Abstract
Nuclear factor-κB essential modulator (NEMO), the regulatory subunit of the IκB kinase complex, is a critical component of the NF-κB pathway. Hypomorphic mutations in the X-linked human NEMO gene cause various forms of anhidrotic ectodermal dysplasia with immunodeficiency (EDA-ID). All known X-linked EDA-ID–causing mutations impair NEMO protein expression, folding, or both. We describe here 2 EDA-ID–causing missense mutations that affect the same residue in the CC2-LZ domain (D311N and D311G) that do not impair NEMO production or folding. Structural studies based on pull-down experiments showed a defect in noncovalent interaction with K63-linked and linear polyubiquitin chains for these mutant proteins. Functional studies on the patients' cells showed an impairment of the classic NF-κB signaling pathways after activation of 2 NEMO ubiquitin-binding–dependent receptors, the TNF and IL-1β receptors, and in the CD40-dependent NF-κB pathway. We report the first human NEMO mutations responsible for X-linked EDA-ID found to affect the polyubiquitin binding of NEMO rather than its expression and folding. These experiments demonstrate that the binding of human NEMO to polyubiquitin is essential for NF-κB activation. They also demonstrate that the normal expression and folding of NEMO do not exclude a pathogenic role for NEMO mutations in patients with EDA-ID.
Introduction
NF-κB essential modulator (NEMO) is a regulatory subunit of the inhibitor of NF-κB (IκB) kinase.1 NEMO is a scaffold protein essential for the formation of the IκB kinase protein complex that catalyzes the phosphorylation of IκBs. In resting cells, NF-κB is sequestered in the cytoplasm through physical interaction with IκBs (IκBα, IκBβ, and IκBϵ). After stimulation of the cell with diverse molecules, such as proinflammatory cytokines (TNF-α, IL-1β), endotoxins (lipopolysaccharide), or CD40L, the canonical NF-κB signaling pathway is activated.1 The phosphorylation of IκB leads to its dissociation from NF-κB, which results in its ubiquitination and degradation in the proteasome. The released NF-κB dimers (p50, p52, c-rel, p65 [also called relA]), and RelB) are translocated to the nucleus, where they induce the transcription of various target genes that encode proinflammatory molecules.2 It has recently been shown that the NEMO protein consists essentially of a series of coiled-coil domains: CC1 in the N-terminal segment, HLX2 in the middle segment, and the CC2-LZ regulatory domain in the C-terminal segment. NEMO also has a zinc finger (ZF) domain at its C-terminal end.3 The function of NEMO depends on its dimerization and its ability to interact with linear or K63-linked polyubiquitin chains.4-7 This function requires the CC2-LZ domain, which is involved in NEMO dimerization and contains a ubiquitin-binding site called NOA/UBAN/NUB, and the ZF domain, which bears a second ubiquitin-binding site.8,9
Diverse mutations of NEMO are responsible for various rare X-linked diseases. Amorphic NEMO mutations that lead to a lack of NEMO-dependent NF-κB activation are associated with X-linked dominant incontinentia pigmenti (XD-IP) in females10 and with in utero lethality in males.10,11 Hypomorphic NEMO mutations that lead to the impairment of NF-κB signaling, but not its abolition, are associated with the syndrome of X-linked recessive anhidrotic ectodermal dysplasia with immunodeficiency (XR-EDA-ID) in males.11-19 The developmental features of EDA-ID include a typical facies (frontal bossing, saddle nose), aberrant development of skin appendages such as hair and teeth, and dry skin. Immunodeficiency results in susceptibility to a wide range of pathogens (pyogenic bacteria, mycobacteria, and viruses, in particular).15,17,20 The extent and severity of the ectodermal phenotype define several different clinical diseases: EDA-ID with osteopetrosis and/or lymphedema (XR-EDA-ID-OL), classic XR-EDA-ID, XR with mild-EDA-ID (eg conical incisors only), and immunodeficiency without EDA.16,21-23
A tremendous diversity of immunologic receptors signal via NEMO and NF-κB, each potentially contributing to the immunodeficiency of patients with XR-EDA-ID, depending on the specific NEMO mutation. Impaired cellular responses to TNF-α that result in a lack of inflammatory cytokine production are found in at least 80% of patients.24,25 Approximately half of the patients display defective responses to IL-1β and at least some TLRs; however, most NF-κB–dependent immunologic pathways have yet to be tested in patients with XR-EDA-ID. In routine immunologic examinations, some patients present with various immunoglobulin production defects. Some patients present with a deficiency in B-cell switching in response to CD40 activation.11-13,26 Interestingly, most if not all patients lack polysaccharide-specific antibodies, despite infection with and/or vaccination against encapsulated bacteria. Allohemagglutinin levels are also typically low in these patients. Only a few of the links between NEMO genotype, cellular phenotype, and clinical phenotype have been deciphered, such as the lack of EDA-R signaling and the developmental phenotype of EDA,27 the lack of CD40-dependent induction of IL-12 and mycobacterial disease,20 and the lack of CD40-dependent B-cell switching and hyper IgM syndrome.26
The mechanisms by which some mutations affect the structural and functional integrity of NEMO have been investigated more thoroughly. Mutations in exon 1B or the 5′ untranslated region result in lower levels of NEMO mRNA and much lower levels of protein,23 with a clinical phenotype of recurrent sinopulmonary infections and dysgammaglobulinemia without EDA.23 The EDA-ID–associated A288G mutation of NEMO, which affects the CC2 domain, has no effect on protein expression but destabilizes NEMO oligomers (dimers and higher-order oligomers), thereby altering assembly of the IκB kinase complex and impairing the canonical activation of NF-κB.28 The immunodeficiency-associated 110_111insC mutation of NEMO creates a premature translation termination codon in codon position 49.16 A methionine codon downstream from the premature stop codon allows the reinitiation of translation. The production of a small amount of residual NH2-truncated NEMO results in a phenotype of immunodeficiency without EDA. The C417F mutation that affects the ZF domain of NEMO is responsible for EDA-ID.11,12 The production of the C417F protein has not been investigated, but the structures of NEMO ZF and its C417F mutant were recently determined by nuclear magnetic resonance.3 The C417F substitution modifies the structure of the C-terminal end of the ZF α-helix and decreases stability, which leads to a defect in NF-κB activation. Another substitution, C417R, does not affect expression of the NEMO protein but impairs c-Rel activation in response to CD40 ligation.29 The EDA-ID-OL–associated X420W allele includes a new translation termination codon and encodes a protein 27 amino acids longer than the wild-type (WT) protein,30 the expression of which is impaired. Two other mutations that affect the LZ (leucine zipper) domain of NEMO, E315A and R319Q, disrupt the formation of the salt bridge normally formed between residues E315 and R31920 without affecting NEMO protein production.20,31 The folding defect of the E315A mutant is responsible for the defect in binding to ubiquitin chains.32 Thus, some previously characterized NEMO mutations interfere simply with the levels of protein expression, and others are associated with impairment of the folding and stability of NEMO domains, in some cases leading to an impairment of NEMO production. We describe here the structural and functional characterization of the D311N and D311G mutations, which affect the NOA ubiquitin-binding site of NEMO and mediate their deleterious effects by impairing NEMO-ubiquitin binding, with no detectable effect on NEMO expression and folding.
