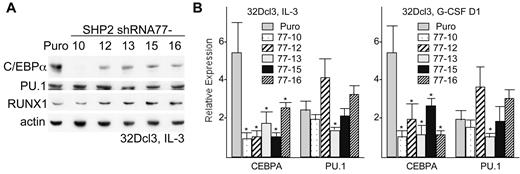
Western blot analysis demonstrated several-fold reduction in C/EBPα protein expression, but no change in PU.1 or RUNX1 protein levels, in all 5 subclones expressing SHP2 shRNA-77, compared with puromycin-resistant cells transduced with empty pLKO.1 vector (Figure 2A). Reduction in C/EBPα protein levels was greatest in line 77-10, which notably also had the greatest reduction in MPO and PR3 expression. Similar reduction in C/EBPα protein expression was seen also in several additional assessments (not shown). CEBPA mRNA was also reduced in all 5 subclones compared with the control cell line, either in IL-3 or after transfer to G-CSF for one day, with 5-fold reduction evident in several subclones (Figure 2B). PU.1 RNA was only reduced significantly, by approximately 2-fold, in line 77-13.
SHP2 knockdown reduces C/EBPα expression in 32Dcl3 cells. (A) Total cellular protein samples from from vector-transduced 32Dcl3 cells (Puro) and from subclones expressing SHP2 shRNA-77 were subjected to Western blotting for C/EBPα, PU.1, RUNX1, and β-actin. (B) Total cellular RNAs prepared from these same cell lines proliferating in IL-3 (left) or after transfer to G-CSF for one day (right) were converted to first-strand cDNA and the relative expression of CEBPA and PU.1 were analyzed by real-time PCR analysis. Shown are means and SE from 3 independent RNA preparations. * indicates P < .05 for each RNA level relative to expression in the vector-transduced cells.
SHP2 knockdown reduces C/EBPα expression in 32Dcl3 cells. (A) Total cellular protein samples from from vector-transduced 32Dcl3 cells (Puro) and from subclones expressing SHP2 shRNA-77 were subjected to Western blotting for C/EBPα, PU.1, RUNX1, and β-actin. (B) Total cellular RNAs prepared from these same cell lines proliferating in IL-3 (left) or after transfer to G-CSF for one day (right) were converted to first-strand cDNA and the relative expression of CEBPA and PU.1 were analyzed by real-time PCR analysis. Shown are means and SE from 3 independent RNA preparations. * indicates P < .05 for each RNA level relative to expression in the vector-transduced cells.
Rescue of CEBPA and granulopoiesis by exogenous SHP2 after SHP2 knockdown
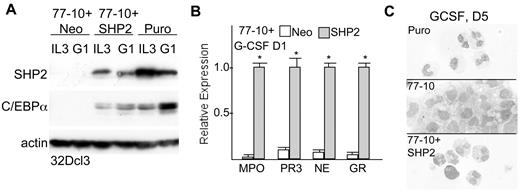
Modest knockdown of SHP2 by shRNA-75 or shRNA-78 did not reduce C/EBPα protein expression or impair granulopoiesis (not shown). As an alternative to verify that SHP2 knockdown and not an off-target effect of shRNA-77 mediates these effects, we generated a SHP2 cDNA resistant to knockdown by altering 7 nucleotides within the 21 bp targeted sequence within the protein-encoding open-reading frame, leaving the encoded amino acids unchanged. 32Dcl3(77-10) cells were then transduced with pBabeNeo expressing this resistant SHP2 cDNA or with the empty pBabeNeo vector. The resulting cell lines were compared with 32D-Puro cells, transduced with the pLKO.1 vector, for SHP2 and C/EBPα protein expression in IL-3 or after 1 day in G-CSF (Figure 3A). Exogenous, shRNA-resistant SHP2 partially rescued SHP2 expression and, strikingly, allowed increased C/EBPα protein expression. Similar transduction of 32Dcl3(77-16) cells, which have weaker initial SHP2 knockdown, allowed almost complete rescue of both SHP2 and C/EBPα expression (supplemental Figure 2). G-CSF induction of several granulocytic markers was then assessed in 32Dcl3(77-10)+Neo or 32Dcl3(77-10)+SHP2 cells. In addition to rescuing C/EBPα protein expression, exogenous SHP2 markedly enhanced expression of the mRNAs encoding MPO, PR3, neutrophil elastase (NE), or G-CSF receptor (GR) one day after G-CSF addition (Figure 3B) and also restored cell surface Mac-1 and Gr-1 expression to approximately 50% of that seen in control cells (supplemental Figure 1A). Morphologic analysis confirmed formation of neutrophils by 32D-Puro or 32Dcl3(77-10)+SHP2 cells after 5 days in G-CSF (Figure 3C, top and bottom panels). Similar morphologic analysis of 32Dcl3(77-10) or 32Dcl3(77-10)+Neo cells is difficult because of poor cell survival after 5 days in G-CSF in most experiments, but surviving 32Dcl3(77-10) cells did not demonstrate neutrophilic forms but rather immature, blastic cells (Figure 3C center panel), and neither of these control lines showed any morphologic evidence of granulocytic differentiation at earlier times after G-CSF addition (not shown).
