Abstract
When Glu-plasminogen binds to cells, its activation to plasmin is markedly enhanced compared with the reaction in solution, suggesting that Glu-plasminogen on cell surfaces adopts a conformation distinct from that in solution. However, direct evidence for such conformational changes has not been obtained. Therefore, we developed anti-plasminogen mAbs to test the hypothesis that Glu-plasminogen undergoes conformational changes on its interaction with cells. Six anti-plasminogen mAbs (recognizing 3 distinct epitopes) that preferentially recognized receptor-induced binding sites (RIBS) in Glu-plasminogen were obtained. The mAbs also preferentially recognized Glu-plasminogen bound to the C-terminal peptide of the plasminogen receptor, Plg-RKT, and to fibrin, plasmin-treated fibrinogen, and Matrigel. We used trypsin proteolysis, immunoaffinity chromatography, and tandem mass spectrometry and identified Glu-plasminogen sequences containing epitopes recognized by the anti-plasminogen-RIBS mAbs: a linear epitope within a domain linking kringles 1 and 2; a nonlinear epitope contained within the kringle 5 domain and the latent protease domain; and a nonlinear epitope contained within the N-terminal peptide of Glu-plasminogen and the latent protease domain. Our results identify neoepitopes latent in soluble Glu-plasminogen that become available when Glu-plasminogen binds to cells and demonstrate that binding of Glu-plasminogen to cells induces a conformational change in Glu-plasminogen distinct from that of Lys-Pg.
Introduction
When Glu-plasminogen (Glu-Pg), the native circulating form of the zymogen, binds to cell surfaces, its activation is markedly enhanced, compared with the reaction in solution (reviewed in Miles et al1 ). This results in arming cell surfaces with the proteolytic activity of plasmin (Pm) that regulates physiologic and pathologic processes in which cells must degrade an extracellular matrix to migrate.2,3 The underlying basis for the enhancement in activation of Glu-Pg on the cell surface is that proteolysis of cell-associated Glu-Pg by Pm, to yield the more readily activated Lys-Pg form, is markedly enhanced when Glu-Pg is associated with the cell surface.4-6 These results suggest that Glu-Pg on cell surfaces adopts a conformation distinct from its conformation in solution. However, direct evidence for such conformational changes and how they are related to the Lys-Pg conformation has not been obtained previously.
In the current study, we developed anti-plasminogen (Pg) mAbs to test the hypothesis that such conformational changes can be detected when Glu-Pg interacts with cells and to identify specific domains that become surface exposed when Glu-Pg binds to cells. Furthermore, as Pg activation is also markedly promoted when Glu-Pg is bound to fibrin and to other regulatory molecules,7 we wished to test the hypothesis that similar conformational changes are induced in Glu-Pg on binding to these proteins.
Previously, a mAb that detects a conformationally altered state of fibrinogen that is induced when fibrinogen is bound to its receptor, GpIIb-IIIa, was described.8,9 The mAb thus detects receptor-induced binding sites (RIBS) and has been designated as an anti-fibrinogen-RIBS mAb.
In the present study, we demonstrate that binding of Glu-Pg to cells induces at least 3 distinct anti-Pg RIBS epitopes that are latent in soluble Glu-Pg but become available when Glu-Pg is bound to cell surfaces. Furthermore, these neoepitopes are also induced when Glu-Pg is adsorbed either to the Pg receptor, Pg-RKT, fibrin, Pm-treated fibrinogen or to the model extracellular matrix, Matrigel. These data provide direct evidence that a conformational change is induced in Glu-Pg (that is distinct from that of Lys-Pg) when Glu-Pg is bound to cells and to other regulatory molecules.
Methods
Proteins
Glu-Pg was purified from fresh human blood as described.10,11 Lys-Pg was from Enzyme Research Laboratories. Elastase degradation products of Pg: residues Tyr79-Val337 or Tyr79-Val353 of Pg containing Pg kringles 1-3 (K1-3), residues Val354-Ala439 of Pg, containing kringle 4 (K4), and residues Val442-Asn790 of Pg containing kringle 5 (K5) and the latent Pm active site within the latent protease domain region (K5-PD) were prepared and characterized as described.12,13
Cells
U937 monocytoid cells were cultured as described.14
ELISA for Glu-Pg
Glu-Pg (11nM in 50 μL) was immobilized on wells of microtiter plates at 22°C for 18 hours. The wells were postcoated with 1% BSA in PBS. Then IgG fractions of mAbs (180nM) were incubated with the wells in the presence of either PBS, Glu-Pg, or Lys-Pg in a final volume of 75 μL. The plates were washed 3 times with PBS containing 0.05% Tween 20 and the bound mAbs were detected with alkaline phosphatase–conjugated goat anti-mouse IgG (Southern Biotechnology Associates).
ELISA for Glu-Pg immobilized on regulatory molecules
For Glu-Pg binding to fibrin, fibrinogen (Enzyme Research Laboratories) 100 μL at 1 mg/mL was dried in mirotiter wells at 37°C for 2 hours and then incubated with thrombin (Sigma-Aldrich) 100 μL at 1 U/mL for 60 minutes at 22°C. The thrombin was inactivated with hirudin and the wells were also washed with trasylol. The wells were incubated with 20μM Glu-Pg or buffer (2 hours at 22°C), washed, and then mAbs were added in the presence or absence of Glu-Pg in solution. The wells were washed 3 times with PBS containing 0.05% Tween 20 and the bound mAbs were detected with alkaline phosphatase–conjugated goat anti–mouse IgG. In other experiments, immobilized fibrinogen was incubated with Pm (Kabi; 67pM) for 30 minutes at 37°C. Pm was inactivated with p'nitrophenyl p'guanidinobenzoate (0.1mM) and aminoethylbenzenesulfonyl fluoride.HCl (0.5mM) followed by washing. Bound Glu-Pg was detected when bound to fibrin.
Matrigel (Collaborative Biomedical Products; BD Biosciences; 100 μL at 0.5 mg/mL) was coated on microtiter wells and incubated for 18 hours at 4°C. The binding of mAbs to Glu-Pg bound to Matrigel was determined when bound to fibrin.
The C-terminal peptide of Pg-RKT with a C-terminal cysteine added for coupling (CEQSKLFSDK coupled to BSA) was coated onto wells of microtiter plates as described.15 The binding of mAbs to Glu-Pg bound to the peptide was determined when bound to fibrin.
Mixed gangliosides (Calbiochem) 100 μL at 2.5 μg/mL, were coated onto microtiter wells as described.16 The binding of mAbs to Glu-Pg bound to the mixed gangliosides was determined when bound to fibrin.
mAbs
BALB/c mice were immunized with Glu-Pg: 3 IP injections of 100 μg were given at 3-week intervals. Three days after the final injection, spleen cells were harvested and fused with P3 Ag 8.653 myeloma cells at a 6:1 ratio. Culture supernatants of the hybridomas were screened in wells of microtiter plates coated with Glu-Pg and selected hybridomas were cloned 3 times by limiting dilution. Ascites fluid was produced by injecting 1-2 × 106 hybridoma cells per mouse and mAbs were purified from the ascites fluid by chromatography on protein A Sepharose (Pharmacia). MOPC21 was from BD Biosciences.
