Abstract
Diffuse large B-cell lymphoma (DLBCL) prognostication requires additional biologic markers. miRNAs may constitute markers for cancer diagnosis, outcome, or therapy response. In the present study, we analyzed the miRNA expression profile in a retrospective multicenter series of 258 DLBCL patients uniformly treated with chemoimmunotherapy. Findings were correlated with overall survival (OS) and progression-free survival (PFS). miRNA and gene-expression profiles were studied using microarrays in an initial set of 36 cases. A selection of miRNAs associated with either DLBCL molecular subtypes (GCB/ABC) or clinical outcome were studied by multiplex RT-PCR in a test group of 240 cases with available formalin-fixed, paraffin-embedded (FFPE) diagnostic samples. The samples were divided into a training set (123 patients) and used to derive miRNA-based and combined (with IPI score) Cox regression models in an independent validation series (117 patients). Our model based on miRNA expression predicts OS and PFS and improves upon the predictions based on clinical variables. Combined models with IPI score identified a high-risk group of patients with a 2-year OS and a PFS probability of < 50%. In summary, a precise miRNA signature is associated with poor clinical outcome in chemoimmunotherapy-treated DLBCL patients. This information improves upon IPI-based predictions and identifies a subgroup of candidate patients for alternative therapeutic regimens.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common type of non-Hodgkin lymphoma in adults, accounting for > 80% of aggressive lymphomas.1 DLBCL is a heterogeneous group of tumors with different genetic abnormalities, clinical features, responses to treatment, and prognosis.2 This heterogeneity hinders outcome prediction based on clinical and/or molecular parameters.
Combination therapy that associates CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone) with rituximab (R-CHOP) has become a standard treatment for DLBCL, leading to complete remission rates of 75%-80% and a 3- to 5-year PFS of 50%-60%.3-8 Nevertheless, patients who fail to respond to first-line therapy or relapse continue to pose a challenge, and identification at diagnosis of poor-outcome cases is crucial for deciding between alternative treatment schemes.
The International Prognostic Index (IPI) has been the primary clinical tool for predicting the outcome of patients with aggressive non-Hodgkin lymphoma.9 Original IPI factors were redistributed in patients treated with R-CHOP to give a revised score (R-IPI) that distinguishes 3 prognostic categories, with 4-year survival rates ranging from 94%-55% for poor-risk patients.7 Nevertheless, the R-IPI does not discriminate patients with < 50% probability of survival, which restricts its clinical value.7
The biologic heterogeneity of DLBCL has been shown substantially to reflect the cell origin of these tumors from germinal center or activated B cells. These differences are significant independently of IPI stratification, showing that identifying4,8 cell or origin signatures captures features other than IPI and can refine outcome prediction.10 These differences between GC and ABC DLBCL remain significant in patients treated with combined immunochemotherapy including rituximab.11 This classification system can be accurately reproduced using immunohistochemistry against GCET1, CD10, bcl6, MUM1, and FOXP1 in formalin-fixed, paraffin-embedded (FFPE) samples,12 thereby providing a simple tool for evaluating the protein-expression profile of the tumor at the time of diagnosis. However, although semiquantitative immunohistochemistry for subclassifying DLBCL is feasible and reproducible, the concordance rates of the different markers vary.13 Different methods of gene-expression profiling based on quantitative RT-PCR have been developed as alternative or complementary strategies for patient subclassification.14,15
Recently, miRNAs, small noncoding RNAs that fine-tune the expression of multiple genes,16,17 have been shown to be excellent biomarkers for cancer diagnosis and prognosis,18-21 including hematologic malignancies.22-27 Furthermore, recent evidence demonstrates that they may constitute markers of differentiation stage, malignant transformation, or sensitivity or resistance to specific drugs.22,28-31
The main goal of the present study was to search for an miRNA signature associated with clinical outcome in DLBCL patients treated with R-CHOP using FFPE tissue samples. We also investigated whether miRNA expression profiling can identify particular miRNA species that are differentially expressed between DLBCL subtypes according to the cell of origin (COO) classification.10,32
Methods
The experimental procedures are summarized in Figure 1.