Exogenous SHP2 rescues C/EBPα expression and granulopoiesis after SHP2 knockdown. (A) 32Dcl3(shRNA77-10) cells were transduced with pBabeNeo (Neo) or pBabeNeo expressing the SHP2 cDNA modified to resist targeting the shRNA-77 without altering the encoded protein sequence (SHP2). These cells and 32Dcl3 cells transduced with the empty pLKO.1 vector (Puro) were cultured in IL-3 or in G-CSF for 1 day (G1). Total cellular proteins were collected and subjected to Western blotting for SHP2, C/EBPα, and β-actin. (B) Total cellular RNAs prepared from 32Dcl3(shRNA-77)-10 cells transduced with exogenous SHP2 or the empty Neo vector were prepared 1 day after transfer to G-CSF and analyzed for relative expression of MPO, PR3, NE, and G-CSFR (GR) RNAs. The mean relative expression of each mRNA after SHP2 rescue was set to 1.0. Results shown are mean and SE of 3 repetitions. * indicates P < .05 for each RNA after SHP2 rescue compared with mock rescue. (C) 32Dcl3-Puro, 32Dcl3(shRNA-77)-10, or 32Dcl3(shRNA-77)-10+SHP2 cells cultured in G-CSF for 5 days were cytospun and subjected to Wright-Giemsa staining.
Exogenous SHP2 rescues C/EBPα expression and granulopoiesis after SHP2 knockdown. (A) 32Dcl3(shRNA77-10) cells were transduced with pBabeNeo (Neo) or pBabeNeo expressing the SHP2 cDNA modified to resist targeting the shRNA-77 without altering the encoded protein sequence (SHP2). These cells and 32Dcl3 cells transduced with the empty pLKO.1 vector (Puro) were cultured in IL-3 or in G-CSF for 1 day (G1). Total cellular proteins were collected and subjected to Western blotting for SHP2, C/EBPα, and β-actin. (B) Total cellular RNAs prepared from 32Dcl3(shRNA-77)-10 cells transduced with exogenous SHP2 or the empty Neo vector were prepared 1 day after transfer to G-CSF and analyzed for relative expression of MPO, PR3, NE, and G-CSFR (GR) RNAs. The mean relative expression of each mRNA after SHP2 rescue was set to 1.0. Results shown are mean and SE of 3 repetitions. * indicates P < .05 for each RNA after SHP2 rescue compared with mock rescue. (C) 32Dcl3-Puro, 32Dcl3(shRNA-77)-10, or 32Dcl3(shRNA-77)-10+SHP2 cells cultured in G-CSF for 5 days were cytospun and subjected to Wright-Giemsa staining.
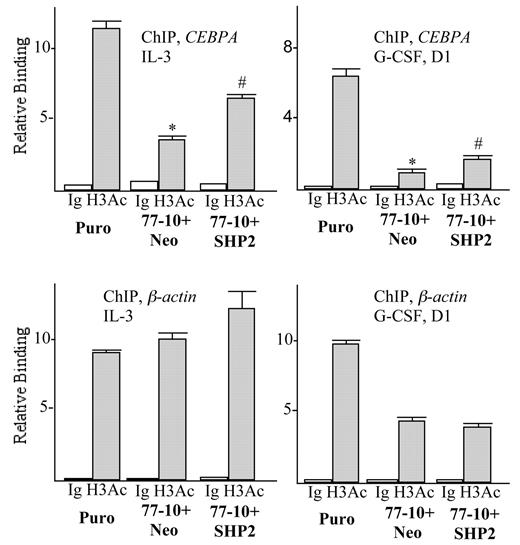
Knockdown of SHP2 reduces CEBPA RNA levels in 32Dcl3 cells, suggesting reduced transcription of the CEBPA gene. Lending support to this idea, ChIP analysis demonstrates that SHP2 knockdown reduces histone H3 acetylation, a histone modification reflective of transcriptionally active chromatin, by > 3-fold in the vicinity of the CEBPA promoter in IL-3 and by approximately 6-fold in G-CSF (Figure 4). In contrast, acetylation of the β-actin promoter was not affected by expression of the SHP2 77-Neo shRNA in IL-3 and was reduced only 2-fold in G-CSF. Modest reduction of β-actin promoter H3 acetylation in G-CSF likely reflects the finding that both G-CSF and SHP2 knockdown reduce the rate of 32Dcl3 proliferation and thus the general rate of transcription. In addition, coexpression of the 77-10 shRNA and a SHP2 cDNA resistant to this shRNA partially rescued CEBPA promoter histone H3 acetylation, in either IL-3 or G-CSF, just as it rescued C/EBPα levels, without altering β-actin promoter H3 acetylation. These findings indicate that SHP2 regulates C/EBPα expression, at least in part, via regulation of CEBPA gene transcription.
SHP2 regulates CEBPA promoter histone H3 acetylation. Chromatin immunoprecipitation was conducted using an antiserum that recognizes acetylated H3 histone or rabbit Ig control, using 32Dcl3-Puro, 32Dcl3(shRNA-77)-10+Neo, or 32Dcl3(shRNA-77)-10+SHP2 cells proliferating in IL-3 (left) or after transfer to G-CSF for one day (right). Shown are mean and SE for relative binding to the CEBPA or β-actin promoters, normalized to signals obtained using input samples, from 3 determinations. * indicates P < .05 comparing Puro with 77-10+Neo cells; # indicates P < .05 comparing 77-10+Neo with 77-10+SHP2 cells.