Pairwise epitope mapping
Pairwise epitope mapping was performed using surface plasmon resonance (SPR) on the Biacore 3000 biosensor. Rabbit anti–mouse Fc IgG (100 μg/mL in sodium acetate pH 4.0) was coupled in 3 channels of a carboxymethyl dextran (CM) chip. As a control, normal rabbit IgG was coupled in one channel of the chip and the response unit (RU) values obtained using this control channel subtracted from the test samples. The reactions were performed at a flow rate of 5 μL/min in 0.01M HEPES pH 7.4, 0.15M NaCl, 3mM EDTA and 0.005% (vol/vol) surfactant P20 (HBS-EP; Biacore). The primary mAb to be analyzed (200 μg/mL in HBS-EP) was captured on the chip, followed by blocking with normal mouse IgG (Sigma-Aldrich). Pg (2.2μM) was injected over each of the 4 flow cells. This was followed by challenge with each secondary mAb (at 200 μg/mL), sequentially. The chip was regenerated with 100mM HCl to remove the primary mAb, leaving the anti–mouse Fc IgG remaining on the chip so that a new cycle could be started. MOPC21 was used as isotype control. Over 49 runs, the baseline remained stable after regeneration.
FACS analysis
Cells were washed with PBS containing 1% BSA and 0.1% sodium azide (PBA), incubated for 10 minutes at 22°C with PBA containing 10% heat-inactivated normal rabbit serum, washed again, and incubated with mAbs for 30 minutes at 4°C. After washing with PBA, the cells were stained for 30 minutes at 4°C with FITC-goat anti–mouse IgG at a 1/100 dilution. Bound IgG was detected in a flow cytometry analyzer (Coulter EPICS XL-MCL).
Western blotting
Proteins were subjected to SDS-PAGE, transferred to nitrocellulose (Amersham Pharmacia Biotech), and incubated with mAbs. The membranes were incubated with an anti–mouse Ab-HRP conjugate, developed using an ECL substrate (Pierce), and subjected to autoradiography using Kodak Biomax MR Film (Fisher).
Epitope determination
ELISA analysis of trypsin digests of Pg.
Trypsin digests of Pg (111nM) were coated onto microtiter wells and blocked with 5% BSA in coating buffer (0.1M NaHCO3). The wells were washed 3 times in 10mM TrisHCl (150mM NaCl) followed by incubation with IgG fractions of mAbs or isotype control (33nM for 1 hour) and washed 4 times. The wells were incubated with HRP-conjugated goat-anti-mouse IgG (Pierce) for 1 hour. The wells were washed and then incubated with HRP substrate for 10 minutes in the dark. H2SO4 (2N) was added to stop the reaction and the plate was read at 490nM.
Reduction, alkylation, and digestion of Pg.
Pg was denatured in 6M urea followed by reduction with 200mM DTT and alkylation with 200mM iodoacetimide in the dark. Then 17mM DTT was added to quench the iodoacetimide. The mixture was made 19mM in NH4HCO3, pH 8.3, and digested with trypsin (mass spectrometry grade; Promega) at 37°C. The weight ratios used were 1/50, 1/200, 1/400 trypsin to Pg for mAbs 51, 109, and 116, respectively, and digestion was for 1 hour. A concentration of soy bean trypsin inhibitor (Sigma-Aldrich), equivalent to the trypsin concentration was added to stop the reaction.
Separation of peptides and mass spectrometry.
The peptides were analyzed by reversed-phase chromatography before mass spectrometry analysis using the following method. Nano-electrospray capillary column tips were made in house using a P-100 laser puller (Sutter Instruments). The columns were packed with Zorbax SB-C18 stationary phase (Agilent) purchased in bulk (5-mm particles, 15-cm length, 75-mm inner diameter). The reversed-phase gradient separation was performed using water and acetonitrile (0.1% formic acid) as the mobile phases. The gradient started at 5% acetonitrile and was ramped to 8% acetonitrile over 10 minutes. The acetonitrile was ramped to 35% acetonitrile over 20 minutes, then increased to 90% acetonitrile for another 20 minutes, and maintained for another 10 minutes before re-equilibration to 5% acetonitrile.
The data-dependent MS/MS data were obtained on an LTQ linear ion trap mass spectrometer using a home built nanoelectrospray source at 2 KV at the tip. The instrument was used in data-dependent MS/MS mode. One MS spectrum was followed by 6 MS/MS scans on the most abundant ions after the application of the dynamic exclusion list. Protein identification was performed using Mascot (Matrix Science; Version 2.1.04) at the 95% confidence level with a calculated false-positive rate of < 1% as determined using a reversed concatenated protein database.
Tandem mass spectra were extracted by the Xcalibur software. All MS/MS samples were analyzed using Mascot (Matrix Science; Version 2.1.04). Mascot was set up to search human proteins contained in the NCBInr protein database assuming the digestion enzyme trypsin. Mascot was searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 2.0 Da. The iodoacetamide derivative of cysteine was specified in Mascot as a fixed modification. Oxidation of methionine was specified in Mascot as a variable modification.
Scaffold (Version Scaffold-01_06_03; Proteome Software Inc) was used to additionally validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm17 Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm.17 Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Statistics
Data are presented as means ± SEM. Results were analyzed by ANOVA followed by Student-Newman-Keuls post-hoc tests for multiple comparisons.
Results
mAbs that distinguish immobilized Glu-Pg from soluble Glu-Pg
To test whether mAbs that distinguish surface-bound from soluble Pg could be elicited, we modified the method that was used to produce mAbs specific for conformationally altered states of fibrinogen.8 BALB/c mice were injected with Glu-Pg, the native form of Pg with N-terminal Glu. After fusion of spleen cells, hybridoma culture supernatants were screened for reactivity with Glu-plasminogen, directly bound to microtiter wells, in an ELISA assay as described in “ELISA for Glu-Pg immobilized on regulatory molecules.” Of 1710 hybridoma culture supernatants screened, 315 reacted with immobilized Glu-Pg. In a second screen of the 315 positive supernatants, 52 supernatants also reacted with immobilized Glu-Pg in the presence of 1μM soluble Glu-Pg (> 50-fold molar excess), while binding of the other 263 culture supernatants to immobilized Pg was not detected in the presence of soluble Glu-Pg. The 6 culture supernatants having the greatest ratio of reactivity in the second screen compared with the first screen were selected for further study and cloned 3 times by limiting dilution and monoclonality was established. All were of the IgG1 subclass. We then verified that the IgG fractions of the mAbs recognized surface-bound Glu-Pg in the presence of soluble Glu-Pg. The reaction of each of the mAbs with immobilized Glu-Pg was not significantly different in the absence compared with the presence of soluble Glu-Pg (Table 1). Therefore, mAbs that reacted preferentially with surface-bound Glu-Pg compared with soluble Glu-Pg were elicited.