Flowchart with experimental design. The whole series of patients was divided into 2 major groups: a discovery group (36 patients with available frozen tissue for profiling using array technologies) and a test group (240 patients with available FFPE tissue from the diagnostic pathologic sample). The test group was further divided systematically into 2 sets of patients: the training group (123 patients) and the validation group (117 patients). The clinical characteristics according to sex, age, stage, extranodal disease, serum lactate dehydrogenase levels, electrocorticogram, and IPI score of the different sets of patients are summarized in Table 1.
Flowchart with experimental design. The whole series of patients was divided into 2 major groups: a discovery group (36 patients with available frozen tissue for profiling using array technologies) and a test group (240 patients with available FFPE tissue from the diagnostic pathologic sample). The test group was further divided systematically into 2 sets of patients: the training group (123 patients) and the validation group (117 patients). The clinical characteristics according to sex, age, stage, extranodal disease, serum lactate dehydrogenase levels, electrocorticogram, and IPI score of the different sets of patients are summarized in Table 1.
Patients and treatments
The study population consisted of a retrospective series of 258 de novo cases of DLBCL obtained from various centers in Spain, 1 in Italy, and 3 in the United States. The study was reviewed and approved as being of minimal/no risk or as exempt by each of the participating institutional review boards, and the overall collaborative study was approved by the institutional review board at the Spanish National Cancer Research Center (CNIO) in Madrid, Spain. The study protocol and sampling methods were approved by the Instituto de Salud Carlos III institutional review board in de-identified anonymous format. Cases associated with HIV or HCV infections or previous immunosuppressive treatments were excluded. All histologic samples corresponded to initial diagnostic biopsies before treatment. Histologic criteria used for diagnosis and classification were those of the World Health Organization.1 All cases positively stained for CD20. Cases diagnosed as T-cell histiocyte-rich B-cell lymphoma, primary mediastinal B-cell lymphoma cases, cutaneous LBCL, intravascular LBCL, and those histologically associated with a follicular lymphoma component were excluded.
All patients were treated as part of their routine care with standard treatment protocols using a combination of anthracycline-based regimens (6-8 cycles in most cases) and immunotherapy including rituximab; the majority were treated with R-CHOP (n = 243). Other regimens included R-EPOCH and R-MegaCHOP. Responses to treatment were determined by a computed tomography scan in most cases (as recorded in the clinical recovery data sheet) and following the response criteria for lymphoma as defined by Cheson et al.33
Array-based expression analysis and in silico prediction of the miRNA regulatory network
miRNA and gene-expression hybridization were carried out using Agilent Technologies microarrays. RNA and DNA extraction methods, details of microarrays and hybridization procedures, and miRNA and gene-expression profiling array normalization are described in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
The differential miRNA expression profile between DLBCL subtypes according to the COO signature was studied after GEP-based classification of the cases32 (for details, see supplemental Methods).
miRNA expression data for all 36 DLBCL cases from the discovery set of patients were examined in a univariate (gene-by-gene) Cox model using SignS.34 miRNA and gene-expression data have been deposited in the Gene Expression Omnibus35 and are accessible through GEO series accession number GSE21849 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21849).
Targets were predicted using available databases (miRBase Version 11.0, MICROCOSM, and TargetScan release 5.1) and a Pearson correlation test based on gene-expression and miRNA-expression data from cases in the discovery set (details and additional references are provided in supplemental Methods).
Real-time PCR for relative miRNA quantification using RNA from FFPE tissue
miRNA expression in FFPE tissues was analyzed using the Applied Biosystems 384-well multiplexed real-time PCR assay with 250 ng of total RNA. Details of the methods, including the selection of endogenous miRNAs, are described in supplemental Methods.
Immunohistochemistry and tissue microarray construction
All cases of DLBCL with available FFPE tissue were histologically reviewed. Representative areas were selected to construct tissue microarrays. Immunohistochemical staining was performed after standard automated protocols using antibodies against CD10, bcl6, MUM-1/IRF4, GCET1, and FOXP1. Immunohistochemical markers were scored on the basis of the cutoffs used by Choi et al12 (see details in supplemental Methods).