SHP2 regulates CEBPA promoter histone H3 acetylation. Chromatin immunoprecipitation was conducted using an antiserum that recognizes acetylated H3 histone or rabbit Ig control, using 32Dcl3-Puro, 32Dcl3(shRNA-77)-10+Neo, or 32Dcl3(shRNA-77)-10+SHP2 cells proliferating in IL-3 (left) or after transfer to G-CSF for one day (right). Shown are mean and SE for relative binding to the CEBPA or β-actin promoters, normalized to signals obtained using input samples, from 3 determinations. * indicates P < .05 comparing Puro with 77-10+Neo cells; # indicates P < .05 comparing 77-10+Neo with 77-10+SHP2 cells.
Rescue of granulopoiesis by C/EBPα and of C/EBPα by RUNX1 after SHP2 knockdown
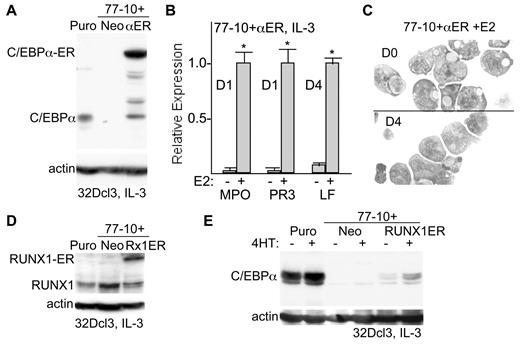
To determine whether reduction of CEBPA expression contributes to impaired granulopoiesis resulting from SHP2 knockdown, 32Dcl3(77-10) cells were transduced with pBabeNeo-C/EBPα-ER, encoding C/EBPα linked to the ligand-binding domain of the estradiol receptor. Stable expression of C/EBPα is markedly cell cycle inhibitory, requiring inducible expression to obtain stable 32Dcl3 cell lines. We previously demonstrated that activation of C/EBPα-ER by estradiol (E2) induces 32Dcl3 cells proliferating in IL-3 to differentiate to neutrophils.26 Expression of C/EBPα-ER (αER) as well as endogenous C/EBPα was assessed in the 32Dcl3-Puro, 32Dcl3(77-10)+Neo and 32Dcl3(77-10)+αER lines (Figure 5A). Cells were grown in phenol-red free media to minimize leakiness of the ER fusion construct. Western blot analysis indicated that in the absence of estradiol C/EBPα-ER was entirely cytoplasmic and entered the nucleus on release from heat shock protein by addition of estradiol (not shown). 32Dcl3-Puro cells express endogenous C/EBPα, SHP2 knockdown eliminates C/EBPα expression, and the C/EBPα-ER transgene is expressed at a several-fold higher level than endogenous C/EBPα. In addition, several minor breakdown products of C/EBPα-ER are noted, as we have seen previously, including one comigrating with C/EBPα. These break-down products likely occur during cell lysis, as our ability to maintain C/EBPα-ER 32Dcl3 lines for extended periods argues against intracellular release of cell cycle-inhibitory C/EBPα. Addition of estradiol to 32Dcl3(77-10)+Neo cells proliferating in IL-3 did not induce MPO or PR3 expression or induce morphologic changes (not shown), whereas addition of estradiol for 1 day to 32cl3(77-10)+αER cells led to marked induction of these early granulocytic markers (Figure 5B). In addition, addition of estradiol for 4 days to this cell line induced LF expression, a later granulocytic marker. Furthermore, activation of C/EBPα-ER in cells expressing SHP2 shRNA-77 also markedly increased cell surface Mac-1 and Gr-1 expression (supplemental Figure 1B). In contrast to our previous findings with 32D-C/EBPα-ER cells, addition of estradiol to 32Dcl3(77-10)+αER cells did not induce formation of neutrophilic forms, although nuclear indentation was evident after estradiol addition (Figure 5C). Thus, exogenous C/EBPα rescues the block to granulopoiesis that results from SHP2 knockdown, although SHP2 may have activities beyond facilitating C/EBPα expression required for full nuclear maturation or for the survival of terminal granulocytes.
Exogenous C/EBPα rescues granulopoiesis and exogenous RUNX1 rescues C/EBPα expression after SHP2 knockdown. (A) 32Dcl3(shRNA-77)-10 cells were transduced with pBabeNeo (Neo) or pBabeNeo-C/EBPα-ER (αER). Total cellular proteins prepared from these cells or from 32Dcl3-Puro cells were subjected to Western blotting for C/EBPα or β-actin. The positions of C/EBPα or C/EBPα-ER are shown. (B) Total cellular RNAs prepared from 32Dcl3(shRNA-77)-10+αER cells 0 or 1 day (D1) or 4 days (D4) after addition of estradiol (E2) were analyzed for relative expression of the mRNAs encoding MPO, PR3 or LF. The mean relative expression of each mRNA after estradiol addition rescue was set to 1.0. Results shown are mean and SE of 3 repetitions. * indicates P < .05 for each RNA comparing expression in the presence versus absence of estradiol. (C) 32Dcl3(shRNA-77)-10+αER cells were cultured with estradiol for 0 or 4 days, cytospun, and subjected to Wright-Giemsa staining. (D) 32Dcl3(shRNA-77)-10 cells were transduced with or pBabeNeo-RUNX1-ER (Rx1ER). Total cellular proteins prepared from these cells, from 32Dcl3-Puro cells, or from 32D(shRNA-77)-10+Neo cells were subjected to Western blotting for RUNX1 or β-actin. (E) Total cellular proteins prepared from these same 3 cell lines cultured ± 4HT for 24 hours were subjected to Western blotting for C/EBPα or β-actin.