Methods
Case reports
The patient (P) is a boy born in 1992 to nonconsanguineous parents originating from and living in the French West Indies (Figure 1A). He was vaccinated with M bovis-BCG and received standard immunizations in infancy, with no side effects. At 17 months of age, he developed bilateral purulent otitis and pneumococcal pneumonia that required hospitalization. He was subsequently hospitalized again for acute gastroenteritis. The patient presented with upper respiratory tract infections that required adenoidectomy at the age of 2 years. At the age of 3 years, osteoarthritis of the right femur was diagnosed and treated, with relapse 1 month later. The patient was hospitalized at the age of 4 years for pelvic osteotomy to treat postinfectious coxa magna. At the same age, he developed multiple cervical lymphoadenopathies, which required surgical excision of the lymph nodes. Histologic analysis revealed necrotizing epithelioid granulomatous adenitis, with M avium growing in culture. Since the age of 6 years, the patient has had left submaxillary adenopathy and skin lesions of the trunk, with an annular granuloma. Routine inflammatory and immunologic evaluations were unremarkable. The patient's lymphocytes proliferated in response to lectins (eg PHA) and antigens (candidin, tuberculin, and tetanic anatoxin). Serum immunoglobulin isotype levels were normal for IgG (11.3 mg/mL; age-matched reference values 6.4-12.4 mg/mL), IgA (4.23 mg/mL; age-matched reference values 0.8-3.4 mg/mL), IgM (0.54 mg/mL; age-matched reference values 0.5-1.5 mg/mL), and IgE (69 IU/mL). IgG subclasses were also assessed: 8.90 mg/mL IgG1 (age-matched reference values > 4 mg/mL), 0.90 mg/mL IgG2 (age-matched reference values > 0.60 mg/mL), 0.33 mg/mL IgG3 (age-matched reference values > 0.17 mg/mL), and 1.11 mg/mL IgG4 (age-matched reference values, no lower limit). The patient had normal (1/16) anti-A serum allohemagglutinin levels and low anti-B serum allohemagglutinin levels (1/2) at the age of 6 years. The granulocyte respiratory burst was normal. This patient received antimycobacterial antibiotics (clarithromycin, rifabutin, and ethambutol) for 3 months, followed by clarithromycin and rifabutin for 6 months and clarithromycin alone for 5 months. There was a partial clinical response. From March 2003 to August 2003, this treatment was combined with ethambutol, which led to recovery. In 2008, the patient presented with an episode of rash associated with multiple hypochromic papules of the face, limbs, and trunk. A biopsy revealed the presence of an intracellular mycobacterial infection, and the patient was treated with clarithromycin, rifabutin, ethambutol, and moxifloxacin for 2 months, followed by clarithromycin and rifabutin for 5 months. He presented with a new episode of polyarthritis and was prescribed rifabutin, ethambutol, and moxifloxacin for 5 months, with a partial response. The patient is now 19 years old and has a lesion of the skin cheek caused by M abscessus. He sweats normally and has all his adult teeth except the maxillary lateral incisors and the first upper premolars on the right. The agenesis of these teeth was confirmed by mandibular radiograph but went unrecognized for some time (Figure 1B). The mother and sister of the patient have no immunodeficiency or incontinentia pigmenti. Informed consent was obtained from all family members, and the study was approved by the Necker institutional review boards.
Pedigree, clinical features, and protein expression for the patient. (A) Pedigree of patient (P) with a NEMO mutation in position 311 (P, D311G). The different generations are indicated by roman numerals (I and II), and each individual is indicated by an Arabic numeral. Male subjects are represented by squares and female subjects by circles. Solid (black) symbols indicate affected individuals; open (white) symbols, healthy individuals; ⊡, female carriers. Individuals whose genetic status could not be determined are indicated by the symbol “E?” (B) Photograph and radiograph of P taken at 15 years, showing the agenesis of 3 adult teeth (14, 12, and 22). (C) Automated sequencing profile showing the NEMO D311G mutation in cDNA extracted from EBV-B cells from the proband (P) and comparison with the sequence obtained from a control subject (C+). The A→G mutation led to the replacement at residue 311 of aspartic acid (D) by glycine (G). NEMO protein was detected by Western blotting with the FL-419 antibody (D) in monocytes and granulocytes from a healthy control subject (C+) and the proband [P (D311G)]; in EBV-B cells from a healthy control subject (C+), a 110_111insC NEMO patient already reported to produce smaller than normal amounts of NEMO protein, an NEMO EDA-ID patient with another mutation in position 311 (D311N) and the proband [P (D311G)]; and in SV40-transformed fibroblasts from a healthy control subject (C+), an XD-IP patient (IP), and the proband [P (D311G)]. (E) In SV40-transformed fibroblasts from the XD-IP NEMO patient transfected or not transfected (IP) with expression vectors carrying NEMO alleles (wild-type [IP+NEMO+WT], D311G [IP+NEMO+D311G], D311N [IP+NEMO+D311N]) or with insert-free vector (IP+Empty), cells were cotransfected with cyan fluorescent (CFP) vector, as a control for transfection efficiency control. GAPDH antibody was used to normalize protein levels in panels D and E. These results are representative of 2 independent experiments. WB indicates Western blot.
Pedigree, clinical features, and protein expression for the patient. (A) Pedigree of patient (P) with a NEMO mutation in position 311 (P, D311G). The different generations are indicated by roman numerals (I and II), and each individual is indicated by an Arabic numeral. Male subjects are represented by squares and female subjects by circles. Solid (black) symbols indicate affected individuals; open (white) symbols, healthy individuals; ⊡, female carriers. Individuals whose genetic status could not be determined are indicated by the symbol “E?” (B) Photograph and radiograph of P taken at 15 years, showing the agenesis of 3 adult teeth (14, 12, and 22). (C) Automated sequencing profile showing the NEMO D311G mutation in cDNA extracted from EBV-B cells from the proband (P) and comparison with the sequence obtained from a control subject (C+). The A→G mutation led to the replacement at residue 311 of aspartic acid (D) by glycine (G). NEMO protein was detected by Western blotting with the FL-419 antibody (D) in monocytes and granulocytes from a healthy control subject (C+) and the proband [P (D311G)]; in EBV-B cells from a healthy control subject (C+), a 110_111insC NEMO patient already reported to produce smaller than normal amounts of NEMO protein, an NEMO EDA-ID patient with another mutation in position 311 (D311N) and the proband [P (D311G)]; and in SV40-transformed fibroblasts from a healthy control subject (C+), an XD-IP patient (IP), and the proband [P (D311G)]. (E) In SV40-transformed fibroblasts from the XD-IP NEMO patient transfected or not transfected (IP) with expression vectors carrying NEMO alleles (wild-type [IP+NEMO+WT], D311G [IP+NEMO+D311G], D311N [IP+NEMO+D311N]) or with insert-free vector (IP+Empty), cells were cotransfected with cyan fluorescent (CFP) vector, as a control for transfection efficiency control. GAPDH antibody was used to normalize protein levels in panels D and E. These results are representative of 2 independent experiments. WB indicates Western blot.
MDDC stimulation
Monocyte-derived dendritic cells (MDDCs) were obtained from PBMCs. They were incubated for 2-3 hours in RPMI 1640 supplemented with 10% heat-inactivated pooled FBS (GIBCO), referred to as complete RPMI 1640 medium, as reported previously.20 After 9 days of culture, 200 000 MDDCs were activated with 1 μg/mL human recombinant MegaCD40L Soluble (Enzo Life Sciences) in 250 μL of complete RPMI medium at 37°C under an atmosphere that contained 5% CO2. Supernatants were collected 24 hours after activation.
Culture and stimulation of cell lines
B lymphocytes were immortalized with Epstein-Barr virus (EBV-B cells) and cultured in complete RPMI 1640 medium. We stimulated 106 EBV-B cells with phorbol 12,13-dibutyrate (PDBu 10−7M; Sigma-Aldrich) in 24-well plates for 24 hours at 37°C under an atmosphere containing 5% CO2. The supernatants were then recovered.
Immunoblotting and DNA-binding activity
For EMSA, nuclear extracts were prepared from EBV-B cells incubated with or without 100 ng/mL soluble MegaCD40L for 20 and 40 minutes and cytosolic extracts for 20, 40, 60, and 120 minutes.
Pull-down analysis
We analyzed the interaction between His-CC2-LZ (WT, D311N, and D311G) and K63-linked or linear tetraubiquitin by performing pull-down experiments with NI-NTA magnetic agarose beads (QIAGEN). CC2-LZ (8μM) was incubated with various concentrations of K63-linked tetraubiquitin (0.2μM, 0.4μM, 0.7μM, 0.9μM, 1.7μM, and 3.5μM) or linear tetraubiquitin (0.5μM, 1μM, 2μM, 3μM, 4μM, 5μM, 6μM, and 7μM). Two nonspecific binding controls were also performed, one with 8μM His-DARPin (a His-tagged control protein that does not interact with ubiquitin) and the other with beads only. All of these experiments were performed in 25mM Tris-HCl buffer pH 8 that contained 50mM KCl, 0.1mM DDM, and 10mM imidazole. We washed 50 μL of magnetic beads twice in water and twice in buffer and then incubated them for 30 minutes at room temperature with either 100 μL CC2-LZ or DARPin or without protein. The samples were then washed twice with 300 μL of buffer, and the beads were again incubated for 30 minutes with various concentrations of tetraubiquitin (K63-linked or linear). The beads were washed twice in buffer, and bound material was eluted in Laemmli loading buffer supplemented with 6M urea (15 minutes at 95°C) for SDS-PAGE analysis. The gel was stained with Coomassie blue, and protein bands were quantified by densitometry with Quantity One software (Bio-Rad). Dissociation constants (Kd) were calculated by nonlinear least-squares fitting of the binding curves with the simple rectangular hyperbolic binding equation.