Effects of soluble Glu-Pg and Lys-Pg on the binding of anti-Pg mAbs to immobilized Glu-Pg
| . | Competitor, absorbance OD 405 . | ||
|---|---|---|---|
| mAb . | Buffer . | Glu-Pg . | Lys-Pg . |
| 20 | 0.76 ± 0.02 | 0.78 ± 0.05 | 0.78 ± 0.05 |
| 109 | 0.83 ± 0.06 | 0.82 ± 0.02 | 0.75 ± 0.06 |
| 49 | 0.57 ± 0.04 | 0.54 ± 0.01 | 0.66 ± 0.02 |
| 51 | 0.68 ± 0.03 | 0.60 ± 0.03 | 0.73 ± 0.06 |
| 53 | 0.69 ± 0.05 | 0.63 ± 0.03 | 0.69 ± 0.01 |
| 116 | 0.55 ± 0.03 | 0.50 ± 0.05 | 0.52 ± 0.02 |
| Buffer | 0.13 ± 0.01 | 0.13 ± 0.01 | 0.13 ± 0.01 |
| . | Competitor, absorbance OD 405 . | ||
|---|---|---|---|
| mAb . | Buffer . | Glu-Pg . | Lys-Pg . |
| 20 | 0.76 ± 0.02 | 0.78 ± 0.05 | 0.78 ± 0.05 |
| 109 | 0.83 ± 0.06 | 0.82 ± 0.02 | 0.75 ± 0.06 |
| 49 | 0.57 ± 0.04 | 0.54 ± 0.01 | 0.66 ± 0.02 |
| 51 | 0.68 ± 0.03 | 0.60 ± 0.03 | 0.73 ± 0.06 |
| 53 | 0.69 ± 0.05 | 0.63 ± 0.03 | 0.69 ± 0.01 |
| 116 | 0.55 ± 0.03 | 0.50 ± 0.05 | 0.52 ± 0.02 |
| Buffer | 0.13 ± 0.01 | 0.13 ± 0.01 | 0.13 ± 0.01 |
Binding of anti-Pg mAbs (180nM) to immobilized Glu-Pg was determined in the presence of either buffer, soluble Glu-Pg (1μM), or soluble Lys-Pg (1μM) as described in “ELISA for Glu-Pg.” Results are mean ± SEM; n = 3 for each experimental group.
Pairwise epitope mapping and Western blotting
We tested whether the 6 mAbs recognized the same or spatially proximal epitopes within the Pg molecule using SPR to perform pairwise epitope mapping. Rabbit anti–mouse Fc IgG was coupled to a CM chip and used to capture the primary mAb to be analyzed. Pg was then injected followed by challenge with each secondary mAb sequentially. Using this method, reaction of the secondary mAb (measured as an increase in response units (RUs) with Pg indicated that the epitope recognized by the first mAb was distinct from that recognized by the second mAb (Table 2). In controls to ensure that the conditions were appropriate, the mAbs used for coupling were also tested as second Ab and did not provide an increase in RU, compared with the isotype control, MOPC21 (Table 2). For confirmation, each mAb was used as the primary Ab. Table 2 shows that our panel of 6 mAbs could be divided into 3 groups based on pairwise epitope mapping: group I, comprising mAbs 49, 51, and 53; group II comprising mAbs 20 and 109; and group III, comprising mAb 116. These results suggest that the mAbs in each group recognize either the same or spatially proximal epitopes within Glu-Pg.
Pairwise epitope mapping of anti-Pg RIBS mAbs
| Captured mAb . | Second mAb, response units . | ||||||
|---|---|---|---|---|---|---|---|
| 20 . | 109 . | 49 . | 51 . | 53 . | 116 . | MOPC21 . | |
| Group I | |||||||
| 49 | 69.3 | 71.2 | 17.5 | 13.7 | 13.7 | 57.1 | 17 |
| 51 | 68.8 | 70.3 | 14.5 | 12.8 | 14.3 | 54.8 | 18.6 |
| 53 | 67.4 | 68.4 | 16.1 | 14.5 | 14.3 | 53.7 | 18.5 |
| Group II | |||||||
| 20 | 9.7 | 15.4 | 43 | 41.4 | 40 | 72.1 | 7.7 |
| 109 | 26 | 25.1 | 62.1 | 60.7 | 61.1 | 100.4 | 17.2 |
| Group III | |||||||
| 116 | 86.3 | 89.9 | 60.8 | 59.7 | 59.2 | 13.4 | 19.3 |
| Control | |||||||
| MOPC21 | 26.5 | 30 | 21.5 | 19.7 | 20.2 | 16.7 | 20.1 |
| Captured mAb . | Second mAb, response units . | ||||||
|---|---|---|---|---|---|---|---|
| 20 . | 109 . | 49 . | 51 . | 53 . | 116 . | MOPC21 . | |
| Group I | |||||||
| 49 | 69.3 | 71.2 | 17.5 | 13.7 | 13.7 | 57.1 | 17 |
| 51 | 68.8 | 70.3 | 14.5 | 12.8 | 14.3 | 54.8 | 18.6 |
| 53 | 67.4 | 68.4 | 16.1 | 14.5 | 14.3 | 53.7 | 18.5 |
| Group II | |||||||
| 20 | 9.7 | 15.4 | 43 | 41.4 | 40 | 72.1 | 7.7 |
| 109 | 26 | 25.1 | 62.1 | 60.7 | 61.1 | 100.4 | 17.2 |
| Group III | |||||||
| 116 | 86.3 | 89.9 | 60.8 | 59.7 | 59.2 | 13.4 | 19.3 |
| Control | |||||||
| MOPC21 | 26.5 | 30 | 21.5 | 19.7 | 20.2 | 16.7 | 20.1 |
Pairwise epitope mapping was performed and compared with the isotype control, MOPC21, as described in “Pairwise epitope mapping.” Results are representative of three profiling experiments.
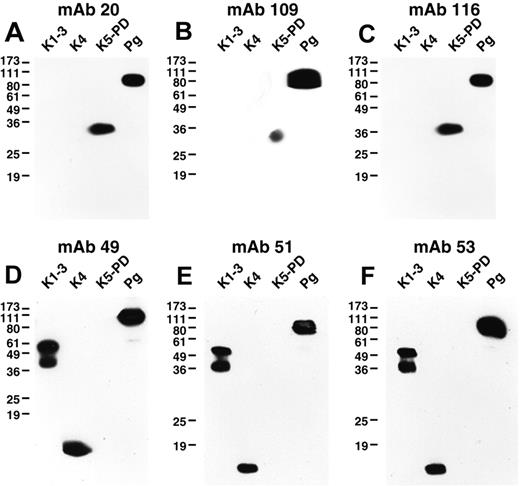
We explored the similarities within the above mAb groups in Western blotting against the well-defined elastase degradation products of Pg, the K1-3 fragment, residues Y79-V337 or Y79-V353, the K4 fragment, residues V354-A439, and the K5-PD fragment, residues V354-A439.12 All of the mAbs reacted with denatured Glu-Pg in Western blotting (Figure 1). In addition, all mAbs of group I detected both K1-3 and K4, but not K5-PD (Figure 1). The mAbs of group II detected K5-PD, but not K1-3 and K4 (Figure 1). The mAb of group III recognized K5-PD, but did not detect K1-3 or K4 (Figure 1). These results are consistent with the pairwise epitope mapping because the mAbs within each group recognized the same Pg fragments.
Western blotting of anti-Pg mAbs. Glu-Pg, K1-3, K4, and K5-PD were each electrophoresed on 12.5% SDS gels under nonreducing conditions and Western blotted with the indicated mAbs (panels A-F). In controls, no bands were detected when normal mouse IgG was used as first Ab (data not shown).
Western blotting of anti-Pg mAbs. Glu-Pg, K1-3, K4, and K5-PD were each electrophoresed on 12.5% SDS gels under nonreducing conditions and Western blotted with the indicated mAbs (panels A-F). In controls, no bands were detected when normal mouse IgG was used as first Ab (data not shown).