Exogenous C/EBPα rescues granulopoiesis and exogenous RUNX1 rescues C/EBPα expression after SHP2 knockdown. (A) 32Dcl3(shRNA-77)-10 cells were transduced with pBabeNeo (Neo) or pBabeNeo-C/EBPα-ER (αER). Total cellular proteins prepared from these cells or from 32Dcl3-Puro cells were subjected to Western blotting for C/EBPα or β-actin. The positions of C/EBPα or C/EBPα-ER are shown. (B) Total cellular RNAs prepared from 32Dcl3(shRNA-77)-10+αER cells 0 or 1 day (D1) or 4 days (D4) after addition of estradiol (E2) were analyzed for relative expression of the mRNAs encoding MPO, PR3 or LF. The mean relative expression of each mRNA after estradiol addition rescue was set to 1.0. Results shown are mean and SE of 3 repetitions. * indicates P < .05 for each RNA comparing expression in the presence versus absence of estradiol. (C) 32Dcl3(shRNA-77)-10+αER cells were cultured with estradiol for 0 or 4 days, cytospun, and subjected to Wright-Giemsa staining. (D) 32Dcl3(shRNA-77)-10 cells were transduced with or pBabeNeo-RUNX1-ER (Rx1ER). Total cellular proteins prepared from these cells, from 32Dcl3-Puro cells, or from 32D(shRNA-77)-10+Neo cells were subjected to Western blotting for RUNX1 or β-actin. (E) Total cellular proteins prepared from these same 3 cell lines cultured ± 4HT for 24 hours were subjected to Western blotting for C/EBPα or β-actin.
RUNX1-ETO represses the CEBPA promoter,27 and we find that RUNX1 binds and activates the CEBPA promoter and that in vivo deletion of RUNX1 in adult mice reduces CEBPA mRNA expression and impairs granulopoiesis.16 In addition, another group has found that SHP2 dephosphorylates RUNX1 and increases its ability to stimulate megakaryopoiesis.28 We therefore also transduced 32Dcl3(77-10) cells with pBabeNeo-RUNX1-ER.29 Expression of RUNX1-ER as well as endogenous RUNX1 was assessed in the 32Dcl3-Puro, 32Dcl3(77-10)+Neo and 32Dcl3(77-10)+RUNX1-ER lines (Figure 5D). The RUNX1-ER transgene is expressed at approximately2-fold higher level than endogenous RUNX1. These cells were then cultured without or with 4HT, to activate RUNX1-ER, for 24 hours, and total cellular proteins were then subjected to Western blotting for C/EBPα or β-actin (Figure 5E). Addition of 4HT did not induce C/EBPα in 32Dcl3(77-10)+Neo cells but did increase C/EBPα expression in the 32Dcl3(77-10)+RUNX1-ER line. A slight increase was already evident before 4HT addition, perhaps reflecting mild leakiness of transgene nuclear localization. After 4HT addition, C/EBPα levels remained several-fold below that seen in the 32Dcl3-Puro cells, potentially reflecting partial inhibition of RUNX1-ER activity by the SHP2 shRNA.