Results
A novel missense germline mutation in NEMO with no impact on protein expression
We studied a boy with recurrent infections, including mycobacterial diseases in particular (Figure 1A). This patient has conical incisors and agenesis of the lateral incisors and the upper right first premolar, which suggests a mild form of EDA-ID (Figure 1B). Molecular genetic analysis revealed a hemizygous nucleotide substitution at position 932 (A > G; Figure 1C) in exon 8 of NEMO, which led to the substitution of asparagine by glycine at residue 311 (designated D311G). This mutation was detected in cDNA from EBV-B cells and SV40-transformed fibroblasts from the patient (P), which indicates that it was a mutation in NEMO itself (and not its pseudogene). This mutation was not found in any of 1300 chromosomes from 52 ethnic groups in the HGDP-CEPH database (a collaboration of the Human Genome Diversity Project and the Centre d'Etude du Polymorphisme Humain), and it was therefore not an irrelevant polymorphism. The mutation in P was inherited from his mother (Figure 1A). We previously reported another missense mutation that affected the same residue, D311N, and conferred a typical clinical phenotype of EDA associated with multiple and recurrent infectious diseases (XR-EDA-ID12 ; supplemental data, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). However, the impact of the D311N mutant protein on the NF-κB pathway was not investigated by functional characterization in human cells. The D311 residue of NEMO is conserved in the 33 species in which the NEMO gene has been sequenced to date. This residue is located in the CC2-LZ domain (supplemental Figure 1). We investigated the impact of these mutations on protein levels by performing Western blotting on cell lines derived from the patient's cells and from the cells of different published patients with NEMO deficiency. We found that the D311G-NEMO mutant protein was produced in the patient's monocytes and granulocytes in similar amounts to the WT NEMO protein in a healthy control subject (Figure 1D). We also analyzed NEMO production in cell lines from patients carrying the D311G or D311N allele. The amounts of NEMO protein generated from the D311G and D311N mutant alleles were similar to the amounts of protein generated from the WT allele in EBV-B cells (Figure 1D; supplemental Figure 2). NEMO protein production was also normal in SV40-transformed fibroblasts carrying the D311G allele (Figure 1D; supplemental Figure 2). SV40-transformed fibroblasts carrying the D311N allele were not available. Western blotting showed the D311G protein to be produced in normal amounts in fresh cells from the patient and cell lines, including hematopoietic and nonhematopoietic cells. We evaluated production of the D311G and D311N mutant NEMO proteins by transiently transfecting SV40-transformed fibroblasts from an XD-IP NEMO patient with WT, D311G, and D311N vectors. Similar amounts of protein were detected for the D311G and D311N mutant proteins as for the WT NEMO protein (Figure 1E); thus, these 2 mutations do not affect the amount of protein produced.
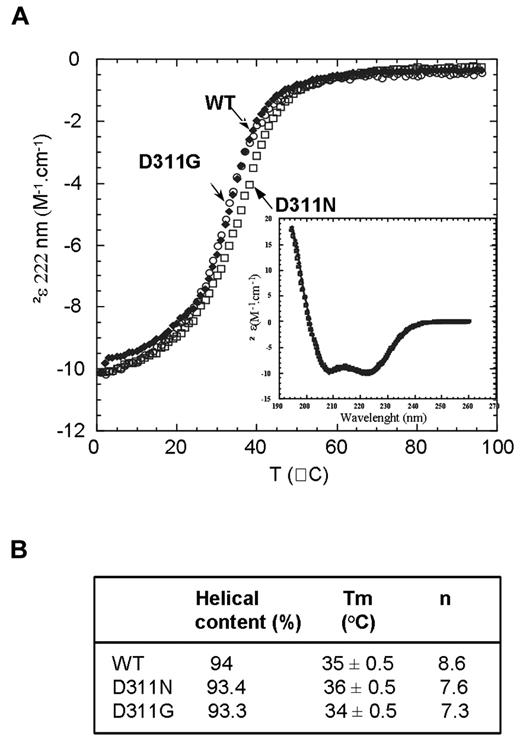
Folding and dimeric stability of the WT and mutant CC2-LZ domains bearing D311N and D311G mutations
We recently showed that most disease-associated mutations that affect the CC2-LZ domain impair polyubiquitin binding indirectly by disturbing the folding and stability of the CC2-LZ dimer.33 Despite the demonstration of a lack of polyubiquitin binding in 4 studies of the D311N mutant protein, no rigorous study has yet been performed to determine whether the D311N mutation also affects the folding of the CC2-LZ ubiquitin-binding domain of NEMO.32,34-36 In addition to investigating the effects of the new D311G mutation, we also studied the aspartate-to-asparagine substitution at position 311 within the NOA/UBAN ubiquitin-binding site of the CC2-LZ domain by generating and purifying fragments of NEMO (CC2-LZ258-344 domain) that bore this substitution (supplemental Figure 3). Sedimentation equilibrium, sedimentation velocity (SV), and dynamic light scattering were used for the complete characterization of CC2-LZ258-344 WT and mutant oligomers. Neither D311N nor D311G affected the oligomeric status of the CC2-LZ domain, because both mutants formed dimers with dimerization constants similar to those of the WT (Table 1). Furthermore, SV data confirmed that D311N and D311G behaved as homogeneous elongated dimers with a sedimentation coefficient of 1.4 ± 0.1 S (S20,w = 1.8 S) and a high frictional coefficient of 2.4 ± 0.2. No significant aggregates or higher-order oligomers were observed, as shown by SV (supplemental Figure 4) and dynamic light scattering (data not shown). We then performed temperature-induced denaturation of the dimer followed by circular dichroism (CD) spectroscopy to determine the thermal stability of the mutants (Figure 2A-B). Consistent with previous data, thermal denaturation showed a complete 2-state transition, which reflects the dissociation of native dimers into unfolded monomers.33 The melting curves for D311N and D311G gave melting temperatures (Tm) of 36 ± 0.5°C and 34 ± 0.5°C, respectively, similar to that for the WT (Tm = 35 ± 0.5°C). Cooperativities (n) were slightly lower for D311N (7.6) and D311G (7.3) than for the WT (8.6). However, this decrease may be because of a slight difference in the enthalpy (ΔHTm) and heat capacity values (ΔCp) of dimer denaturation, although these differences were not statistically significant. The far-UV spectra of the 2 mutants were similar to that of the WT (Figure 2A inset), and the deconvolution of CD spectra gave similar high α-helix contents, which indicates an absence of effect of the D311N and D311G substitutions on the dimeric coiled-coil structure. Thus, the D311N and D311G mutations do not affect the folding and dimeric stability of the CC2-LZ domain of NEMO.
Sedimentation equilibrium data for the His-tagged CC2-LZ WT, D311N, and D311G mutants
| . | Calculated dimer masses, Da . | Mean molecular masses, Da . | Monomer-dimer equilibrium, Kd, μM . |
|---|---|---|---|
| WT | 24 880 | 22 420 | 32 ± 3 |
| D311N | 24 878 | 24 960 | 43 ± 4 |
| D311G | 24 764 | 23 990 | 37 ± 3 |
| . | Calculated dimer masses, Da . | Mean molecular masses, Da . | Monomer-dimer equilibrium, Kd, μM . |
|---|---|---|---|
| WT | 24 880 | 22 420 | 32 ± 3 |
| D311N | 24 878 | 24 960 | 43 ± 4 |
| D311G | 24 764 | 23 990 | 37 ± 3 |
Mean molecular masses and monomer-dimer equilibrium constants (Kd) were determined at 10°C with an initial protein concentration of 15μM, as described in Methods. The theoretical molecular masses of the dimers are indicated for each construct.
Folding and dimeric stability of the WT and mutant CC2-LZ domains bearing D311N and D311G mutations. (A) Thermal denaturation curves of WT (●), D311N (□), and D311G (○), followed by CD at 222 nm, at a concentration of 8μM (0.1 mg/mL). (Inset) Far-UV CD spectra of WT, D311N, and D311G recorded at 1°C and at 8μM. (B) Secondary structure contents deduced from the deconvolution of the spectra at 1°C, melting temperatures (Tm), and cooperativities (n) calculated from the thermal denaturation profiles with a 2-state model.