Expression of neopitopes by cell-bound Pg
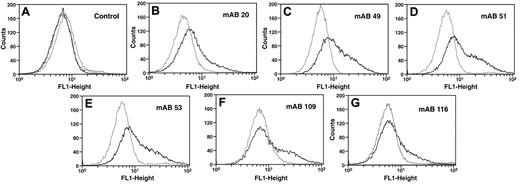
We tested whether the 6 mAbs could detect cell-associated Pg in the presence of a large molar excess of soluble Glu-Pg. U937 monocytoid cells were incubated with the anti-Pg mAbs in the presence of 20μM Glu-Pg and positive cells were detected by FITC-goat anti–mouse IgG in FACS analysis. With the 6 mAbs, a fluorescent population of cells was clearly detected following reaction with cells in the presence of Glu-Pg, compared with cells in the absence of Glu-Pg (Figure 2B-G). In controls, in the presence of Glu-Pg the isotype control Ab provided the same signal obtained in the absence of first Ab and the signal obtained in the absence of first Ab was not increased in the presence of Glu-Pg (Figure 2A). The reactivity of the mAbs with cell-bound Pg in the presence of 20μM soluble Glu-Pg suggests that these antibodies react preferentially with cell-bound Pg. These results also suggest that there are epitopes latent in soluble Glu-Pg that become available when Glu-Pg is bound to cells and suggest that binding of Glu-Pg to cells induces a conformational change in Glu-Pg. Based on these results, we will refer to these Abs as anti-Pg receptor induced binding site (RIBS) Abs, for example, anti-Pg-RIBS mAbs, using the terminology developed for mAbs that recognize fibrinogen bound to platelets.9
Reaction of anti-Pg mAbs with cell-bound Pg. U937 cells (3 × 107/mL) were incubated without (gray) or with (black) 20μM Pg for 30 minutes at 37°C. Then anti-Pg mAbs were added and detection was with FITC-labeled goat anti–mouse IgG (B-G). Controls with second Ab only without (gray) or with (black) 20μM Pg or with isotype control IgG in the presence of 20μM Pg (dashed lines) are in panel A.
Reaction of anti-Pg mAbs with cell-bound Pg. U937 cells (3 × 107/mL) were incubated without (gray) or with (black) 20μM Pg for 30 minutes at 37°C. Then anti-Pg mAbs were added and detection was with FITC-labeled goat anti–mouse IgG (B-G). Controls with second Ab only without (gray) or with (black) 20μM Pg or with isotype control IgG in the presence of 20μM Pg (dashed lines) are in panel A.
Reaction of anti-Pg RIBS mAbs with Glu-Pg bound to substrates and regulatory molecules
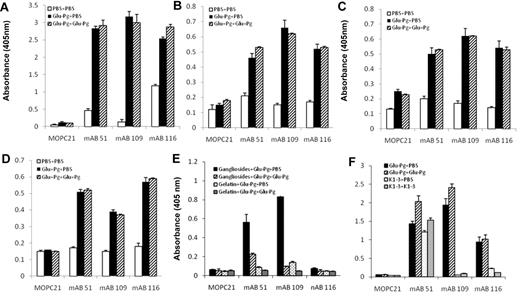
Glu-Pg interacts with the C-terminus of the Pg receptor, Plg-RKT.15,18 Therefore, we examined whether its interaction with the Plg-RKT C-terminus would mimic the effect of the interaction of Glu-Pg with cells with respect to induction of neoepitope expression. Glu-Pg binding to the C-terminal peptide of Plg-RKT, was detected with representative anti-Pg RIBS mAbs from each epitope group and the binding was not inhibited in the presence of soluble Glu-Pg (Figure 3A). Therefore, binding of Glu-Pg to the C-terminus of Plg-RKT mimicked the interaction of Glu-Pg with the cell surface.
Reaction of anti-Pg RIBS mAbs with Glu-Pg bound to substrates and regulatory molecules. Binding of RIBS mAbs (180nM) or MOPC21 isotype control (180nM) to Glu-Pg bound to a synthetic nonapeptide corresponding to the C-terminus of Pg-RKT (A), to fibrin (B), to Pm-treated fibrinogen (C), to Matrigel (D), to mixed gangliosides (E), or binding of RIBS mAbs and MOPC21 to either K1-3 or Glu-Pg bound to a synthetic nonapeptide corresponding to the C-terminus of Pg-RKT (F) was determined in the presence of either buffer (■, Glu-Pg + PBS) or soluble Glu-Pg (1μM; ▨, Glu-Pg + Glu-Pg) and in panel E, binding of mAbs to gelatin preincubated with Glu-Pg in the presence of either buffer ( , Gelatin + Glu-Pg + PBS) or soluble Glu-Pg (▨, Gelatin + Glu-Pg + Glu-Pg). Panel F, buffer (
, Gelatin + Glu-Pg + PBS) or soluble Glu-Pg (▨, Gelatin + Glu-Pg + Glu-Pg). Panel F, buffer ( , K1-3 + PBS) or soluble K1-3 (
, K1-3 + PBS) or soluble K1-3 ( , K1-3 + K1-3). In controls, the substrates and regulatory molecules were not preincubated with Glu-Pg and mAb binding was determined in the absence of soluble Glu-Pg (□, PBS + PBS).
, K1-3 + K1-3). In controls, the substrates and regulatory molecules were not preincubated with Glu-Pg and mAb binding was determined in the absence of soluble Glu-Pg (□, PBS + PBS).
Reaction of anti-Pg RIBS mAbs with Glu-Pg bound to substrates and regulatory molecules. Binding of RIBS mAbs (180nM) or MOPC21 isotype control (180nM) to Glu-Pg bound to a synthetic nonapeptide corresponding to the C-terminus of Pg-RKT (A), to fibrin (B), to Pm-treated fibrinogen (C), to Matrigel (D), to mixed gangliosides (E), or binding of RIBS mAbs and MOPC21 to either K1-3 or Glu-Pg bound to a synthetic nonapeptide corresponding to the C-terminus of Pg-RKT (F) was determined in the presence of either buffer (■, Glu-Pg + PBS) or soluble Glu-Pg (1μM; ▨, Glu-Pg + Glu-Pg) and in panel E, binding of mAbs to gelatin preincubated with Glu-Pg in the presence of either buffer ( , Gelatin + Glu-Pg + PBS) or soluble Glu-Pg (▨, Gelatin + Glu-Pg + Glu-Pg). Panel F, buffer (
, Gelatin + Glu-Pg + PBS) or soluble Glu-Pg (▨, Gelatin + Glu-Pg + Glu-Pg). Panel F, buffer ( , K1-3 + PBS) or soluble K1-3 (
, K1-3 + PBS) or soluble K1-3 ( , K1-3 + K1-3). In controls, the substrates and regulatory molecules were not preincubated with Glu-Pg and mAb binding was determined in the absence of soluble Glu-Pg (□, PBS + PBS).
, K1-3 + K1-3). In controls, the substrates and regulatory molecules were not preincubated with Glu-Pg and mAb binding was determined in the absence of soluble Glu-Pg (□, PBS + PBS).