SHP2 does not stimulate granulopoiesis via ERK activation
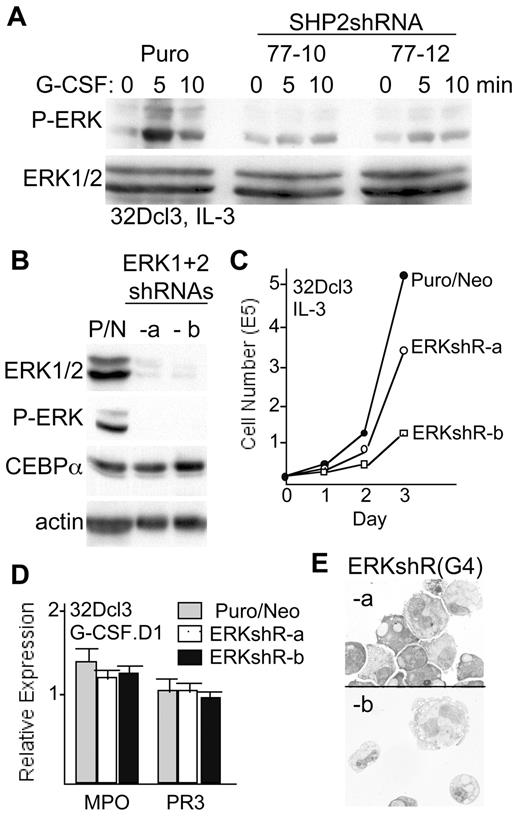
We previously found that a chemical inhibitor of SHP1 and SHP2 prevents ERK activation by G-CSF but not M-CSF in lineage-negative marrow cells.18 Confirming a key role for SHP2 in G-CSF-mediated ERK activation, in 32Dcl3 cells G-CSF leads to rapid ERK phosphorylation, and SHP2 knockdown markedly reduces ERK phosphorylation in two 32Dcl3-shRNA77 subclones (Figure 6A). In addition, expression of exogenous, shRNA-resistant SHP2 in 32Dcl3(77-10) cells rescued ERK activation (not shown). There are 2 isoforms of ERK, ERK1 and ERK2. Using separate lentiviral vectors targeting each isoform, we generated two 32Dcl3 subclones, designated ERKshR-a and ERKshR-b, with efficient knockdown of both isoforms (Figure 6B). Despite greatly reduced ERK levels, C/EBPα protein levels were unaffected in these lines compared with 32Dcl3-Puro/Neo, double vector-transduced cells (Figure 6B). ERKshR-a cells grew at a mildly slower rate than did control Puro/Neo cells, and ERKshR-b cells had a markedly reduced prolifertion rate (Figure 6C). On transfer to G-CSF, both ERK-knockdown subclones showed expression of MPO and PR3 mRNAs similar to the Puro/Neo control line on day 1 (Figure 6D), and formation of morphologic band forms and neutrophils was preserved on day 4 (Figure 6E). In addition, these lines expressed high levels of cell surface Mac-1 and Gr-1, with G-CSF strongly inducing Gr-1 expression (supplemental Figure 1C). Thus, suppression of ERK expression and slowed proliferation resulting from SHP2 knockdown does not impair C/EBPα protein expression or granulopoiesis.
ERK knockdown does not reduce C/EBPα expression or impair granulopoiesis. (A) 32Dcl3-Puro (32D), 32Dcl3(shRNA-77)-10, or 32Dcl3(shRNA-77)-12 cells were removed from IL-3 for 3 hours and then exposed to G-CSF for 0, 5, or 10 minutes. Total cellular proteins were then subjected to Western blotting for phosphorylated ERK (P-ERK) or for total ERK levels. (B) Total cellular proteins from 32Dcl3 cells transduced with the pLKO.1-Puro and pLKO.1-Neo vectors (P/N) or from 2 subclones of 32Dcl3 cells (ERKshR-a and ERKshR-b) expressing shRNAs targeting ERK-1 and ERK-2 were subjected to Western blotting for ERK, P-ERK, C/EBPα, and β-actin. (C) Growth curves show daily viable cell counts for the cells transduced with empty vectors and for the 2 subclones with knockdown of ERK-1 and ERK-2. (D) Total cellular RNAs prepared from these 3 cell lines 1 day after transfer to G-CSF were analyzed for relative expression of MPO and PR3 RNAs. The average expression of PR3 RNA was set to 1.0 in the 32Dcl3-Puro/Neo cells. Results shown are mean and SE from 3 determinations. (E) 32Dcl3-ERK shRNA subclones ERKshR-a and ERKshR-b were cultured in G-CSF for 4 days, cytospun, and subjected to Wright-Giemsa staining.
ERK knockdown does not reduce C/EBPα expression or impair granulopoiesis. (A) 32Dcl3-Puro (32D), 32Dcl3(shRNA-77)-10, or 32Dcl3(shRNA-77)-12 cells were removed from IL-3 for 3 hours and then exposed to G-CSF for 0, 5, or 10 minutes. Total cellular proteins were then subjected to Western blotting for phosphorylated ERK (P-ERK) or for total ERK levels. (B) Total cellular proteins from 32Dcl3 cells transduced with the pLKO.1-Puro and pLKO.1-Neo vectors (P/N) or from 2 subclones of 32Dcl3 cells (ERKshR-a and ERKshR-b) expressing shRNAs targeting ERK-1 and ERK-2 were subjected to Western blotting for ERK, P-ERK, C/EBPα, and β-actin. (C) Growth curves show daily viable cell counts for the cells transduced with empty vectors and for the 2 subclones with knockdown of ERK-1 and ERK-2. (D) Total cellular RNAs prepared from these 3 cell lines 1 day after transfer to G-CSF were analyzed for relative expression of MPO and PR3 RNAs. The average expression of PR3 RNA was set to 1.0 in the 32Dcl3-Puro/Neo cells. Results shown are mean and SE from 3 determinations. (E) 32Dcl3-ERK shRNA subclones ERKshR-a and ERKshR-b were cultured in G-CSF for 4 days, cytospun, and subjected to Wright-Giemsa staining.