Statistics
The statistical analyses are fully described in supplemental Methods. In brief, outcome-related indices (overall survival [OS] and PFS) were calculated as defined by Cheson et al.33 Survival distributions were estimated using the Kaplan-Meier method36 and compared with the log-rank test.37 The percentage of patients alive at the median follow-up time (and 95% confidence intervals) were noted. The χ2 test was used to assess differences in the proportions of individual prognostic factors between series.
Cox regression analysis38 was used to derive 3 independent survival models based on IPI score, miRNA expression (of 9 selected miRNAs), and GC-ABC classification based on immunohistochemistry12 for both OS and PFS in the training set. More complex models composed of combinations of the 3 individual predictor models were examined. We assessed the improved model fit using χ2 values to check for significant changes in log likelihood.
Only the combination of miRNAs and IPI score gave a significantly better fit than the individual models for both OS and PFS (P < .05 in all comparisons), which justified our fitting a large model with these variables. In this model, significant variables were determined by backward stepwise selection using AUC as the criterion. In this way, we derived definitive combined models for both OS and PFS. These were then validated in the test series using the (integrated) area under the ROC curve, the concordance index, and the Brier score (for details, see supplemental Methods).
Both miRNA-based and combined survival models (functions) based on IPI score and miRNAs after variable selection were applied to the entire test group of patients. For details, see survival functions hi(t)(OS) and hi(t)(PFS) in supplemental Methods.
Models were constructed and validated using the R statistical program, specifically with the packages survival (T. Therneau), pec (T. Gerds), and survcomp (B. Haibe-Kains, C. Sotiriou, and G. Bontempi). Additional analyses were conducted with SPSS Version 15.0.0.
Results
Clinical characteristics of the series
A summary of the clinical characteristics of the entire set of patients used in this study can be found in Table 1. Complete clinical and histopathologic data were available for all 258 patients. The median follow-up time for all patients was 21.3 months. The median follow-up time among patients alive at last follow-up was 27 months (range, 2-105 months). The estimated 2-year OS was 74.7% ± 3% and the estimated PFS was 67.5% ± 3% (supplemental Figure 1). Because the number of events at the median follow-up time comprised 75% (44 of 58) and 92% (66 of 77) of the total number of events during follow-up for OS and PFS, respectively, we considered that the series was suitable for further statistical analysis despite the limited follow-up.
Clinical features of the series
| . | Screening set . | Test set (n = 240 patients) . | ||||
|---|---|---|---|---|---|---|
| Training . | P . | Validation . | Relative risk of death, OS (95%CI) . | Relative risk of progression, PFS (95% CI) . | ||
| Number of patients | 36 | 123 | 117 | |||
| IPI factors | ||||||
| Age, y | .004 | 2.406 (1.298-4.462); P = .005 | 1.238 (0.758-2.021); P = .394 | |||
| ≤ 60 | 13 | 61 | 35 | |||
| > 60 | 23 | 62 | 79 | |||
| Stage | .5 | 2.221 (1.213-4.067); P = .01 | 3.287 (1.823-5.927); P = .000 | |||
| I-II | 8 | 55 | 43 | |||
| III-IV | 28 | 68 | 65 | |||
| Lactate dehydrogenase | .112 | 3.13 (1.53-6.43); P = .001 | 3.051 (1.66-5.608); P = .000 | |||