Folding and dimeric stability of the WT and mutant CC2-LZ domains bearing D311N and D311G mutations. (A) Thermal denaturation curves of WT (●), D311N (□), and D311G (○), followed by CD at 222 nm, at a concentration of 8μM (0.1 mg/mL). (Inset) Far-UV CD spectra of WT, D311N, and D311G recorded at 1°C and at 8μM. (B) Secondary structure contents deduced from the deconvolution of the spectra at 1°C, melting temperatures (Tm), and cooperativities (n) calculated from the thermal denaturation profiles with a 2-state model.
D311N and D311G substitutions disrupt monoubiquitin, K63-linked, and linear polyubiquitin binding
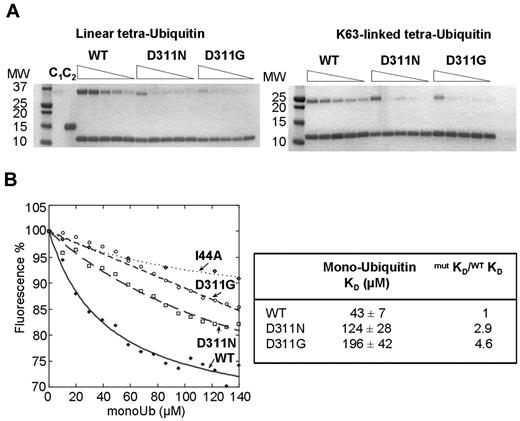
We next investigated the effects of mutation on polyubiquitin binding. We performed pull-down experiments to assess the ability of the WT and mutant His-tagged CC2-LZs to bind K63-linked and linear tetraubiquitin chains. Unlike previous studies,32 we compared the interactions of oligomers with 4 ubiquitins, which have higher affinity than diubiquitin chains.5,33 Note also that pull-down experiments were performed in a dose-dependent manner with His-tag versions of CC2-LZ but not with GST-tagged proteins,5,34-36 because it has recently been shown that the dimeric GST moiety can artificially influence polyubiquitin binding properties.37 The D311N substitution decreased the affinities of the protein for K63-linked and linear tetraubiquitin chains by factors of 14 and 15.6, respectively (Figure 3; Table 2). A similar decrease was observed for the novel D311G mutant protein, because its affinities for K63-linked and linear tetraubiquitin chains were lower than those of the WT by factors of 28 and 32, respectively. The affinity of CC2-LZ for monoubiquitin is too low for easy detection of the complex in a pull-down assay. We therefore used a more sensitive fluorescence assay based on the F312W mutant.33 This mutant has a single tryptophan residue in the ubiquitin-binding site that can be used as a fluorescent probe for monitoring monoubiquitin binding under equilibrium conditions. The F312W mutant has a dimeric stability similar to that of WT, as shown by SV, sedimentation equilibrium, and thermal denaturation followed by CD (not shown), and the corresponding full-length NEMO mutant fully rescues the TNF-α–induced NF-κB activation observed in NEMO-deficient T lymphocytes.7 We purified F312W/D311N and F312W/D311G mutant proteins for the quantification of monoubiquitin binding by fluorescence (supplemental Figure 3). Moreover, the binding constants of the F312W/D311N and F312W/D311G mutant proteins were lower than that of F312W by factors of 2.9 and 4.6, respectively, which indicates that both mutations also alter monoubiquitin binding (Figure 3B).
K63-linked, linear polyubiquitin, and monoubiquitin binding of the D311N and D311G CC2-LZ domains. (A) Pull-down activity of His-tagged CC2-LZ WT, D311N, and D311G mutants for linear (left) and K63-linked (right) tetraubiquitin chains. His-tagged CC2-LZ WT, D311N, and D311G mutants were incubated for 1 hour at a concentration of 8μM with various concentrations of linear (7μM, 2μM, 1μM, 0.5μM, and 0.25μM) or K63-linked tetraubiquitin chains (3.5μM, 1.7μM, 0.9μM, 0.7μM, and 0.4μM). After 3 washes, the amount of bound tetraubiquitin chains was determined by Coomassie staining followed by densitometry. C1 and C2 indicate controls with beads alone and immobilized with a His-tagged protein control, respectively (both represent nonspecific ubiquitin binding); MW, molecular weight (in kilodaltons). (B) Fluorescence titration of F312W, F312W/D311N, and F312W/D311N mutants (10μM) in the presence of monoubiquitin (mono-Ub) or I44A monoubiquitin. Lines represent the fit to each binding curve, giving the affinity (Kd) or the relative affinity with respect to the WT (mutKD/WTKD) for mono-Ub (right panel).
K63-linked, linear polyubiquitin, and monoubiquitin binding of the D311N and D311G CC2-LZ domains. (A) Pull-down activity of His-tagged CC2-LZ WT, D311N, and D311G mutants for linear (left) and K63-linked (right) tetraubiquitin chains. His-tagged CC2-LZ WT, D311N, and D311G mutants were incubated for 1 hour at a concentration of 8μM with various concentrations of linear (7μM, 2μM, 1μM, 0.5μM, and 0.25μM) or K63-linked tetraubiquitin chains (3.5μM, 1.7μM, 0.9μM, 0.7μM, and 0.4μM). After 3 washes, the amount of bound tetraubiquitin chains was determined by Coomassie staining followed by densitometry. C1 and C2 indicate controls with beads alone and immobilized with a His-tagged protein control, respectively (both represent nonspecific ubiquitin binding); MW, molecular weight (in kilodaltons). (B) Fluorescence titration of F312W, F312W/D311N, and F312W/D311N mutants (10μM) in the presence of monoubiquitin (mono-Ub) or I44A monoubiquitin. Lines represent the fit to each binding curve, giving the affinity (Kd) or the relative affinity with respect to the WT (mutKD/WTKD) for mono-Ub (right panel).
Affinity of His-tagged CC2-LZ WT and of the D311N and D311G mutants for linear and K63-linked tetraubiquitin chains
| . | Tetraubiquitin chains (Kd in μM) . | mutKd/WTKd . | ||
|---|---|---|---|---|
| K63 . | Linear . | K63 . | Linear . | |
| WT | 4 ± 1 | 0.9 ± 0.3 | 1 | 1 |
| D311N | 56 ± 16 | 14 ± 4 | 14 | 15.6 |
| D311G | 112 ± 30 | 29 ± 9 | 28 | 32 |
| . | Tetraubiquitin chains (Kd in μM) . | mutKd/WTKd . | ||
|---|---|---|---|---|
| K63 . | Linear . | K63 . | Linear . | |
| WT | 4 ± 1 | 0.9 ± 0.3 | 1 | 1 |
| D311N | 56 ± 16 | 14 ± 4 | 14 | 15.6 |
| D311G | 112 ± 30 | 29 ± 9 | 28 | 32 |
mutKd/WTKd indicates the difference in affinity with respect to the WT for linear and K63-linked tetraubiquitin chains.
Binding constants were calculated by densitometry, as described in “Methods”, in the pull-down experiments shown in Figure 2.