The major interactive site within Glu-Pg for cells13 and for Plg-RKT15 is contained within the lysine binding sites within the Pg K1-3 domain. We tested whether the K1-3 domain, when bound to the Plg-RKT C-terminus, would preferentially interact with anti-Pg RIBS mAbs compared with soluble K1-3. mAb 51, representing group I, detected immobilized K1-3 and the binding was not competed by soluble K1-3 (Figure 3F). Bound K1-3 was not detected by representative mAbs from groups II and III (Figure 3F). These results are consistent with the abilities of mAbs in group I, but not in groups II and III, to detect K1-3 in Western blotting (Figure 1).
When Glu-Pg binds to fibrin its activation is markedly enhanced because of a large decrease in the Km for the reaction,7 similar to the effect of binding of Pg to the cell surface.1 Therefore, we tested whether the neoepitopes recognized by the 3 epitope groups of mAbs were also induced in Glu-Pg when bound to this substrate. Glu-Pg bound to fibrin was detected by anti-Pg-RIBS mAbs representing each epitope group (Figure 3B). In the presence of a large molar excess of Glu-Pg, binding of the anti-Pg-RIBS mAbs was not inhibited. These results suggest that conformational changes induced in Glu-Pg when bound to fibrin (that are detected by the anti-Pg-RIBS mAbs) are similar to those induced when Glu-Pg binds to cells.
We also tested whether neoepitopes recognized by the 3 epitope groups of mAbs were induced in Glu-Pg when bound to Pm-treated fibrinogen because the Pm-treated fibrinogen assay is representative of the state of fibrin during fibrinolysis, that is, C-terminal lysines are generated by Pm and these C-terminal lysines are new binding sites for Pg. We found that Glu-Pg bound to Pm-treated fibrinogen, as detected with anti-Pg-RIBS mAbs representing each epitope group, and the binding of the mAbs was not inhibited in the presence of soluble Glu-Pg (Figure 3C).
Pg also interacts with the model extracellular matrix Matrigel.19 Matrigel, produced by Engelbreth-Holm-Swarm mouse sarcoma cells20 has been used as a model basement membrane-like extracellular matrix in many studies (eg21-23 ). Therefore, we tested whether neoepitopes recognized by anti-Pg RIBS mAbs were also exposed in Glu-Pg when bound to Matrigel. Glu-Pg bound to Matrigel, as detected with anti-Pg-RIBS mAbs, and the binding of the mAbs was not inhibited in the presence of soluble Glu-Pg (Figure 3D). Taken together, these results suggest that similar conformational changes are induced in Glu-Pg when bound to fibrin, Pm-treated fibrinogen, and extracellular matrix that are also similar to the changes induced when Pg binds to cells and to Plg-RKT.
Glu-Pg also interacts with gangliosides that are present on cell surfaces.16 Therefore, we tested whether neoepitopes recognized by anti-Pg RIBS mAbs were exposed in Glu-Pg when bound to this nonprotein substrate. Glu-Pg bound to mixed gangliosides, as detected with anti-Pg-RIBS mAbs (Figure 3E). Interestingly, the binding of the mAbs was inhibited in the presence of soluble Glu-Pg, suggesting that the conformation of Glu-Pg bound to gangliosides was similar to that of soluble Glu-Pg.
Comparison of surface-induced conformational changes in surface associated Pg with other conformational changes in Pg
Lys-Pg is produced when Pm catalyzes cleavage of Glu-Pg at the carboxyl sides of Lys62, Arg68, or Ly77. 24-26 and at additional minor sites27 to generate new N termini of Pg (Lys Pg has a more open conformation than Glu-Pg28-30 and is more readily activated by Pg activators7,30,31 ). To test whether the conformation adopted by Glu-Pg on its interaction with cells and other surfaces mimicked the conformational change on its cleavage to Lys-Pg, we tested whether soluble Lys-Pg could compete for binding of the mAbs to immobilized Glu-Pg. Soluble Lys-Pg did not compete for binding of the mAbs to immobilized Glu-Pg (Table 1). These results suggest that the conformational changes in immobilized Glu-Pg detected by the anti-Pg-RIBS mAbs were distinct from the conformational change in soluble Lys-Pg compared with soluble Glu-Pg.
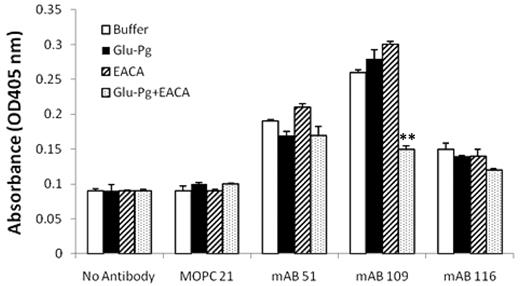
In the presence of ϵ-aminocaproic acid (EACA) Glu-Pg adopts a more flexible state that is more readily activated than in the absence of EACA.26 We tested whether treatment of Glu-Pg with EACA would induce a conformational change that would be detected by the anti-Pg-RIBS mAbs. In the presence of EACA, Glu-Pg did not compete with immobilized Pg for its interaction with mAbs 51 and 116 (Figure 4). Therefore, neoepitopes recognized by these mAbs do not reflect conformational changes in Glu-Pg when bound to EACA. In contrast, binding of mAb 109 to surface-bound Glu-Pg was markedly inhibited by soluble Glu-Pg in the presence of EACA. This result suggests that in the presence of EACA, soluble Glu-Pg exposes an epitope that is also exposed when Glu-Pg is associated with surfaces, cells, substrates, and regulatory molecules.
Effect of EACA on the binding of anti-Pg RIBS mAbs to surface-bound Glu-Pg. Binding of RIBS mAbs or MOPC21 isotype control(180nM) to immobilized Glu-Pg was determined in the presence of either buffer (□), soluble Glu-Pg (1μM) in the absence (■) or presence ( ) of EACA (0.2M), or in the presence of EACA alone (▨). Additional controls show absorbance in the absence of Ab (No Antibody). **P < .001 for soluble Glu-Pg in the presence of EACA compared with soluble Glu-Pg in the absence of EACA.
) of EACA (0.2M), or in the presence of EACA alone (▨). Additional controls show absorbance in the absence of Ab (No Antibody). **P < .001 for soluble Glu-Pg in the presence of EACA compared with soluble Glu-Pg in the absence of EACA.
Effect of EACA on the binding of anti-Pg RIBS mAbs to surface-bound Glu-Pg. Binding of RIBS mAbs or MOPC21 isotype control(180nM) to immobilized Glu-Pg was determined in the presence of either buffer (□), soluble Glu-Pg (1μM) in the absence (■) or presence ( ) of EACA (0.2M), or in the presence of EACA alone (▨). Additional controls show absorbance in the absence of Ab (No Antibody). **P < .001 for soluble Glu-Pg in the presence of EACA compared with soluble Glu-Pg in the absence of EACA.
) of EACA (0.2M), or in the presence of EACA alone (▨). Additional controls show absorbance in the absence of Ab (No Antibody). **P < .001 for soluble Glu-Pg in the presence of EACA compared with soluble Glu-Pg in the absence of EACA.