| low | 10 | 56 | 28 | |||
| high | 26 | 67 | 55 | |||
| Performance status | .87 | 2.265 (1.306-3.929); P = .004 | 1.956 (1.182-3.238); P = .009 | |||
| Ambulatory (0-1) | 29 | 90 | 56 | |||
| Not ambulatory (2-4) | 7 | 33 | 22 | |||
| Extranodal site involvement | .49 | 2.012 (1.135-3.565); P = .017 | 2.197 (1.315-3.670); P = .003 | |||
| ≤ 1 site | 28 | 96 | 57 | |||
| > 1 site | 8 | 27 | 21 | |||
| IPI score (number of IPI factors) | .067 | 1.690 (1.378-2.073); P = .000 | 1.479 (1.244-1.758); P = .000 | |||
| Low risk (0,1) | 6 | 46 | 33 | |||
| Low to intermediate risk (2) | 10 | 24 | 25 | |||
| Intermediate to high risk (3) | 12 | 29 | 26 | |||
| High risk (4,5) | 8 | 22 | 28 | |||
| . | Screening set . | Test set (n = 240 patients) . | ||||
|---|---|---|---|---|---|---|
| Training . | P . | Validation . | Relative risk of death, OS (95%CI) . | Relative risk of progression, PFS (95% CI) . | ||
| Number of patients | 36 | 123 | 117 | |||
| IPI factors | ||||||
| Age, y | .004 | 2.406 (1.298-4.462); P = .005 | 1.238 (0.758-2.021); P = .394 | |||
| ≤ 60 | 13 | 61 | 35 | |||
| > 60 | 23 | 62 | 79 | |||
| Stage | .5 | 2.221 (1.213-4.067); P = .01 | 3.287 (1.823-5.927); P = .000 | |||
| I-II | 8 | 55 | 43 | |||
| III-IV | 28 | 68 | 65 | |||
| Lactate dehydrogenase | .112 | 3.13 (1.53-6.43); P = .001 | 3.051 (1.66-5.608); P = .000 | |||
| low | 10 | 56 | 28 | |||
| high | 26 | 67 | 55 | |||
| Performance status | .87 | 2.265 (1.306-3.929); P = .004 | 1.956 (1.182-3.238); P = .009 | |||
| Ambulatory (0-1) | 29 | 90 | 56 | |||
| Not ambulatory (2-4) | 7 | 33 | 22 | |||
| Extranodal site involvement | .49 | 2.012 (1.135-3.565); P = .017 | 2.197 (1.315-3.670); P = .003 | |||
| ≤ 1 site | 28 | 96 | 57 | |||
| > 1 site | 8 | 27 | 21 | |||
| IPI score (number of IPI factors) | .067 | 1.690 (1.378-2.073); P = .000 | 1.479 (1.244-1.758); P = .000 | |||
| Low risk (0,1) | 6 | 46 | 33 | |||
| Low to intermediate risk (2) | 10 | 24 | 25 | |||
| Intermediate to high risk (3) | 12 | 29 | 26 | |||
| High risk (4,5) | 8 | 22 | 28 | |||
Distribution of IPI factors in the whole series is shown. The χ2 test was used to assess differences in the proportions of individual prognostic factors between the training and validation sets of patients. Relative risk of event (OS and PFS) estimated from univariate Cox regression is shown for each IPI factor in the test set of patients.
No significant differences were found between the IPI variables in the training and validation groups of patients in the test set (240 patients), with the exception of age. All clinical components of IPI were predictive for OS in the univariate analysis and all but age for PFS (relative risks and confidence intervals for each variable are shown in Table 1, and survival estimates according to IPI in supplemental Figure 1).
Confirmation of the prognostic capacity of COO classification based on immunohistochemistry
Immunohistochemistry was performed in all 240 cases with available FFPE tissue. Most cases (232 of 240) could be classified into GC or ABC subtypes according to previously published algorithms.12 There were 106 cases classified as the GC type (46%) and 126 as the ABC type (54%). The estimated 2-year OS for ABC-type DLBCL cases was 69.8% ± 4.5%, significantly worse than for GC-type DLCBL patients (81.4% ± 4.3%; P < .05). Differences were also found for PFS (60.7% ± 4.7% for the ABC type compared with 75.6% ± 4.6% for the GC type, P < .05; supplemental Figure 1).
Identification of a COO miRNA signature.