Impact of the NEMO D311G allele on the cellular response to TNF-α and IL-1β
Many human mutations in the NEMO gene that confer different clinical and cellular phenotypes in humans have been reported. Neither of these 2 mutations has been characterized in cells from patients. We assessed the impact of the NEMO D311G mutation on the NF-κB pathway by studying IκBα degradation by Western blot. As previously shown,30 IκBα degradation was impaired in fibroblasts from a patient with XR-EDA-ID-OL, whereas it was normal in fibroblasts from a patient with an X-linked recessive form of mendelian susceptibility to mycobacterial disease (XR-MSMD). The degradation of IκBα in response to TNF-α stimulation was impaired in fibroblasts from the patient with the D311G NEMO mutation, as shown by comparison with cells from a healthy control subject (Figure 4A). No such clear phenotype was observed in response to IL-1β activation (Figure 4A; supplemental Figure 5A). We also assessed nuclear NF-κB p50/p65 complex DNA-binding activity by EMSA. SV40-transformed fibroblasts from the NEMO D311G patient displayed weaker NF-κB–binding activity than cells from a healthy control subject stimulated with TNF-α or IL-1β (Figure 4B; supplemental Figure 5B-C). These data suggest that the NEMO D311G allele is intrinsically deleterious. NEMO mutations in patients with EDA-ID are usually associated with impairment of the canonical activation of NF-κB.15-17 We therefore evaluated cellular responses to TNF-α in terms of IL-10 induction in whole blood. Low but detectable levels of IL-10 were found in whole blood from the patient with the D311G allele, as for most previously reported patients bearing hypomorphic NEMO mutations (Figure 4C). We then stimulated SV40-transformed fibroblasts from a healthy control subject, patients with XD-IP, XR-EDA-ID-OL, and XR-MSMD, and the patient with the D311G allele (P) with TNF-α and IL-1β. We observed an impairment of IL-6 and IL-8 production by cells from P after stimulation with TNF-α (Figure 4D; supplemental Figure 6A) and IL-1β (Figure 4D; supplemental Figure 6B), as observed with fibroblasts from the XR-EDA-ID-OL patient but not with those from the XR-MSMD patient, which secreted IL-6 and IL-8 normally after activation with TNF-α and IL-1β.20
NF-κB activity in response to TNF-α and IL-1β. (A) IκBα degradation, detected by Western blotting and induced by TNF-α and IL-1β, for a healthy control subject (C+), an XD-IP patient, a hypomorphic NEMO EDA-ID-OL patient (X420W), the proband (P [D311G]), and an XR-MSMD NEMO patient (R319Q). Antibodies against GAPDH were used for normalization of protein levels on the Western blot. (B) NF-κB translocation was observed by EMSA with a labeled probe after exposure to TNF-α and IL-1β for various periods of time (0, 20, 40 minutes) for a healthy control subject (C+), an XD-IP patient, a hypomorphic NEMO EDA-ID-OL patient (X420W), the proband (P [D311G]), and an NEMO XR-MSMD patient (R319Q). (C) IL-10 production after TNF-α activation in whole blood from healthy control subjects, the proband (P [D311G]), and an NEMO XR-MSMD patient (R319Q). (D) IL-6 and IL-8 production, determined by ELISA after 24 hours of activation with TNF-α and IL-1β, for SV40-transformed fibroblast cell lines from a healthy control subject (C+), an XD-IP patient, an NEMO EDA-ID-OL patient (X420W), the proband (P [D311G]), and an NEMO XR-MSMD patient (R319Q). These results are representative of 2 independent experiments.
NF-κB activity in response to TNF-α and IL-1β. (A) IκBα degradation, detected by Western blotting and induced by TNF-α and IL-1β, for a healthy control subject (C+), an XD-IP patient, a hypomorphic NEMO EDA-ID-OL patient (X420W), the proband (P [D311G]), and an XR-MSMD NEMO patient (R319Q). Antibodies against GAPDH were used for normalization of protein levels on the Western blot. (B) NF-κB translocation was observed by EMSA with a labeled probe after exposure to TNF-α and IL-1β for various periods of time (0, 20, 40 minutes) for a healthy control subject (C+), an XD-IP patient, a hypomorphic NEMO EDA-ID-OL patient (X420W), the proband (P [D311G]), and an NEMO XR-MSMD patient (R319Q). (C) IL-10 production after TNF-α activation in whole blood from healthy control subjects, the proband (P [D311G]), and an NEMO XR-MSMD patient (R319Q). (D) IL-6 and IL-8 production, determined by ELISA after 24 hours of activation with TNF-α and IL-1β, for SV40-transformed fibroblast cell lines from a healthy control subject (C+), an XD-IP patient, an NEMO EDA-ID-OL patient (X420W), the proband (P [D311G]), and an NEMO XR-MSMD patient (R319Q). These results are representative of 2 independent experiments.
Impact of the NEMO D311G allele on the IL-12/IFN-γ pathway
Most patients with NEMO mutations are susceptible to mycobacterial infections, and those carrying the R319Q and E315A mutations appear to be specifically susceptible to mycobacteria (XR-MSMD).12,16,20 The mechanism underlying this susceptibility involves the impairment of CD40-dependent IL-12 production.20 The patients carrying the D311G and D311N mutations had mycobacterial diseases. We thus evaluated the impact of the NEMO D311G mutation on the IL-12/IFN-γ pathway. Unfortunately, whole blood and fresh leukocytes could not be obtained from the patient with the D311N mutation, who died in 2000. We activated whole blood from the other patient (P) with the D311G mutation with live BCG alone or with IL-12 or IFN-γ. The BCG plus IL-12 response was normal in terms of IFN-γ production (Figure 5A). IL-12p40 production was also similar to that of the positive control after stimulation with BCG plus IFN-γ (Figure 5B). This immunologic result, obtained with fresh blood cells, is consistent with those obtained for XR-MSMD patients with NEMO mutations,20 which demonstrates the maintenance of the microbe-dependent, T cell– and CD40-independent pathway of IL-12 production. We also investigated the impact of the D311G and D311N mutations on IL-12 production by EBV-B cells stimulated with a phorbol ester (PDBu). EBV-B cells from healthy control subjects (n = 9), patients with IL-12B deficiency (n = 2), a patient with complete CD40 deficiency, patients with XR-EDA-ID (n = 5), patients with the D311N and D311G mutations (n = 2), and XR-MSMD NEMO patients (n = 2) were tested. EBV-B cells bearing the D311N and D311G mutations displayed impaired IL-12p40 production in response to PDBu stimulation (Figure 5C; supplemental Figure 7A). This defect is observed in some EDA-ID patients and has already been reported for the first XR-MSMD patients described.20 TNF-α production in response to PDBu was studied as a control and found to be normal in D311N and D311G cells (supplemental Figure 7B).
IL-12/IFN-γ pathway. (A) Determination by ELISA of IFN-γ secretion before (NS) and after stimulation with BCG or BCG plus IL-12 in whole blood from healthy control subjects, the patient studied [P (D311G)], and an NEMO XR-MSMD patient (R319Q). (B) IL-12p40 production before (NS) and after activation with BCG or BCG plus IFN-γ in whole blood from healthy control subjects, patients with a complete IL-12B defect (IL12B−/−), the patient studied (P [D311G]), and an NEMO XR-MSMD patient (R319Q). NS indicates not stimulated. (C) Determination, by ELISA, of the production of IL-12p40 by EBV-B cells after 24 hours of activation with 10-7M PDBu for healthy control subjects (C+; n = 9); patients with complete IL-12B deficiency (IL-12B−/−; n = 2); a patient with complete CD40 deficiency (CD40−/−); patients with an EDA phenotype with a hypomorphic mutation of NEMO susceptible to pyogenic infections, associated or not associated with mycobacterial infection (NEMO EDA-ID; n = 5); an NEMO EDA-ID patient with a different mutation affecting the same position in the protein as affected in the proband; the proband (P [D311G]); and NEMO patients susceptible to mycobacterial infections (NEMO E315A and R319Q; XR-MSMD; n = 2). These results are representative of 3 independent experiments.
IL-12/IFN-γ pathway. (A) Determination by ELISA of IFN-γ secretion before (NS) and after stimulation with BCG or BCG plus IL-12 in whole blood from healthy control subjects, the patient studied [P (D311G)], and an NEMO XR-MSMD patient (R319Q). (B) IL-12p40 production before (NS) and after activation with BCG or BCG plus IFN-γ in whole blood from healthy control subjects, patients with a complete IL-12B defect (IL12B−/−), the patient studied (P [D311G]), and an NEMO XR-MSMD patient (R319Q). NS indicates not stimulated. (C) Determination, by ELISA, of the production of IL-12p40 by EBV-B cells after 24 hours of activation with 10-7M PDBu for healthy control subjects (C+; n = 9); patients with complete IL-12B deficiency (IL-12B−/−; n = 2); a patient with complete CD40 deficiency (CD40−/−); patients with an EDA phenotype with a hypomorphic mutation of NEMO susceptible to pyogenic infections, associated or not associated with mycobacterial infection (NEMO EDA-ID; n = 5); an NEMO EDA-ID patient with a different mutation affecting the same position in the protein as affected in the proband; the proband (P [D311G]); and NEMO patients susceptible to mycobacterial infections (NEMO E315A and R319Q; XR-MSMD; n = 2). These results are representative of 3 independent experiments.