Identification of Pg domains containing RIBS epitopes
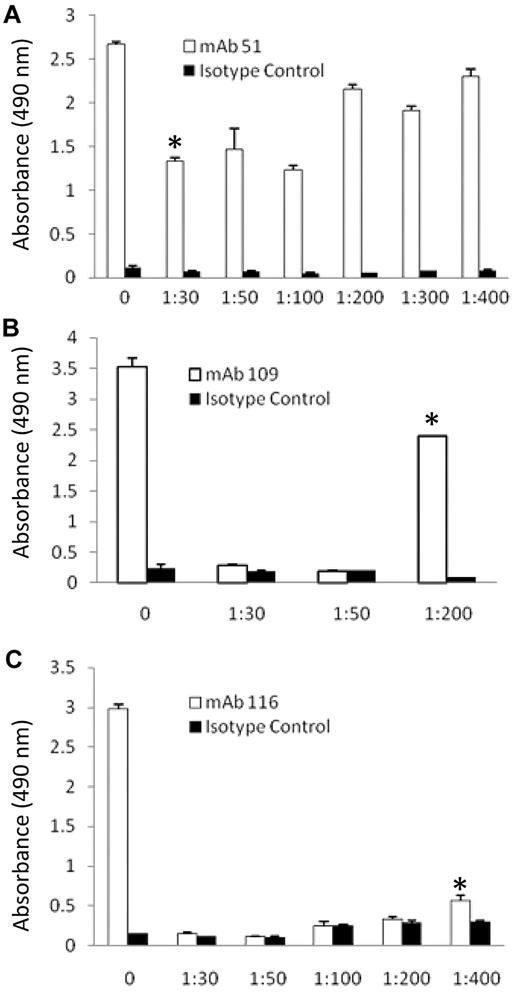
To identify Pg domains containing the epitopes recognized by the anti-Pg-RIBS mAbs, we used an approach combining trypsin proteolysis and immunoaffinity chromatography, followed by tandem mass spectrometry (LC-MS-MS). Glu-Pg was reduced, alkylated, and denatured and then digested with increasing concentrations of trypsin. The digestion products were analyzed in an ELISA with anti-Pg-RIBS mAbs representing each epitope recognition group. For each mAb, we selected a trypsin concentration and digestion time resulting in maximal proteolysis that could still be detected with each anti-Pg-RIBS mAbs in the ELISA (Figure 5). The weight ratios we selected were 1/50, 1/200, 1/400 trypsin to Pg for mAbs 51, 109, and 116, respectively, and digestion was for 1 hour. For identification of domains containing epitopes recognized by anti-Pg-RIBS mAbs, a quantity of 0.5 mg of reduced, alkylated, and denatured Glu-Pg was digested with trypsin and the digest subjected to immunoaffinity chromatography on either mAb 51-Sepharose, mAb 109-Sepharose, or mAb 116-Sepharose, or isotype control MOPC21-Sepharose, separately. The columns were washed an 10mM Tris HCl, pH 8.0, and when no protein was detected at 280nM, the columns were eluted with 0.2M Glycine HCl, pH 2.6.
Concentration dependence of trypsin digestion of Glu-Pg. Glu-Pg was reduced and alkylated and either untreated (0) or digested with the indicated weight ratio of trypsin to Glu-Pg for 1 hour and coated onto wells of microtiter plates as described in “Methods.” Reactivity with mAb 51 (□; A), mAb 109 (□; B) and mAb 116 (□; C) was determined by ELISA as described in “Epitope determination: ELISA analysis of trypsin digests of Pg” and compared with the isotype control, MOPC21 (■ in all panels). *The trypsin:Glu-Pg ratio selected for study of each mAb.
Concentration dependence of trypsin digestion of Glu-Pg. Glu-Pg was reduced and alkylated and either untreated (0) or digested with the indicated weight ratio of trypsin to Glu-Pg for 1 hour and coated onto wells of microtiter plates as described in “Methods.” Reactivity with mAb 51 (□; A), mAb 109 (□; B) and mAb 116 (□; C) was determined by ELISA as described in “Epitope determination: ELISA analysis of trypsin digests of Pg” and compared with the isotype control, MOPC21 (■ in all panels). *The trypsin:Glu-Pg ratio selected for study of each mAb.
Peptides eluted from the immunoaffinity columns were subjected to reversed-phase chromatography, followed by tandem mass spectrometry. The criteria used for selection of the peptides was a probability based Mowse score > 50 and expectation value < 0.05 (The Mowse scoring alogrithm32 uses empirically determined factors to assign a statistical weight to each individual peptide match and accurately models the behavior of a proteolytic enzyme [http://snowwhite.scripps.edu/mascot/help_index.html]. The expectation value is the number of matches with equal or better scores that are expected to occur by chance alone. An expectation value of 0.05 provides 95% confidence that the peptide has been correctly identified). Tandem mass spectra for peptides meeting these criteria are shown for each representative anti-Pg-RIBS mAb (Figure 6). No peptides meeting these criteria were obtained when trypsin digests of Pg were subjected to affinity chromatography on MOPC21-Sepharose (data not shown).
LC/MS/MS data for Pg fragments bound by anti-Pg-RIBS mAbs. (A) The tandem mass spectrum obtained from the [M+2H+]2+ ion from the peptide YDYCDILECEECMHCSGENYDGK, which bound to mAb 51-Sepharose produced characteristic sequence ions which were used for its positive identification. (B) The tandem mass spectrum obtained from the [M+2H+]2+ ion from the peptide LSSPAVITDK, which bound to mAb 109-Sepharose, produced characteristic sequence ions which were used for its positive identification. (C) The tandem mass spectrum obtained from the [M+2H+]2+ ion from the peptide HSIFTPETNPR, which bound to mAb 109-Sepharose, produced characteristic sequence ions which were used for its positive identification. (D) The tandem mass spectrum obtained from the [M+2H+]2+ ion from the peptide QLGAGSIEECAAKCEEDEEFTCR, which bound to MAb 116-Sepharose. (E) The tandem mass spectrum obtained from the [M+2H+]2+ ion from the peptide, VILGAHQEVNLEPHVQEIEVSR, which bound to mAb 116-Sepharose.
LC/MS/MS data for Pg fragments bound by anti-Pg-RIBS mAbs. (A) The tandem mass spectrum obtained from the [M+2H+]2+ ion from the peptide YDYCDILECEECMHCSGENYDGK, which bound to mAb 51-Sepharose produced characteristic sequence ions which were used for its positive identification. (B) The tandem mass spectrum obtained from the [M+2H+]2+ ion from the peptide LSSPAVITDK, which bound to mAb 109-Sepharose, produced characteristic sequence ions which were used for its positive identification. (C) The tandem mass spectrum obtained from the [M+2H+]2+ ion from the peptide HSIFTPETNPR, which bound to mAb 109-Sepharose, produced characteristic sequence ions which were used for its positive identification. (D) The tandem mass spectrum obtained from the [M+2H+]2+ ion from the peptide QLGAGSIEECAAKCEEDEEFTCR, which bound to MAb 116-Sepharose. (E) The tandem mass spectrum obtained from the [M+2H+]2+ ion from the peptide, VILGAHQEVNLEPHVQEIEVSR, which bound to mAb 116-Sepharose.