After gene expression–based classification using the gene set classifier of Wright et al,32 11 cases were classified as the GC type and 18 cases as the ABC type. Eight miRNAs were found to be differentially expressed between these subtypes (FDR < 0.03; see supplemental Table 2). These included miR-331, miR-151, miR-28, and miR-454-3p, which were up-regulated in the GC-type DLBCL, whereas miR-222, miR-144, miR-451, and miR-221 were up-regulated in the ABC-type DLBCL. We searched for the putative gene targets of the 8 miRNAs that were differentially expressed among the subtypes. Selected target pairs were miR151-5p and miR28-5p targeting FOXP1, miR144 targeting LRMP1, and miR451 targeting MME (CD10; see supplemental Table 2). Moreover, Gene Set Enrichment Analysis (GSEA Version 239 ) demonstrated that the GC–B-cell pathway was the main target set of genes that could be modulated by this set of miRNAs (see supplemental Table 2).
Identification of a set of candidate miRNAs associated with outcome in DLBCL
To search for miRNAs related to outcome but not associated with the previously described COO signatures, miRNA expression data for all 36 DLBCL cases from the discovery set of patients were subjected to a univariate (gene-by-gene) Cox analysis after FCMS using SignS.34 Fifty-seven human miRNAs were correlated positively or negatively with OS (P < .05; see supplemental materials). None of these 57 miRNAs formed part of the COO signature, suggesting that this method may add some complementary information to the previous approach.
After the previously described procedures, a final set of 9 miRNAs was subjected to relative RT-PCR quantification in the entire test group of 240 patients for whom FFPE tissue was available (see details in supplemental Methods). Seven of these miRNAs (miR-221, miR-222, miR-331, miR-451, miR-28, miR-151, and miR-148a) were identified using the COO-signature approach and 2 additional miRNAs (miR-93 and miR-491) were obtained from the univariate (gene-by-gene) Cox analysis using SignS.34
Generation of a miRNA-based predictor model using FFPE tissue samples
After Cox regression analysis,38 3 independent survival models based on IPI score, miRNA expression (of 9 selected miRNAs), and GC-ABC classification based on immunohistochemistry12 were derived for OS and PFS in the training set.
miRNA expression-based models using the expression of 9 miRNAs as a continuous variable were able to predict both OS and PFS. Their predictive performance in the validation group of patients was confirmed by 3 different statistical methods including the (integrated) area under the ROC curve, the concordance index, and the Brier score (for details, see supplemental Methods). When evaluating the Brier score, we used a conditional weighting scheme from a Cox model of the censoring distribution,40 including all miRNAs and the IPI score as predictors. The results for all tests are shown in supplemental Tables 2 and 3 and in supplemental Figure 2.
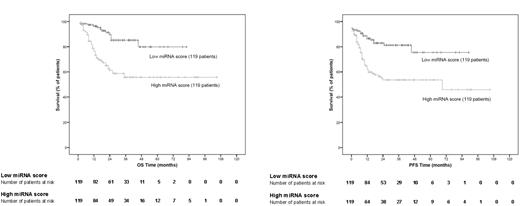
After confirming the predictive performance in the validation set of patients, we plotted Kaplan-Meier curves using the whole test group (training and validation sets) of patients. After median stratification of the continuous score obtained from the miRNA-based models, all patients from the test group were classified as having either a low miRNA score (below median) or a high miRNA score (above median). Significant differences in OS and PFS were found between the 2 groups of patients (log-rank test, P < .001 for both; Figure 2).
Kaplan-Meier representation of the miRNA-based survival model. miRNA-based survival scores were calculated for each patient in the test group according to the survival function obtained in the training set of patients. After median stratification, Kaplan-Meier estimates were plotted for OS and PFS (log-rank test, P < .001 for both OS and PFS).
Kaplan-Meier representation of the miRNA-based survival model. miRNA-based survival scores were calculated for each patient in the test group according to the survival function obtained in the training set of patients. After median stratification, Kaplan-Meier estimates were plotted for OS and PFS (log-rank test, P < .001 for both OS and PFS).
Only the combination of miRNAs and IPI score was significantly better than the individual models for both OS and PFS (χ2 test for the change in log likelihood, P < .05 for all comparisons), justifying our fitting a large model with these variables. The model was derived by backward stepwise selection using area under the curve as the criterion. This yielded combined models for both OS and PFS (details of survival functions can be found in the supplemental Methods). Because the construction of the combined models involves a multivariate analysis (variable selection step), the prediction based on the expression of the miRNAs present in the combined models was independent of the IPI score (the miRNAs included in these combined models together with IPI score and its associated hazard ratios are shown in Figure 3).