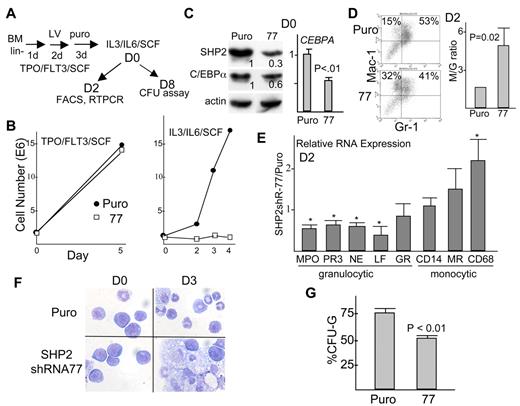
SHP2 knockdown impairs marrow granulopoiesis
As diagrammed (Figure 7A), marrow mononuclear cells were lineage-depleted and placed in TPO, FL, and SCF. After one day of pre-stimulation, cells were transduced with a control lentiviral vector expressing puromycin resistance or with a vector expressing SHP2 shRNA-77. After an additional 2 days, puromycin was added and selection was allowed to proceed for 3 days. At this point dead cells were removed by density gradient centrifugation, and viable cells were then transferred to IL-3, IL-6, and SCF in liquid or methylcellulose culture to induce myeloid differentiation. Notably, SHP2 knockdown did not reduce cell expansion during the 5-day period in TPO/FL/SCF after virus addition, whereas cells expressing SHP2 shRNA-77 showed subsequent inhibition of proliferation in IL-3/IL-6/SCF (Figure 7B). Just before transfer to IL-3/IL-6/SCF, Western blot analysis, representative of 3 independent experiments, indicates effective SHP2 knockdown and almost 2-fold reduction in C/EBPα protein expression (Figure 7C left). CEBPA mRNA expression on Day 0 was also reduced approximately 2-fold in marrow cells transduced with SHP2 shRNA-77 compared with empty Puro vector (Figure 7C right). After 2 days in liquid culture with IL-3/IL-6/SCF, SHP2 shRNA knockdown resulted in a significant reduction in the proportion of Mac-1+Gr-1+ granulocytes, with a corresponding increase in Mac-1+Gr-1− monocytes, as indicated by a representative FACS analysis (Figure 7D left). In 3 separate experiments the ratio of monocytes to granulocytes assessed by this assay increased significantly, by 4.9 ± 1.4 fold, P < .01 (Figure 7D right). In addition, mRNAs encoding the MPO, PR3, NE, LF, and GR granulocytic markers were reduced while those encoding the monocytic markers CD14, M-CSF receptor (MR), and CD68, were elevated (Figure 7E). These changes in RNA abundance reflect a shift in the cell population from a predominance of granulocytes to a predominance of monocytes. In particular, morphologic evaluation just before transfer to IL-3/IL-6/SCF showed many immature cells as well as a small proportion of granulocytes or monocytes (Figure 7F left panels), whereas Wright-Giemsa staining 3 days later showed a strikingly increased proportion of monocyte/macrophage cells in the population subjected to SHP2 knockdown (Figure 7F right panels). After SHP2 knockdown, the intensity of Mac-1 or Gr-1 did not change in Mac-1+Gr-1− monocytes or in Mac-1+Gr-1+ granulocytes, as seen in Figure 7D, indicating that SHP2 knockdown did not alter intrinsic expression of these differentiation markers. Finally, CFU-G and CFU-M were enumerated after 8 days of methylcellulose culture; SHP2 shRNA-77 significantly reduced the proportion of CFU-G, from 78%-52% (Figure 7G).
SHP2 knockdown suppresses granulopoiesis and favors monopoiesis in murine marrow cells. (A) Diagram of experimental protocol. Marrow cells were lineage-depleted, placed in TPO, FL, and SCF, transduced with pLKO.1 (Puro) or pLKO.1-shRNA-77, and then exposed to puromycin for 3 days. Viable cells were then transferred to IL-3, IL-6, and SCF on day 0 (D0), in liquid culture for FACS or RNA analyses or in methylcellulose for assay of myeloid colony-forming units (CFU). (B) Growth curves show viable cell counts over a 5 day period in TPO, FL, and SCF and daily after transfer to IL-3, IL-6, and SCF. (C) Western blot analysis of SHP2, C/EBPα, and β-actin expression on Day 0 (left). Relative intensity of SHP2 or C/EBPα bands is shown, normalized to β-actin. Expression of CEBPA RNA on day 0 (right), normalized to ribosomal protein S16 mRNA expression (mean and SE from 3 determinations). (D) Representative FACS analysis for Mac-1 and Gr-1 expression on Day 2 and the increase in the Mac-1+Gr-1− monocyte (M)/Mac-1+Gr-1+ granulocyte (G) ratio (mean and SE, with the M/G ratio set to 1 in each of 3 experiments). (E) Total cellular RNAs prepared on day 2 were analyzed for expression of the indicated mRNAs, normalized to S16 mRNA expression. The ratio of expression of each mRNA in cells transduced with SHP2 shRNA-77 relative to expression in cells transduced with empty vector is shown (mean and SE from 3 determinations). * indicates P < .05 relative to a ratio of 1.0. (F) Cells on day 0 or day 3 after transfer to IL-3, IL-6, or SCF were cytospun and subjected to Wright-Giemsa staining. (G) The proportion of CFU-G relative to CFU-G + CFU-M after culture of cells transduced with empty vector or shRNA-77 is shown (mean and SE from 3 determinations). Average number of CFU-G+CFU-M was 113/10 000 cells in the vector group and 30/10 000 cells in the shRNA-77 group. P < .01 for shRNA-77–expressing cells relative to vector transduced cells.