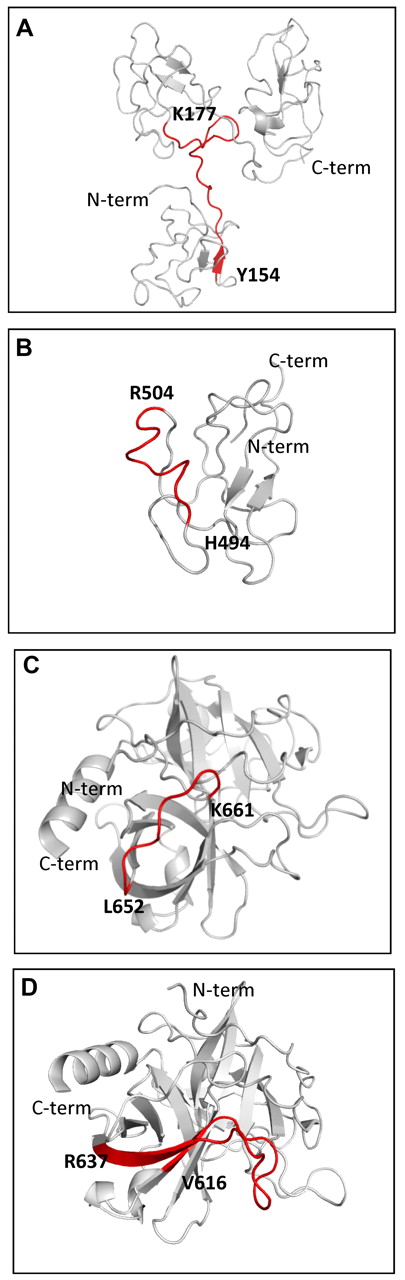
For mAb 51, a single peptide meeting the selection criteria was identified (Figure 6A). The peptide corresponded to Pg K177-Y154, spanning a domain linking Pg kringles 1 and 2 (Figure 7A). This identification was consistent with the ability of mAb 51 to recognize K1-3 in Western blotting (Figure 1) as well as the ability of mAb 51 to recognize K1-3 bound to the C-terminus of Plg-RKT (Figure 3F). Although mAb 51 also recognized K4 in Western blotting (Figure 1) and when immobilized onto plastic (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), we did not detect an epitope in K4 using this method. This could be because the epitope is nonlinear or that the peptide sequence within K4 did not ionize sufficiently for detection by mass spectrometry.
Ribbon diagram of epitopes recognized by anti-Pg-RIBS mAbs. (A) Pg kringles 1-3 (PDB ID 1KIO, gray) showing mAb 51 binding residues Y154-K177 (red ribbon and bold characters). (B) Pg kringle 5 (PDB ID 5HPG, gray) showing mAb 109 binding residues H494-R504 (red ribbon and bold characters). (C) Pg protease domain (PBD ID 1L4D, gray) showing mAb 109 binding residues L652-K661 (red ribbon and bold characters). (D) Pg protease domain (PBD ID 1L4D, gray) showing mAb 116 binding residues V616-R637 (red ribbon and bold characters).
Ribbon diagram of epitopes recognized by anti-Pg-RIBS mAbs. (A) Pg kringles 1-3 (PDB ID 1KIO, gray) showing mAb 51 binding residues Y154-K177 (red ribbon and bold characters). (B) Pg kringle 5 (PDB ID 5HPG, gray) showing mAb 109 binding residues H494-R504 (red ribbon and bold characters). (C) Pg protease domain (PBD ID 1L4D, gray) showing mAb 109 binding residues L652-K661 (red ribbon and bold characters). (D) Pg protease domain (PBD ID 1L4D, gray) showing mAb 116 binding residues V616-R637 (red ribbon and bold characters).
For mAb 109, 2 peptides meeting the selection criteria were identified (Figure 6B-C). Two peaks with masses of 1298 Atomic Mass Unit (AMU) and 1030 AMU corresponding to a nonlinear epitope contained within H494-R504 within the K5 domain (Figure 7B) and L652-K661 in the PD (Figure 7C), respectively, were obtained. This result is consistent with the ability of mAb 109 to recognize K5-PD in Western blotting (Figure 1). However, the epitope was not detected when K5 was directly bound to plastic (supplemental Table 1), suggesting that the epitope is expressed differently when intact Pg versus isolated K5 is bound to plastic.
For mAb 116, 2 peptides meeting the selection criteria were identified (Figure 6D-E). Two peaks with masses of 2688 AMU and 2495 AMU, corresponding to a nonlinear epitope contained within Q21-R43 within the N-terminal domain of Glu-Pg and V616-R637 within the latent PD (Figure 7D), respectively, were identified. This result is consistent with the ability of mAb 116 to recognize K5-PD in Western blotting (Figure 1) as well as when K5-PD was immobilized onto plastic (supplemental Table 1).
Discussion
When Glu-Pg binds to cells, it acquires functional properties distinct from the soluble zymogen. Glu-Pg bound to cell surfaces is more efficiently cleaved by Pm, compared with soluble Glu-Pg, to yield the more readily activated Lys-Pg form.4-6 The enhanced stimulation of Glu-Pg activation on the cell surface, thus, arises via its transition through the Lys-Pg form.4,5 These functional changes have been ascribed to changes in the conformation of cell-associated Glu-Pg,4,33 but have not been directly demonstrated. In this study, we provide direct evidence for conformational changes in Glu-Pg when bound to the cell surface using mAbs that detect changes in Glu-Pg conformation, via exposure of specific neoepitopes, when Glu-Pg is bound to cell surfaces. Furthermore, these mAbs have allowed us to identify specific domains that are poorly accessible in soluble Glu-Pg but become exposed when Glu-Pg binds to the cell surface.
We developed 6 anti-Pg mAbs that reacted with Glu-Pg bound to microtiter wells in the presence of a > 50-fold molar excess of soluble Glu-Pg. Three distinct epitopes were recognized by this panel of 6 mAbs. This type of approach was first used to address conformational changes in fibrinogen8 when a mAb that recognized an epitope that was latent in soluble fibrinogen, but became available when fibrinogen was adsorbed onto microtiter wells was identified.8 The same epitope was exposed when fibrinogen interacted with its receptor, GPIIb-IIIa, and was designated a RIBS.8,9 Here, we have developed anti-Pg mAbs that recognize 3 distinct neopitopes in Glu-Pg on the cell surface in the presence of a large molar excess of soluble Glu-Pg. Thus, the epitopes that became available when Glu-Pg was adsorbed onto microtiter wells also became available when Glu-Pg was bound to the cell surface. We have designated these Abs as anti-Pg-RIBS mAbs, according to the terminology of Zamarron and colleagues.9
To further address the relationships between changes in Glu-Pg conformation and cellular binding sites, we found that Glu-Pg binding to the C-terminal peptide of the recently described Pg receptor, Plg-RKT,18 was detected with the anti-Pg RIBS mAbs and the binding was not inhibited in the presence of soluble Glu-Pg. Therefore, binding of Glu-Pg to the C-terminus of Plg-RKT mimicked the interaction of Glu-Pg with the cell surface, with respect to conformational changes detected by the anti-Pg RIBS mAbs. Moreover, when bound to the C-terminus of Plg-RKT, the K1-3 domain (that provides the major interactive site within Glu-Pg for cells13 and for Plg-RKT15 ) was detected by anti-Pg RIBS mAb 51 (that recognized an epitope within K1-3) and the binding was not inhibited in the presence of soluble K1-3. Thus, isolated K1-3 and K1-3 within the intact Glu-Pg molecule appear to adopt a similar conformation when bound to the C-terminus of Plg-RKT.
The anti-Pg RIBS mAbs also preferentially recognized Glu-Pg bound to substrates and regulatory molecules that also stimulate Glu-Pg activation: fibrin, Pm-treated fibrinogen, and the representative extracellular matrix, Matrigel. These results suggest that there are common conformational changes induced when Glu-Pg is bound to these regulatory molecules and to the cell surface. In contrast, although the anti-Pg RIBS mAbs detected Glu-Pg bound to nonprotein gangliosides, soluble Glu-Pg competed for this binding. These results suggest that the conformation of Glu-Pg when bound to gangliosides is similar to the conformation of Glu-Pg in solution.
Using an approach combining trypsin proteolysis, immunoaffinity chromatography and mass spectrometry, we identified domains in Glu-Pg that contained sequences comprising the 3 epitopes identified by our panel of anti-Pg RIBS mAbs. The epitope for anti-Pg RIBS mAb 51 was contained within Pg K177-Y154, within a domain linking Pg kringles 1 and 2 (Figure 7A). This result is consistent with this region behaving as a flexible domain, linking kringles that function independently.34 As the K1 lysine binding site mediates Glu-Pg binding to the cell surface,13,35 changes in the conformation of Glu-Pg when K1 interacts with the cell are likely to influence the mobility and, hence, surface exposure of this linker region. The epitope for anti-Pg RIBS mAb 109 was composed of a nonlinear epitope contained within H494-R504 within the K5 domain (Figure 7B) and within L652-K661 in the latent protease domain (PD; Figure 7C). This result suggests that, on binding of Glu-Pg to the cell surface, these regions become surface exposed and proximal. This result was corroborated in the identification of a component of the nonlinear epitope of mAb 116, which resided within V616-R637, also within the PD domain which is likely to be spatially proximal to the epitope within the PD recognized by mAb 109 as shown in the ribbon diagram (Figure 7D). Interestingly, the other component of the nonlinear epitope recognized by anti-Pg RIBS mAb 116 was contained within the N-terminal domain (Q21-R43). Formation of Lys-Pg occurs by plasmic cleavage of Glu-Pg at the carboxyl sides of Lys62, Arg68, and Lys77.24-26 Therefore, exposure of this segment of the N-terminal domain on binding to the cell surface is likely to promote its ability to be cleaved by Pm to Lys-Pg; a cleavage that is enhanced when Glu-Pg binds to the cell surface.4,5 Furthermore, a transition between an interaction of the N-terminal domain with K5 in the tight (T) conformation of Glu-Pg [as previously reported36 ] to an interaction with the PD of cell-associated Glu-Pg is suggested. It is also noteworthy that none of the anti-Pg RIBS mAbs recognized domains contained within the lysine binding structures of the Pg kringles. This result is consistent with occupancy of these binding sites by cell-surface receptors,13 so that these regions of Glu-Pg are not accessible to the anti-Pg RIBS mAbs.
It has been generally accepted that Glu-Pg adopts a Lys-like conformation when bound to the cell surface.37 In contrast, our results suggest that the conformation induced in cell-bound Glu-Pg is distinct from the conformation of Lys-Pg because soluble Lys-Pg did not compete for the binding of the anti-Pg RIBS mAbs to surface-associated Glu-Pg. These results are consistent with our previous data suggesting that the enhancement of Glu-Pg activation on the cell surface requires cleavage of Glu-Pg by Pm to yield the more readily activated Lys-Pg.4,5 Furthermore, conformational changes in Glu-Pg induced by EACA were detected by anti-Pg RIBS mAb 109, suggesting that the relaxed (R) form of Glu-Pg induced by its interaction with EACA33 is distinct from the conformation of Lys-Pg. Thus, in the presence of EACA, soluble Glu-Pg exposes an epitope that is also exposed when Glu-Pg is associated with cells, consistent with binding of Glu-Pg to cell-surface proteins exposing C-terminal lysines, as exemplified by Plg-RKT.18,38
These anti-Pg RIBS mAbs have many potential uses for future investigations of structure/function relationships in the field of plasminogen activation. For example, they could be of great use in imaging studies of cell- or fibrin-associated Pg. They could be used to assess the relative contributions of surface-associated and soluble Glu-Pg to overall plasminogen activation capacity in a recently described fibrinolytic cross-talk mechanism.37 The mAbs could be used in studies to assess the contribution of Pg bound to different fibrin structures that differentially stimulate plasminogen activation and fibrinolysis.39
In summary, in this study we have shown that the interaction of Glu-Pg with cell surfaces and with Plg-RKT induces a conformational change in this zymogen and have defined this conformational change and shown that it is distinct from that of Lys-Pg. Furthermore, similarities in the conformation of Glu-Pg are induced on its binding to fibrin and to other regulatory molecules on which Glu-Pg activation is promoted, suggesting that these conformational changes globally influence Pg activation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Michael A. Green for excellent technical assistance.
This work was supported by National Institutes of Health Grants (HL38272, Hl45934, and HL 081046 [L.A.M.], HL50398 [R.J.P.], and HL013423 [F.J.C.]) and Department of Veterans Affairs (R.J.P.).
This is publication no. 20933 from The Scripps Research Institute.
National Institutes of Health
Authorship
Contribution: J.H., N.B., K.-H.K., J.-M.Y., G.W.H., Y.G., M.J., and J.F. designed experiments, performed research, analyzed data, and participated in manuscript drafting; F.J.C. and R.J.P. analyzed data and participated in manuscript drafting; and L.A.M. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lindsey A. Miles, PhD, Department of Cell Biology, SP30-20, The Scripps Research Institute, 10550 N Torrey Pines Rd, La Jolla, CA 92037; e-mail: lmiles@scripps.edu.

![Figure 6. LC/MS/MS data for Pg fragments bound by anti-Pg-RIBS mAbs. (A) The tandem mass spectrum obtained from the [M+2H+]2+ ion from the peptide YDYCDILECEECMHCSGENYDGK, which bound to mAb 51-Sepharose produced characteristic sequence ions which were used for its positive identification. (B) The tandem mass spectrum obtained from the [M+2H+]2+ ion from the peptide LSSPAVITDK, which bound to mAb 109-Sepharose, produced characteristic sequence ions which were used for its positive identification. (C) The tandem mass spectrum obtained from the [M+2H+]2+ ion from the peptide HSIFTPETNPR, which bound to mAb 109-Sepharose, produced characteristic sequence ions which were used for its positive identification. (D) The tandem mass spectrum obtained from the [M+2H+]2+ ion from the peptide QLGAGSIEECAAKCEEDEEFTCR, which bound to MAb 116-Sepharose. (E) The tandem mass spectrum obtained from the [M+2H+]2+ ion from the peptide, VILGAHQEVNLEPHVQEIEVSR, which bound to mAb 116-Sepharose.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/6/10.1182_blood-2010-11-316943/4/m_zh89991176110006.jpeg?Expires=1769110689&Signature=tYT44V8USQE1yBPiM2vWKr~2pNpWzwVo50oqXfcMPQhrCuFeVb2UjjjpdXNvI98jlumsaeCrW~tEMWiubzba0fkCiJVp9L31K201f30HK0~XpI3~nZV4r1qs-Up8crnelp7qxe9R~RJN8eZrqzSmYzzN85-B8rpkmWseYcsCKgATljhZODFmhITD2KJ6NvSEerX5X7T~j9Mm3OQE27qpx359Bup4a4Mrtjk0UBXKErXkBjaM8BOrDap1YHkol4EHlNnDIbrLEo-49~ixxCOLUDhbDh1M7TPv-1N0TE~7Pcx8FovQNoPiHab7741lEdno3AIX1Wk52wdHA06xJIjE1g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)