Impact of the NEMO D311G allele on the CD40-dependent NF-κB pathway
In previous studies with XR-MSMD patients, we reported an NF-κB defect that affected NF-κB/c-Rel binding activity in response to CD40 stimulation.20 We investigated the CD40-triggered NF-κB pathway by generating EBV-B cell lines from the frozen PBMCs of the patients with D311G and D311N mutations. We assessed IκBα degradation in EBV-B cells from a healthy control subject, a patient with a complete CD40 deficiency, an XR-EDA-ID-OL NEMO patient (X420W), an XR-MSMD NEMO patient (R319Q), and the D311N and D311G NEMO patients. The NEMO D311N and D311G cells displayed normal IκBα degradation, resembling that observed in the healthy control subject and the XR-MSMD patient, whereas the XR-EDA-ID-OL NEMO patient did not (Figure 6A). However, NF-κB complex DNA-binding activity involving p50, p65, c-Rel, and RelB subunit (Figure 6B; supplemental Figure 8), measured by EMSA, was much weaker for the NEMO D311G patient, an XR-EDA-ID-OL NEMO patient (X420W), an XR-MSMD NEMO patient (R319Q), and the D311N NEMO patient than for the healthy control subject (C+). No induction was observed in cells from a patient with complete CD40 deficiency (Figure 6B; supplemental Figure 8). XR-MSMD NEMO patients display impaired IL-12 production by MDDCs.20 We therefore assessed the response to CD40 pathway activation using MDDCs from 8 healthy control subjects, a patient with a complete CD40 deficiency, an XR-MSMD NEMO patient (R319Q), and patients with the D311N and D311G NEMO mutations. MDDCs generated from frozen PBMCs were activated by incubation for 24 hours with trimeric soluble CD40L. Impaired IL-12p40 production was observed in MDDCs from both D311N and D311G NEMO patients as in XR-MSMD NEMO patients and the CD40−/− patient (Figure 6C). IL-6 production in response to stimulation with lipopolysaccharide plus IL-1β was studied as a control and was normal in D311N and D311G cells (supplemental Figure 9). These findings are consistent with a defect of the CD40-NEMO-NF-κB signaling pathway and provide an explanation for the susceptibility to mycobacterial diseases observed in this patient. The mechanism by which some NEMO mutations impair the PDBu-dependent induction of IL-12 in EBV-B cells whereas others do not is unknown but appears to be correlated with NEMO-dependent impairment of the CD40-dependent induction of IL-12 in MDDCs.
Impact of the NEMO D311G substitution on the CD40-dependent NF-κB pathway. (A) IκBα degradation was observed by Western blot analysis on cytoplasmic protein extracts. Antibodies against GAPDH were used for normalization of protein levels on Western blots, and (B) NF-κB translocation was observed by EMSA on nuclear protein extracts from EBV-B cells activated with CD40L (100 ng/mL) for various times from a healthy control subject (C+), a patient with complete CD40 deficiency (CD40−/−), a hypomorphic NEMO EDA-ID-OL patient (X420W), an NEMO EDA-ID patient with another mutation affecting position 311 (D311N), the proband (P [D311G]), and an NEMO XR-MSMD patient (R319Q). These results are representative of 2 independent experiments. (C) The production of IL-12p40, as assessed by ELISA, in MDDCs activated or not (NS) for 24 hours with trimeric soluble CD40L (1 μg/mL) from healthy control subjects (C+; n = 8), a patient with complete CD40 deficiency (CD40−/−), an NEMO EDA-ID patient with another mutation in position 311 (D311N), the proband (P [D311G]), and an NEMO XR-MSMD patient (R319Q). NS indicates not stimulated.
Impact of the NEMO D311G substitution on the CD40-dependent NF-κB pathway. (A) IκBα degradation was observed by Western blot analysis on cytoplasmic protein extracts. Antibodies against GAPDH were used for normalization of protein levels on Western blots, and (B) NF-κB translocation was observed by EMSA on nuclear protein extracts from EBV-B cells activated with CD40L (100 ng/mL) for various times from a healthy control subject (C+), a patient with complete CD40 deficiency (CD40−/−), a hypomorphic NEMO EDA-ID-OL patient (X420W), an NEMO EDA-ID patient with another mutation affecting position 311 (D311N), the proband (P [D311G]), and an NEMO XR-MSMD patient (R319Q). These results are representative of 2 independent experiments. (C) The production of IL-12p40, as assessed by ELISA, in MDDCs activated or not (NS) for 24 hours with trimeric soluble CD40L (1 μg/mL) from healthy control subjects (C+; n = 8), a patient with complete CD40 deficiency (CD40−/−), an NEMO EDA-ID patient with another mutation in position 311 (D311N), the proband (P [D311G]), and an NEMO XR-MSMD patient (R319Q). NS indicates not stimulated.
Discussion
Previously reported NEMO mutations have been shown to impair the production or folding of the protein.3,12,16,20,28,30 We report here that the XR-EDA-ID–causing NEMO mutations D311G and D311N prevent the binding of NEMO to ubiquitin but have no effect on its production and folding. In the present study, we investigated mutant proteins in cells from patients and cell lines derived from patients' cells. By contrast to other studies based on transfected cells or animal models, the protein levels demonstrated here are representative of reality. This is the first study in which folding has been rigorously investigated for these 2 mutant proteins, leading to the conclusion that NEMO folding is perfectly conserved in both cases. The D311G and D311N mutations selectively affect the binding of the protein to ubiquitin. These mutations have no effect on the stability and dimerization of the CC2-LZ NEMO domain or the thermal stability of NEMO ZF, unlike the previously reported mutations.3,28 Residue D311 is located in the NOA ubiquitin-binding site of NEMO. We showed that the mutation of this residue resulted in a strong defect in binding to linear and K63-linked polyubiquitin chains. These findings are consistent with those of previous studies on cell lines for D311N.5,32,34,36 However, this is the first analysis of the D311G mutant protein. We provide here the first report of human patients with immunodeficiency and specific impairment of NEMO binding to polyubiquitin. These findings support the hypothesis that the binding of human NEMO to linear or K63-linked polyubiquitin chains is critical for NF-κB signaling. Furthermore, they demonstrate that the production of normal amounts of NEMO is not sufficient to rule out a diagnosis of NEMO deficiency in a patient with EDA-ID, a coding mutation in NEMO, or both.
Consistent with previous studies,5,7 we observed that the NEMOWT CC2-LZ had a higher affinity for the Lys63-linked tetraubiquitin chain than for K63-diubiquitin. Indeed, a Kd value of 4μM was found for K63-tetraubiquitin chains, whereas those reported for K63-diubiquitin chains were 131μM and 12μM in isothermal titration calorimetry and biolayer interferometry, respectively.32,38 This gain of affinity was not observed with linear chains, for which similar binding constants of approximately 1-2μM were obtained for dimeric, tetrameric, and nonameric polyubiquitin chains.5,7,32 Therefore, the linkage specificity between linear and K63 chains is markedly lower when longer oligomers of ubiquitin (> 2) are considered. One possible reason for this is the distinct rotational flexibility and spacing of ubiquitin units that result from a particular linkage. A K63-linked tetraubiquitin chain, unlike diubiquitin, may wrap around the dimeric NEMO NOA/UBAN and simultaneously bind, in a perpendicular fashion, the 2 distal ubiquitin-binding sites.5,32 By contrast, a linear tetraubiquitin chain may bind in a parallel fashion similar to that of a linear diubiquitin chain (Figure 7C), such that 2 adjacent ubiquitin moieties can bind both proximal and distal ubiquitin-binding sites on the CC2-LZ dimer.5
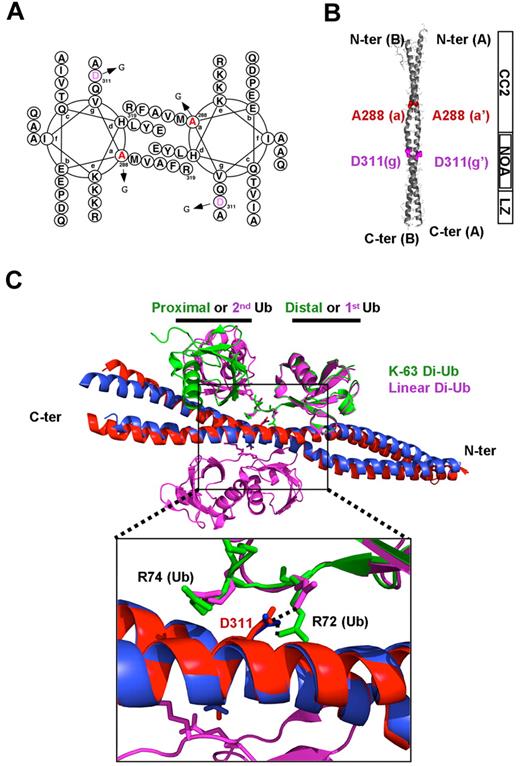
Crystal structures of the NEMO CC2-LZ domain. (A) Helical wheel representation of the NEMO CC2-LZ structure, starting from residue A288 at position a and ending at residue R319 at position a. The residues A288 and D311 are shown in red and purple, respectively. (B) Cartoon representation of the entire NEMO CC2-LZ structure showing domain organization (residues 258-344). Residues A288 and D311, which are located in the CC2 and NOA-LZ domains, respectively, are shown as ball and stick representations. CC2 indicates coiled coil 2; NOA, ubiquitin-binding site; and LZ, leucine zipper. (C) Structural comparison of the CC2-LZ domain in complex with a K63-linked diubiquitin chain (Di-Ub; green) or a linear diubiquitin chain (magenta). Ub indicates ubiquitin. The superimposition was achieved by Ca alignment of residues 300-322 (RMSD: 0.603 Å). NEMO helices in the complex with linear diubiquitin are shown in blue, whereas those in complex with K63-linked diubiquitin are shown in red. (Bottom) Zoom view of the NOA ubiquitin-binding site showing the interaction between the D311 residue and the R72 residue located in the distal ubiquitin C-terminal tail. In both complexes, the D311 side chain of NEMO forms a specific intermolecular salt bridge with the R72 side chain of the K63-linked (green) or linear (magenta) diubiquitin linker.
Crystal structures of the NEMO CC2-LZ domain. (A) Helical wheel representation of the NEMO CC2-LZ structure, starting from residue A288 at position a and ending at residue R319 at position a. The residues A288 and D311 are shown in red and purple, respectively. (B) Cartoon representation of the entire NEMO CC2-LZ structure showing domain organization (residues 258-344). Residues A288 and D311, which are located in the CC2 and NOA-LZ domains, respectively, are shown as ball and stick representations. CC2 indicates coiled coil 2; NOA, ubiquitin-binding site; and LZ, leucine zipper. (C) Structural comparison of the CC2-LZ domain in complex with a K63-linked diubiquitin chain (Di-Ub; green) or a linear diubiquitin chain (magenta). Ub indicates ubiquitin. The superimposition was achieved by Ca alignment of residues 300-322 (RMSD: 0.603 Å). NEMO helices in the complex with linear diubiquitin are shown in blue, whereas those in complex with K63-linked diubiquitin are shown in red. (Bottom) Zoom view of the NOA ubiquitin-binding site showing the interaction between the D311 residue and the R72 residue located in the distal ubiquitin C-terminal tail. In both complexes, the D311 side chain of NEMO forms a specific intermolecular salt bridge with the R72 side chain of the K63-linked (green) or linear (magenta) diubiquitin linker.
No asparagine substitution other than D311N has been reported in NEMO. Interestingly, other glycine substitutions in NEMO have different molecular impacts, despite the similarity of their clinical impact, which results in EDA-ID disease. Indeed, the EDA-ID–related A288G mutation, which affects the dimerization core of the CC2 region of NEMO (outside the NOA ubiquitin-binding site; Figure 7A-B), alters NEMO function by disrupting NEMO dimerization and probably higher-order oligomerization as well. By contrast, the D311G substitution on one side of the dimerization core of the NOA ubiquitin-binding site (Figure 7C) severely impairs polyubiquitin binding without disturbing the dimeric coiled-coil structure. Nevertheless, because dimerization of the NEMO CC2-LZ domain is absolutely required for efficient binding to K63-linked and linear chains,33 both glycine substitutions, at positions 288 and 311, may lead to defective polyubiquitin binding, which accounts for the similarity of the EDA-ID diseases they cause.
It is also interesting to speculate how mutations that affect the NEMO CC2-LZ domain can lead to clinical diseases as radically different as a narrow predisposition to mycobacterial diseases (XR-MSMD; mutations E315A and R319Q)20 and immunodeficiency with mild or classic signs of EDA (mutations A288G and 811_28del).15,28 Recent pull-down assay studies have reported that the R319Q and E315A mutations associated with MSMD result in a milder defect in ubiquitin binding than the D311N mutation associated with EDA-ID.32 The D311G mutation is as deleterious as D311N in human cells (fibroblasts and EBV-B cells) and has a more damaging effect on polyubiquitin binding than the mutations associated with pure MSMD. These differences may account for the normal NF-κB activation observed after activation with TNF-α or IL-1β in the fibroblasts of MSMD patients20 and the significantly lower levels of NF-κB activation in fibroblasts derived from the patient with the D311G mutation associated with an EDA-like immunodeficiency-associated disease. Interesting, the impact of the D311G and D311N mutations differs in different cells (fibroblasts, EBV-B cells, dendritic cells), the subunits associated in NEMO (different IκB and NF-κB subunits), and the level of activation of the pathway (degradation of different IκBs, DNA binding activity, production of cytokines), which suggests that there is a specific requirement for NEMO. Thus, detailed NF-κB–dependent immunologic pathways should be investigated in these patients. In any event, impaired ubiquitin binding is clearly the molecular mechanism of disease in patients with the D311G and D311N mutations of NEMO, and this mechanism may contribute to disease in patients with other mutations with and without effects on protein stability.18,19 The binding of human NEMO to ubiquitin is therefore essential for NF-κB activation in vivo. Furthermore, our findings demonstrate that normal levels of NEMO production and normal folding of this molecule should not be considered sufficient to exclude a diagnosis of NEMO deficiency in patients with XR-EDA-ID and/or NEMO mutations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors warmly thank family members for kindly agreeing to participate in this study. They thank all members of the 2 branches of the laboratory of Human Genetics of Infectious Diseases for discussions and T. Leclerc, Y. Rose, M. de Suremain, and L. Jannière for technical assistance. They also thank M. Kaminska and E. Fontan for assistance with molecular cloning and purification and P. England, B. Raynal, and B. Baron for advice on CD spectroscopy and sedimentation experiments.
This work was supported by BNP-Paribas Foundation, Schlumberger Foundation, The Gerber Fondation, Institut Universitaire de France, the Agence Nationale de la Recherche, the EURO-PADnet HEALTH-F2-2008-201549 (A. Plebani), and The Rockefeller University Center for Clinical and Translational Science grant no. 5UL1RR024143-03, The Rockefeller University.
Authorship
Contribution: M.H. and F.N. performed experiments and contributed to the writing of the manuscript; A. Puel, L.I., J.F., and M.C. performed experiments and analyzed data for the controls; A. Puel, L.I., and C.P. provided important experimental advice; K.B., C.B., I.F., A.F., and A. Plebani contributed to the clinical diagnosis of patients; S.B.-D., C.P., A.I., L.A., and M.V. contributed to the writing and editing of the manuscript; C.P. performed the lymphocyte proliferation assays and phenotyping; and J.-L.C., F.A., and J.B. designed, planned, and supervised the entire study, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Fabrice Agou, PhD, Unit of Structural and Cellular Biochemistry, Institut Pasteur, CNRS, Unité de Recherche Associée 2185, 75015 Paris, France; e-mail: fabrice.agou@pasteur.fr; and Jacinta Bustamante, MD, PhD, Laboratory of Human Genetics of Infectious Diseases, Inserm, U980, Necker Branch, 75015 Paris, France; e-mail: jacinta.bustamante@inserm.fr.
References
Author notes
M.H. and F.N. contributed equally to this study.
J-L.C., F.A., and J.B. contributed equally to this study.

![Figure 1. Pedigree, clinical features, and protein expression for the patient. (A) Pedigree of patient (P) with a NEMO mutation in position 311 (P, D311G). The different generations are indicated by roman numerals (I and II), and each individual is indicated by an Arabic numeral. Male subjects are represented by squares and female subjects by circles. Solid (black) symbols indicate affected individuals; open (white) symbols, healthy individuals; ⊡, female carriers. Individuals whose genetic status could not be determined are indicated by the symbol “E?” (B) Photograph and radiograph of P taken at 15 years, showing the agenesis of 3 adult teeth (14, 12, and 22). (C) Automated sequencing profile showing the NEMO D311G mutation in cDNA extracted from EBV-B cells from the proband (P) and comparison with the sequence obtained from a control subject (C+). The A→G mutation led to the replacement at residue 311 of aspartic acid (D) by glycine (G). NEMO protein was detected by Western blotting with the FL-419 antibody (D) in monocytes and granulocytes from a healthy control subject (C+) and the proband [P (D311G)]; in EBV-B cells from a healthy control subject (C+), a 110_111insC NEMO patient already reported to produce smaller than normal amounts of NEMO protein, an NEMO EDA-ID patient with another mutation in position 311 (D311N) and the proband [P (D311G)]; and in SV40-transformed fibroblasts from a healthy control subject (C+), an XD-IP patient (IP), and the proband [P (D311G)]. (E) In SV40-transformed fibroblasts from the XD-IP NEMO patient transfected or not transfected (IP) with expression vectors carrying NEMO alleles (wild-type [IP+NEMO+WT], D311G [IP+NEMO+D311G], D311N [IP+NEMO+D311N]) or with insert-free vector (IP+Empty), cells were cotransfected with cyan fluorescent (CFP) vector, as a control for transfection efficiency control. GAPDH antibody was used to normalize protein levels in panels D and E. These results are representative of 2 independent experiments. WB indicates Western blot.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/4/10.1182_blood-2010-10-315234/4/m_zh89991174700001.jpeg?Expires=1767701276&Signature=Jz~zr3E1ifz3nJTbLWPpUm9VAdFgQP9tmMQ4-PhdD9AU-TChL7DUNLWA8yDYydSjGXjXXuzJm57z7LqlmD3~3ZEIMMcyxmuZnmyJdlY9BP~rI5JcwFS-7FEsGtwLu9yyueXLvuobcICZsA4N4vdzRYTgA0jWWw~G0ZAvm2Efbh1rLt3Th1-ZNmyz44fVgNwI-CYeitRQN8PXRc3ER65Agn7Lm-b4ox9MPq~VyLQ3dOZHrJXuJGpdATqOtfZvNdPom3fxNME8xyJq8FGTeZcEGxsQYeBZR4HzWFj3e52JMSPZt0RqClam7I6jE6QCeoTIDql5itUubB2-hBbn99M7mQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. NF-κB activity in response to TNF-α and IL-1β. (A) IκBα degradation, detected by Western blotting and induced by TNF-α and IL-1β, for a healthy control subject (C+), an XD-IP patient, a hypomorphic NEMO EDA-ID-OL patient (X420W), the proband (P [D311G]), and an XR-MSMD NEMO patient (R319Q). Antibodies against GAPDH were used for normalization of protein levels on the Western blot. (B) NF-κB translocation was observed by EMSA with a labeled probe after exposure to TNF-α and IL-1β for various periods of time (0, 20, 40 minutes) for a healthy control subject (C+), an XD-IP patient, a hypomorphic NEMO EDA-ID-OL patient (X420W), the proband (P [D311G]), and an NEMO XR-MSMD patient (R319Q). (C) IL-10 production after TNF-α activation in whole blood from healthy control subjects, the proband (P [D311G]), and an NEMO XR-MSMD patient (R319Q). (D) IL-6 and IL-8 production, determined by ELISA after 24 hours of activation with TNF-α and IL-1β, for SV40-transformed fibroblast cell lines from a healthy control subject (C+), an XD-IP patient, an NEMO EDA-ID-OL patient (X420W), the proband (P [D311G]), and an NEMO XR-MSMD patient (R319Q). These results are representative of 2 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/4/10.1182_blood-2010-10-315234/4/m_zh89991174700004.jpeg?Expires=1767701276&Signature=w5dWF6bJSKqT6T5mAl~OUbDZV0eTolkzwsy07Qtcbt9tvU4fU5IbhdHv097XPKpn6iKPIB6Kx0q2TuZKWy66uba1igH7mKQIGbCr1jV6UR7n1mjNMMqoaH3Z6VFrMLEad8s8vqrocqTsWAClrfbrL5OWC7gVS8sRsDbNTeOAN3WIamPspIWW7WyT7CVnDVeuz5F2uwUtLGbuDbQCmYeDRRB~tlA4ev9DHAwdmJFqXbf-YinAeLAi1ReHslMruYFy-HWcc95m-bXJRGDIrsGBjgXCFBwO-MIL3M1Yq5UfaZPlvDLa5CtWDMU~MrdL9JwdxpfRdogVzDX6vATuDDQlyg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. IL-12/IFN-γ pathway. (A) Determination by ELISA of IFN-γ secretion before (NS) and after stimulation with BCG or BCG plus IL-12 in whole blood from healthy control subjects, the patient studied [P (D311G)], and an NEMO XR-MSMD patient (R319Q). (B) IL-12p40 production before (NS) and after activation with BCG or BCG plus IFN-γ in whole blood from healthy control subjects, patients with a complete IL-12B defect (IL12B−/−), the patient studied (P [D311G]), and an NEMO XR-MSMD patient (R319Q). NS indicates not stimulated. (C) Determination, by ELISA, of the production of IL-12p40 by EBV-B cells after 24 hours of activation with 10-7M PDBu for healthy control subjects (C+; n = 9); patients with complete IL-12B deficiency (IL-12B−/−; n = 2); a patient with complete CD40 deficiency (CD40−/−); patients with an EDA phenotype with a hypomorphic mutation of NEMO susceptible to pyogenic infections, associated or not associated with mycobacterial infection (NEMO EDA-ID; n = 5); an NEMO EDA-ID patient with a different mutation affecting the same position in the protein as affected in the proband; the proband (P [D311G]); and NEMO patients susceptible to mycobacterial infections (NEMO E315A and R319Q; XR-MSMD; n = 2). These results are representative of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/4/10.1182_blood-2010-10-315234/4/m_zh89991174700005.jpeg?Expires=1767701276&Signature=ekCRvPzysX7YsibTI9mjACSDbxRckXRDE3JDT5E4nECAcor3qRlmD7rCfW-2UG1Z3lE5RGkfzGO2wo-tNrbBDPLextg2d9Rq2T3ZWWca0uq1fRTk0lcnfTmVPM9TuzKH4FF22Pm-U4tDO3-EBFtH35HRB88yMLv168OJkQ-JJId-A4C~pfgdQVLlBoPSha-G3pAw7-Pynw471ytNF4w24oxfZyBGCV0qS80qbOSX~0Y5hJBfzodjstysZJAjPiFDikB1asjfSfCNachArOiprxXsQUzcAB7JIVy4vQ-mItSt0aOH3bk4TmwHC-GR~xbEzJpRg2I0SHblOBJrGa3ouQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Impact of the NEMO D311G substitution on the CD40-dependent NF-κB pathway. (A) IκBα degradation was observed by Western blot analysis on cytoplasmic protein extracts. Antibodies against GAPDH were used for normalization of protein levels on Western blots, and (B) NF-κB translocation was observed by EMSA on nuclear protein extracts from EBV-B cells activated with CD40L (100 ng/mL) for various times from a healthy control subject (C+), a patient with complete CD40 deficiency (CD40−/−), a hypomorphic NEMO EDA-ID-OL patient (X420W), an NEMO EDA-ID patient with another mutation affecting position 311 (D311N), the proband (P [D311G]), and an NEMO XR-MSMD patient (R319Q). These results are representative of 2 independent experiments. (C) The production of IL-12p40, as assessed by ELISA, in MDDCs activated or not (NS) for 24 hours with trimeric soluble CD40L (1 μg/mL) from healthy control subjects (C+; n = 8), a patient with complete CD40 deficiency (CD40−/−), an NEMO EDA-ID patient with another mutation in position 311 (D311N), the proband (P [D311G]), and an NEMO XR-MSMD patient (R319Q). NS indicates not stimulated.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/4/10.1182_blood-2010-10-315234/4/m_zh89991174700006.jpeg?Expires=1767701276&Signature=NPTV~maNBae9dKYLLj4M7w9oDdhzMllvpvi6kbPKtfn0bvVQ88PRyjijNFegNToJsELt9lDz0uuIn31glqSDWAVE3EwerCj~3l9GEbioxCCijQ6pPSFatOzmqfuRWy09hbh0XZlFX~HcPmcLvbmC1~3tAVhLEBjyjoxilHLrsM3a21PT6ItxpHFtaBKw1yM6FgsKG9NDFQNWjnJR7shHvRkKqNmJh7XnymHvP7VdTmBQxyKlJkF5S46tBix5DV~W1xoePKY1o7c3-3T3LrUz5vP~HORBUxxE5NLY47t-3db-SsaBqaHmouE1cRaD~vkaxK4Un1nnbpiBrCU4ht5Jug__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)