SHP2 knockdown suppresses granulopoiesis and favors monopoiesis in murine marrow cells. (A) Diagram of experimental protocol. Marrow cells were lineage-depleted, placed in TPO, FL, and SCF, transduced with pLKO.1 (Puro) or pLKO.1-shRNA-77, and then exposed to puromycin for 3 days. Viable cells were then transferred to IL-3, IL-6, and SCF on day 0 (D0), in liquid culture for FACS or RNA analyses or in methylcellulose for assay of myeloid colony-forming units (CFU). (B) Growth curves show viable cell counts over a 5 day period in TPO, FL, and SCF and daily after transfer to IL-3, IL-6, and SCF. (C) Western blot analysis of SHP2, C/EBPα, and β-actin expression on Day 0 (left). Relative intensity of SHP2 or C/EBPα bands is shown, normalized to β-actin. Expression of CEBPA RNA on day 0 (right), normalized to ribosomal protein S16 mRNA expression (mean and SE from 3 determinations). (D) Representative FACS analysis for Mac-1 and Gr-1 expression on Day 2 and the increase in the Mac-1+Gr-1− monocyte (M)/Mac-1+Gr-1+ granulocyte (G) ratio (mean and SE, with the M/G ratio set to 1 in each of 3 experiments). (E) Total cellular RNAs prepared on day 2 were analyzed for expression of the indicated mRNAs, normalized to S16 mRNA expression. The ratio of expression of each mRNA in cells transduced with SHP2 shRNA-77 relative to expression in cells transduced with empty vector is shown (mean and SE from 3 determinations). * indicates P < .05 relative to a ratio of 1.0. (F) Cells on day 0 or day 3 after transfer to IL-3, IL-6, or SCF were cytospun and subjected to Wright-Giemsa staining. (G) The proportion of CFU-G relative to CFU-G + CFU-M after culture of cells transduced with empty vector or shRNA-77 is shown (mean and SE from 3 determinations). Average number of CFU-G+CFU-M was 113/10 000 cells in the vector group and 30/10 000 cells in the shRNA-77 group. P < .01 for shRNA-77–expressing cells relative to vector transduced cells.
Discussion
Transcription factors such as C/EBPα provide hematopoietic cells with intrinsic developmental programs by regulating expression of lineage-specific genes. To maintain homeostasis and to respond to environmental stresses, hematopoietic stem and progenitor cells must in addition possess the capacity to respond to environmental signals. Although cytokines stimulate the survival and proliferation of hematopoietic cells expressing their cognate receptors, the role of cytokines in directing lineage fate decisions in multipotent progenitors remains largely uncertain.30 Notably, a recent study demonstrates that G-CSF directs granulocyte lineage-specification while M-CSF directs monocyte lineage-specification from cultured GMP.17 In addition, on comparison of signaling pathways activated by G-CSF versus M-CSF in lineage-negative marrow cells, we found that M-CSF more potently activates ERK via PLCγ, whereas G-CSF more potently activates STAT3 and induces SHP2 phosphorylation on tyrosines 542 and 580.18 SHP2 tyrosine phosphorylation potentially alters its substrate specificity or phosphatase activity, dependent on the growth factor receptor or cellular environment involved.21,31 Moreover, we found that chemical inhibition of MEK to reduce ERK activation suppresses CFU-M but not CFU-G formation from cultured marrow cells, whereas a chemical inhibitor that targets SHP1 and SHP2 preferentially reduces CFU-G formation. We have now further investigated the role of SHP2 in granulopoiesis and find that knockdown of SHP2 mRNA reduces 32Dcl3 or marrow progenitor granulopoiesis via reduction of C/EBPα RNA and protein expression without reducing PU.1 or RUNX1 and independent of diminished ERK activity.
After screening a panel of SHP2 shRNAs we identified one, shRNA-77, which effectively reduces SHP2 protein expression to < 20% of control levels. Striking reduction of C/EBPα protein and RNA was noted in several 32Dcl3 subclones expressing this shRNA. Three-fold reduction in CEBPA promoter histone H3 acetylation suggests that reduction in CEBPA transcription contributes to lower CEBPA mRNA expression. To confirm that these effects were not because of knockdown of an RNA other than that encoding SHP2, we transduced one of these lines with a SHP2 cDNA resistant to the shRNA and indeed rescued SHP2 and C/EBPα expression, CEBPA promoter acetylation, and granulopoiesis. To confirm that reduction in C/EBPα contributed to impaired granulopoiesis in the presence of reduced SHP2 levels, we introduced C/EBPα-ER and demonstrated rescue of granulopoiesis on its activation with estradiol, with expression of both early and late granulocytic markers. Lack of formation of cells with a neutrophilic morphology on expression of exogenous C/EBPα in cells with SHP2 knockdown suggests a requirement for SHP2 to mediate activation of genes or proteins other than C/EBPα required for neutrophil nuclear maturation or for the survival of mature granulocytes. Two recent studies demonstrate that deletion of the SHP2 genes by pIpC injection of SHP2(flox/flox);Mx1-CRE mice markedly reduces HSC and myeloid progenitors due in part to impaired proliferation and survival,32,33 underscoring the utility of our shRNA knockdown strategy to partially but not totally reduce SHP2 in 32Dcl3 cells or in marrow myeloid progenitors. In particular, the role of SHP2 in the granulocyte versus monocyte lineage decision and in subsequent granulopoiesis has not been well characterized.