Hazard ratio charts. The interquartile range of the hazard ratios estimated from the models after variable selection is shown for each continuous variable. For each miRNA, we calculated the log of the hazard ratio, where the difference in that variable was that of the interquartile range (ie, the third to the first quartiles). For example, for miR-222, the first and the third quartiles are 1.515 and 3.945; the bar shows the hazard ratio. For PFS: exp(0.315 * [3.945-1.515] = 2.15) and a 75% and 95% interval. For IPISCORE, a discrete variable, we show the log hazard ratio comparing each of the values of IPISCORE (except 0) with IPISCORE = 0 as a reference.
Hazard ratio charts. The interquartile range of the hazard ratios estimated from the models after variable selection is shown for each continuous variable. For each miRNA, we calculated the log of the hazard ratio, where the difference in that variable was that of the interquartile range (ie, the third to the first quartiles). For example, for miR-222, the first and the third quartiles are 1.515 and 3.945; the bar shows the hazard ratio. For PFS: exp(0.315 * [3.945-1.515] = 2.15) and a 75% and 95% interval. For IPISCORE, a discrete variable, we show the log hazard ratio comparing each of the values of IPISCORE (except 0) with IPISCORE = 0 as a reference.
Kaplan-Meier estimates for OS and PFS in all test group series using the combined models were calculated and plotted according to the distribution of terciles (Figure 4). A high-risk subgroup of patients with OS and PFS below 50% after a 2-year follow-up was identified.
Kaplan-Meier representation of combined survival model based on IPI score and miRNAs. Survival scores according to IPI and miRNAs were calculated for each patient in the test group according to the combined survival function obtained in the training set of patients (see “Generation of a miRNA-based predictor model using FFPE tissue samples”). After tercile stratification, Kaplan-Meier estimates were plotted for OS and PFS (log-rank test, P < .001 for both OS and PFS). Hazard ratios estimated from the models are shown in Figure 3.
Kaplan-Meier representation of combined survival model based on IPI score and miRNAs. Survival scores according to IPI and miRNAs were calculated for each patient in the test group according to the combined survival function obtained in the training set of patients (see “Generation of a miRNA-based predictor model using FFPE tissue samples”). After tercile stratification, Kaplan-Meier estimates were plotted for OS and PFS (log-rank test, P < .001 for both OS and PFS). Hazard ratios estimated from the models are shown in Figure 3.
The miRNA-regulatory network
To gain a deeper insight into the network of genes that might be regulated by the set of outcome predictor miRNAs, we interrogated the current versions of the MiRBase and TargetScan miRNA target prediction databases. The predicted pairs we found included miR-222–CDKN1A (p21), miR-93–BCL66, miR-93–MCL1, and miR-93–MAP3K14 (NIK), among others.
We also examined the correlation between gene expression and miRNA expression in the data from the 29 samples of the discovery set. Because down-regulation of the target mRNA41,42 is considered the main mechanism by which miRNAs modulate protein expression, significant negative correlation pairs were identified, including miR-331–CARD10, miR-331–IRF4, miR-331–PIM2, and miR-331–AID. The complete correlation grid obtained is shown in supplemental Table 2. Interestingly, the absence of any significant negative correlation between many in silico–predicted pairs of miRNAs and mRNAs in this test suggests that additional mechanisms to mRNA down-regulation, such as translation inhibition, may explain the protein-expression modulation afforded by miRNAs.41,43,44
Discussion
miRNAs are emergent biomarkers of disease that have proved useful for cancer diagnosis and prognosis18-21 and in hematologic malignancies.22-27 They have been demonstrated to reflect accurately the differentiation stage of human lymphoid B cells,22,27,28 providing valuable information for tumor classification19,28 and prognostication26,27 that may be added to that available from gene-expression and clinical data.
In the present study, we analyzed miRNA expression profiles in a series of homogeneously treated DLBCL patients, deriving miRNA-based models that correlate with OS and PFS, but which are independent of IPI. We used a 2-step approach to identify a candidate set of miRNAs and then validated this with multiplex RT-PCR using RNA from FFPE tissue in 2 independent sets of patients. This method (real-time PCR) has been found to give reliable measures of gene and miRNA expression14,15 that are alternatives to the classic semiquantitative immunohistochemical approaches.13 Furthermore, the technique can be routinely applied to FFPE samples, because the small size of miRNAs makes them relatively resistant to RNAse degradation and because they can be successfully isolated from routinely processed FFPE tissue.26
After the first step of candidate identification, we found a signature of miRNAs related to the differentiation stage as defined by the COO signature.32 Specifically, we found miR-331, miR-151, miR-28, and miR-454-3p to be up-regulated in the GC-type DLBCL. Conversely, miR-222, miR-144, miR-451, and miR-221 were up-regulated in the ABC-type DLBCL. Our data from patient tissue samples classified according to their gene-expression profile (using the COO classifier) confirm those from other studies, which found miR-222 and miR221 to be up-regulated in the well-known ABC-DLCL cell lines OCI-Ly-10 and OCI-Ly-3.26,27,29,45 Furthermore, our in silico prediction indicates that genes essential as markers of the COO subtype (mainly GCB genes) are putative targets of certain differentially expressed miRNAs.
When we compared the classification of patients according to the miRNA model and the immunochemically based GC-ABC classification, the systems were shown to be nonoverlapping and complementary, thereby establishing different subsets of cases (P < .001, supplemental Table 4 sheet D). These results are particularly interesting because there is not complete agreement about the best method for stratifying patients with DLBCL. Different immunohistochemical algorithms are currently being tested with various results among series of patients, including the one presented here.46,47
Our results also identify a set of miRNAs in which expression is associated with poor clinical outcome in R-CHOP–treated DLBCL patients. miRNA expression–based models using the expression of miRNAs as a continuous variable were able to predict both OS and PFS in 2 independent sets of patients. These models predict OS and PFS and improve IPI-based predictions, which allowed us to generate combined models identifying a high-risk subgroup of patients with OS and PFS below 50% after a 2-year follow-up.
This particular signature, which contains some miRNAs previously shown to be correlated with the outcome of DLBCL patients27,29 and other hematologic malignancies,23 includes miRNAs that target pathways commonly deregulated in DLBCL. These include, for example, genes related to apoptosis (MCL1), the cell cycle (CDKN1A), MAPK and NFkB signaling (MAP3K14, MUM1/IRF4, CARD10, and PIM2), somatic mutation during the GC reaction (AID), and key transcription factors such as BCL6. Specifically, miR-221 and miR-222 have been found to be essential growth-regulatory mediators inhibiting p27 (Kip1), a cell-cycle inhibitor and tumor suppressor.48-51 Other components of the signature, such as the miR-106b-25 cluster (including miR-93), have recently been found to interfere with the expression of CDKN1A (p21) and BCL2L11, thereby impairing the TGFB tumor-suppressor pathway in other cancers.52 Functional experiments will be required to confirm the candidate interactions identified here. Transfection experiments (introducing the miRNA and measuring changes in the target mRNA/protein) and silencing experiments (using shRNAs to inhibit the constitutive expression of the miRNAs) might both be performed to address this matter, but this was beyond the scope of the present study.
It is remarkable that the miRNA signature identified here predicted survival independently of the COO classification. Although the GC-ABC subclassification has a potentially predictive role in the identification of patients likely to respond to specific therapies for ABC-type DLBCL,53 the development of new methods that can capture a set of DLBCL cases with particularly aggressive behavior paves the way for the design of trials using alternative therapeutic strategies (eg, untargeted therapies such as stem cell transplantation or more refined targeted therapies against the substrate genes/pathways involved).
The recent observation that c-MYC rearrangements in DLBCL are associated with poor prognosis in a subset (5%-15%) of rituximab-treated patients54,55 led us to consider the possible relationship between MYC status and the miRNA expression classifier. The biologic basis for such an association might reside in the role of MYC as a transcriptional regulator of the expression of some miRNAs.56-58 None of the miRNAs included in the prognostic signatures described here has been found to be associated with MYC in a wide range of functional studies in lymphoid cell lines and lymphoma animal models.56-58 However, because of the possible combinatorial effect of both MYC rearrangements and miRNA deregulation, additional studies on the combination of MYC and miRNA predictive impact are warranted. Our data identified a set of miRNAs that could be useful outcome prognostic markers in DLBCL treated with R-CHOP, and an integrated model with IPI identified a subset of high-risk patients with a 2-year OS < 50%. Therefore, our approach may serve to refine outcome prediction and to assign a risk-stratified therapy for DLBCL patients.
Presented in part at the United States and Canadian Academy of Pathology Annual Meeting, March 2009, Boston, MA.59
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank María Encarnación Castillo and CNIO's Tumor Bank Unit (María Jesús Artiga, Laura Cereceda, and Manuel Morente) for their skillful retrieval and handling of clinical data and samples from different clinical institutions and Lorena di Lisio for fruitful discussions on methodologic issues. They also acknowledge all of the clinical colleagues who kindly completed the clinical data form, particularly M. Canales, F. Mazorra, M. Cruz, J. Menarguez, and T. Gerds for discussions about Brier scores.
This study was supported by grants from the Ministerio de Sanidad y Consumo (PI051623, PI052800, CP06/00002, RTICC), the Asociación Española contra el Cancer (AECC), and the Ministerio de Ciencia e Innovación Spain (SAF 2008-03871).
Authorship
Contribution: S.M.-M. designed and performed the research, analyzed the data, and wrote the manuscript; N.M. and B.S.-E. performed the research and analyzed the data; A.S., C.M., E.G.-B., A.L., M.M., C.G., and M.A.M. contributed clinical data; M.E.R. performed the research; R.D.U., D.G.P., and G.G. analyzed the data; J.F.G. contributed clinical data and performed the research; C.D., E.D.H., E.C., C.V., and R.S.G. contributed clinical data and skills; G.B.R., Z.X.-M., and J.C. performed the data analysis; K.H.Y. analyzed the data and critically revised and drafted the manuscript; and M.A.P. designed the research, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Miguel A. Piris, Pathology Department, Hospital Universitario Margués de Valdecilla, IFIMAV, Avenida de Valdecilla n 25, 39008 Santander, Spain; e-mail: mapiris@cnio.es.


![Figure 3. Hazard ratio charts. The interquartile range of the hazard ratios estimated from the models after variable selection is shown for each continuous variable. For each miRNA, we calculated the log of the hazard ratio, where the difference in that variable was that of the interquartile range (ie, the third to the first quartiles). For example, for miR-222, the first and the third quartiles are 1.515 and 3.945; the bar shows the hazard ratio. For PFS: exp(0.315 * [3.945-1.515] = 2.15) and a 75% and 95% interval. For IPISCORE, a discrete variable, we show the log hazard ratio comparing each of the values of IPISCORE (except 0) with IPISCORE = 0 as a reference.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/4/10.1182_blood-2010-11-321554/4/m_zh89991174820003.jpeg?Expires=1764957169&Signature=yqq3Odok~WX4leKK~e5nuLTHBTP3l5eqj6l1imqoJ2LTk7GbvrgVJRpp~zh4jnKGmB-QTKYXVY47HlT0LPlxbsdpUWMy4zTJ8V93bjDiA74DA7SSDlqoIrY0Ssw1~d7kCPfcX4XhcPzkXsDSG8xKQnMjLszijysFtsE7h36JpPgpIOgbvbK3q1kR7tDR11S3lUl9565M7y7I~tRWBhGdnh4~4FB8bE09U5LsnbOV3Si~QTzBwdAgPGda-JMtTAhc57u5UE9fy6~ppwb99JvTxkyOGb-5HHWOAakF94rOpS02JBUjfl4z2BQLoNwyaxwjI6NZRb9FEe9zVGe5jaKdQw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