SHP2 knockdown slowed proliferation of 32Dcl3 cells, likely reflecting reduced activation of ERK. Simultaneous knockdown of ERK1 and ERK2 also slowed 32Dcl3 cell proliferation, but in contrast to SHP2 knockdown did not reduce C/EBPα levels or prevent granulopoiesis. Granulocytic differentiation mediated by C/EBPα-ER in 32Dcl3 cells or in marrow cells is also associated with marked slowing of cell proliferation because of transgene expression, further indicating that slowed cell proliferation because of SHP2 knockdown does not account for the observed impairment in granulopoiesis.25,34
ERK activation leads to C/EBPα phosphorylation on S21 and consequent reduced capacity of C/EBPα to direct granulopoiesis.35 There is no evidence that this modification affects C/EBPα protein stability; indeed we previously found that chemical inhibition of MEK reduces S21 phosphorylation of C/EBPα without affecting total C/EBPα protein levels in culture marrow cells,18 and shRNA knockdown of ERK1 and ERK2 in this study again did not alter C/EBPα protein levels. Thus, reduction in ERK activation because of SHP2 knockdown does not alter C/EBPα protein levels because of affects on S21 phosphorylation. Although G-CSF activates ERK via SHP2, M-CSF more potently activates ERK via PLCγ, leading us to postulate that C/EBPα retains greater activity in G-CSF compared with M-CSF because of less frequent S21 modification.18 The finding the ERK knockdown does not impair 32Dcl3 granulopoiesis is consistent with the idea that C/EBPα lacking S21 phosphorylation effectively mediates granulopoiesis and supports our hypothesis that ERK activation is required specifically for monopoiesis and likely only helps mediate granulocytic proliferation.
If SHP2 knockdown does not reduce C/EBPα expression because of its affect on ERK activity, then the direct substrate of SHP2 that allows increased CEBPA transcription remains in question. Our finding that RUNX1 binds the CEBPA promoter and stimulates its transcription,16 including rapid induction of CEBPA mRNA even in the presence of cycloheximide (H. Guo and A. Friedman, unpublished data, February 2011), and the finding that SHP2-mediated, direct tyrosine dephosphorylation of RUNX1 increases its activity during megakaryopoiesis28 led us to ask whether exogenous RUNX1 could rescue C/EBPα expression in 32Dcl3 cells expressing SHP2 shRNA. Partial rescue was observed, consistent with the idea that G-CSF stimulates SHP2 to act on RUNX1 to allow transcriptional induction of the CEBPA gene and thereby granulopoiesis. Partial but not complete rescue of C/EBPα expression by exogenous RUNX1-ER may reflect increased tyrosine phosphorylation in the setting of SHP2 knockdown, reducing its trans-activation strength.
SHP2 knockdown also interfered with granulocytic development from lineage-negative murine marrow cell myeloid progenitors in response to IL-3/IL-6/SCF, as assessed morphologically, by FACS analysis, by assessment of lineage-specific RNA expression, and by CFU assay, correlated again with reduced C/EBPα expression. This cytokine combination was chosen as it mediates both granulopoiesis and monopoiesis, allowing the balance between these 2 lineage pathways to be assessed, and indeed reduced granulopoiesis was associated with an increased proportion of monocytic cells in these assays. Of note, SHP2 knockdown did not impair proliferation of marrow cells in TPO/FL/SCF, minimizing bias of the vector- versus SHP2 shRNA-transduced cells before transfer to IL3/IL6/SCF. Subsequent bias was minimized by FACS and mRNA analysis just 2 days later, the minimum time needed to allow lineage maturation.
Based on these and earlier findings, we propose that G-CSF-mediated SHP2 tyrosine phosphorylation modifies its activity against a substrate other than ERK, potentially RUNX1, to increase CEBPA RNA expression. In a broader sense, these data further support the idea that cytokine signals help direct myeloid lineage specification. Increased PU.1 levels favor monopoiesis whereas reduced levels are sufficient for granulopoiesis, and so it is noteworthy that SHP2 knockdown did not alter PU.1 RNA or protein levels. C/EBPα activates its own promoter,36 but there is no evidence that C/EBPα itself is tyrosine phosphorylated. From a clinical perspective, reduced SHP2 expression or activity might contribute to neutropenic syndromes or to defective granulocytic differentiation associated with myeloid leukemias, and stimulation of SHP2 might assist in recovery from neutropenia or in generation of neutrophils from embryonic or induced pluripotent stem cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grants R01 HL089176 and U01 HL099775/HL100397 to A.D.F.
National Institutes of Health
Authorship
Contribution: L.Z. performed the experiments; and L.Z. and A.D.F. designed the study, analyzed the data, wrote the paper and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alan D. Friedman, Division of Pediatric Oncology, Johns Hopkins University School of Medicine, CRB I, Rm 253, 1650 Orleans St, Baltimore, MD 21231; e-mail: afriedm2@jhmi.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal