Abstract
Increased mast cell burden is observed in the inflamed tissues and affected organs and tissues of patients with mast cell proliferative disorders. However, normal mast cells participate in host defense, so approaches to preferentially target clonally expanding mast cells are needed. We found that mammalian target of rapamycin complex 1 (mTORC1) and 2 (mTORC2) are up-regulated in neoplastic and developing immature mast cells compared with their terminally differentiated counterparts. Elevated mTOR mRNA was also observed in bone marrow mononuclear cells of patients exhibiting mast-cell hyperplasia. Selective inhibition of mTORC1 and mTORC2 through genetic and pharmacologic manipulation revealed that, whereas mTORC1 may contribute to mast-cell survival, mTORC2 was only critical for homeostasis of neoplastic and dividing immature mast cells. The cytostatic effect of mTORC2 down-regulation in proliferating mast cells was determined to be via inhibition of cell-cycle progression. Because mTORC2 was observed to play little role in the homeostasis of differentiated, nonproliferating, mature mast cells, these data provide a rationale for adopting a targeted approaching selectively inhibiting mTORC2 to effectively reduce the proliferation of mast cells associated with inflammation and disorders of mast cell proliferation while leaving normal differentiated mast cells largely unaffected.
Introduction
Mast cells (MCs) are considered to be critical components of both the innate and acquired immune defense systems.1 Central to these functions is the ability of MCs to release a plethora of inflammatory mediators after activation through cell-surface receptors, primarily the high-affinity receptors for IgE (FcϵRI).2 Although these reactions are considered to have evolved to protect host organisms against invading parasites and other microorganisms,3 inappropriate or exaggerated activation of MCs manifests the reactions associated with allergic diseases.
MCs develop from bone marrow (BM) CD13+/CD34+/CD117 (KIT)+ progenitor cells that enter into circulation and mature during migration to and residency in their target tissues.4 MC numbers within tissues appear to be tightly regulated, with a several-fold increase in numbers occurring in inflammatory conditions and even higher numbers in association with parasitic inflammation. Clonal MC disorders may result in 10-fold or greater numbers of MCs in tissues such as the BM, liver, and spleen.5,6 Similarly, a dysregulated increase in MC numbers is also observed in certain types of cancer, and in that context may contribute to cancer progression.7-9
We observed previously that the mammalian target of rapamycin (mTOR) is overexpressed and constitutively phosphorylated in neoplastic MCs regardless of whether activating mutations in the MC growth factor receptor KIT are present.10 MTOR is a serine/threonine kinase that regulates divergent signaling pathways depending on its interactions with 2 regulatory proteins: raptor, a major component of mTOR complex 1 (mTORC1), and rictor, a major component of mTOR complex 2 (mTORC2).11 mTORC1 induces phosphorylation of 4E-BP1 and p70-S6 kinase, leading to transcriptional regulation,12 whereas mTORC2 induces the phosphorylation and feedback activation of AKT.13 Therefore, in the present study, we investigated the hypothesis that mTORC1 and mTORC2 may differentially affect the proliferative and survival potential of neoplastic compared with nonneoplastic (hereafter referred to as “normal”) human MCs (huMCs).
As will be shown, neoplastic and developing MCs have significantly increased mTORC1 and mTORC2 expression/activities compared with terminally differentiated MCs. Furthermore, the BM mononuclear cell fraction from patients with the clonal MC disorder systemic mastocytosis had elevated expression of mRNA for mTOR compared with normal donors. Our studies further revealed that, whereas mTORC1 may be required for MC survival, mTORC2 selectively regulates proliferation in developing and neoplastic human MCs but has little impact on terminally differentiated mature MC homeostasis. These observations indicate that it may be possible to target the rapidly dividing MC populations associated with myeloproliferative and inflammatory disorders while having a minimal impact on normal resident MCs.
Methods
BMMCs from KI mice with disrupted mTOR
The mTOR knock-in (KI) mice were generated as described previously.14 Animals were treated in accordance with the guidelines of the Animal Care and Use Committee of the National Cancer Institute. MCs from BM (BMMCs) were developed from the BM of KI (homozygotes) and wild-type (WT) littermates of the same age and sex, as described previously,15 in medium containing mouse recombinant IL-3 (muR-IL-3; 30 ng/mL) with or without 100 ng/mL of muR-SCF.
HuMCs and shRNA KD
Primary huMCs were prepared from CD34+ peripheral blood progenitors16 isolated from healthy volunteers after informed consent under a protocol (NCT00001756) approved by the National Institutes of Health (NIH) Internal Review Board. Cells were cultured as described previously,17 and 7- to 9-week-old huMCs were used for experiments. LAD2,18 HMC-1.1,19 and HMC-1.220 MCs were cultured as described previously.10 The targeted knockdown (KD) was done as described in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Patients
Thirteen patients with systemic mastocytosis and 10 healthy volunteers underwent BM biopsies as part of the National Institute of Allergy and Infectious Diseases (NIAID) Institutional Review Board–approved research protocols (NCT00044122, NCT00806364, and NCT00090662). All subjects provided informed consent in accordance with the Declaration of Helsinki. Patients with systemic mastocytosis were diagnosed according to World Health Organization criteria. Human BM mononuclear cells from mastocytosis patients and healthy volunteers were obtained and the KIT D816V mutation was detected as described previously.21 RNA isolation and quantitative real-time RT-PCR from BM cells were performed as described in supplemental Methods.
Abs and reagents
The protein-specific Abs were: β-actin (Sigma-Aldrich) and Lyn and Syk (Santa Cruz Biotechnology). Other phosphoprotein- and protein-specific Abs were from Cell Signaling Technology. Human myeloma IgE (Calbiochem) was biotinylated by the NIAID Core Facility. The fluorophore-conjugated Abs and annexin V were from BD Biosciences. Reagents used were: Torin122 (a gift of Dr Nathanael S. Gray, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA), rapamycin (Calbiochem), and imatinib and dasatinib (LC Laboratories). All other chemicals were from Sigma-Aldrich.
Immunoblotting, flow cytometry, and annexin V assay
Samples were prepared and analyzed as described previously.23 To detect externalized phosphatidylserine (PS) by annexin V staining, the cells, processed and labeled as described previously,24 were analyzed by flow cytometry on a FACSCalibur using CellQuest 3.3 software (BD Biosciences). FlowJo 7.6 software (TreeStar) was used for data analysis.
BrdU cell-proliferation assay
Cell proliferation was analyzed by 24 hours incorporation of the thymidine analog bromodeoxyuridine (BrdU) into de novo–synthesized DNA using the BrdU Cell Proliferation Assay (Calbiochem).
MTT assay
The metabolic activity of cells was determined by evaluation of mitochondrial dehydrogenase activity using the MTT-based cell growth determination kit according to the manufacturer's instructions (Sigma-Aldrich).
Cell cycle analysis
Cells were fixed with ice-cold 70% ethanol overnight, washed with PBS supplemented with 0.1% (wt/vol) BSA, treated with RNase A, labeled with propidium iodide, and analyzed by flow cytometry on an LSRII using FACSDiva 6.2 software (BD Biosciences). A single cell population on a pulse-area versus pulse-width dot plot was used for analysis of the cell cycle with the Watson pragmatic mathematical model and FlowJo 7.6 software.
Statistical evaluation
Means and SEM were calculated from the number of indicated experiments. Statistical significance (P < .05) of intergroup differences between 2 sets of data were calculated by unpaired 2-tailed Student t test and among multiple sets of data by 1-way ANOVA with the Tukey posttest. Correlation was calculated with the Spearman correlation test. For statistical evaluations, huMCs from different donors, BMMCs from different mice, and/or independent sample preparations or KD transfections from indicated cell lines were evaluated.
Results
Role of mTOR in homeostasis of developing BMMCs
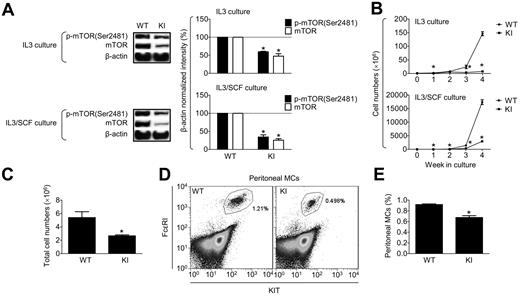
To establish the potential role(s) of mTOR complexes in MC growth and survival, we examined the overall impact of mTOR deficiency on the expansion of MCs from BM of KI mice with neo-disrupted mTOR expression compared with the expansion of MCs from the BM of WT mice. As shown in Figure 1A, both expression and phosphorylation of mTOR was substantially reduced in the BMMCs derived from the KI mice compared with the WT controls. After 4 weeks in culture, there were no gross morphological differences (supplemental Figure 1A) nor substantial changes in KIT or FcϵRI expression (supplemental Figure 1B) between WT and KI MCs, irrespective of whether the cells were grown in IL-3 or IL-3/SCF. However, there was substantially reduced MC expansion from the BM of KI mice compared with their WT counterparts in both IL-3 and IL-3/SCF cultures (Figure 1B). To assess how mTOR down-regulation affects the numbers of MCs and other cells of hematopoietic origin in vivo, we examined blood cells and MCs in skin from the back and peritoneum in the KI mice. As shown in supplemental Table 1, despite the general mTOR down-regulation, there were few consequences for the majority of cell types examined, and any effects that were present were primarily restricted to inflammatory cells. Although we also did not observe any difference in MC density in the back skin, the total number of MCs recovered from peritoneal cavity (Figure 1C), as well as their relative content in the recovered peritoneal cells (Figure 1D-E), were significantly reduced in KI mice. Whereas the reduced total MCs recovered from peritoneum might reflect the generally reduced size of these KI mice,14 the reduced percentage of MCs in the recovered peritoneal cells demonstrate that down-regulation of mTOR can affect MC numbers in vivo. The differential impact of reduced mTOR activity on the skin and peritoneal MCs may reflect the phenotypic differences in these cells or their different rates of growth/turnover combined with the fact that mTOR activity is not completely ablated in these mice. These data support the conclusion that mTOR is required for the expansion and/or survival of developing MC populations. Nevertheless, these studies did not permit us to define the exact role of the individual mTOR complexes in MC homeostasis, particularly in the human.
Role of mTORC1 and mTORC2 in the growth of BMMC progenitors. (A) Four- to 6-week-old BMMCs from WT or KI-disrupted mTOR (KI) mice were analyzed by immunoblotting with mTOR phosphoprotein-specific or protein-specific Abs and data were quantitated (right). (B) One million BM cells from WT or KI mice were cultured for 4 weeks in the presence of IL-3 or IL-3/SCF and cell numbers were determined. (C) Total numbers of peritoneal cells recovered from WT or KI mice. (D) The peritoneal cells recovered in panel C were stained with PE-KIT-, FITC-FcϵRI-, and FITC-IgE–specific Abs and analyzed by flow cytometry. (E) The percentage of peritoneal MC population, FcϵRI, and KIT+ population determined in panel D was evaluated. In panel D, a representative flow cytometry analysis is shown. In panels A through C and E, the data represent means and SEM (A-B, n = 3; C and E, n = 6; note that “n” refers to the number of pairs of mice) and differences between WT and KI (*P < .05 by t test) are indicated.
Role of mTORC1 and mTORC2 in the growth of BMMC progenitors. (A) Four- to 6-week-old BMMCs from WT or KI-disrupted mTOR (KI) mice were analyzed by immunoblotting with mTOR phosphoprotein-specific or protein-specific Abs and data were quantitated (right). (B) One million BM cells from WT or KI mice were cultured for 4 weeks in the presence of IL-3 or IL-3/SCF and cell numbers were determined. (C) Total numbers of peritoneal cells recovered from WT or KI mice. (D) The peritoneal cells recovered in panel C were stained with PE-KIT-, FITC-FcϵRI-, and FITC-IgE–specific Abs and analyzed by flow cytometry. (E) The percentage of peritoneal MC population, FcϵRI, and KIT+ population determined in panel D was evaluated. In panel D, a representative flow cytometry analysis is shown. In panels A through C and E, the data represent means and SEM (A-B, n = 3; C and E, n = 6; note that “n” refers to the number of pairs of mice) and differences between WT and KI (*P < .05 by t test) are indicated.
Expression and activity of mTORC1 and mTORC2 during MC development
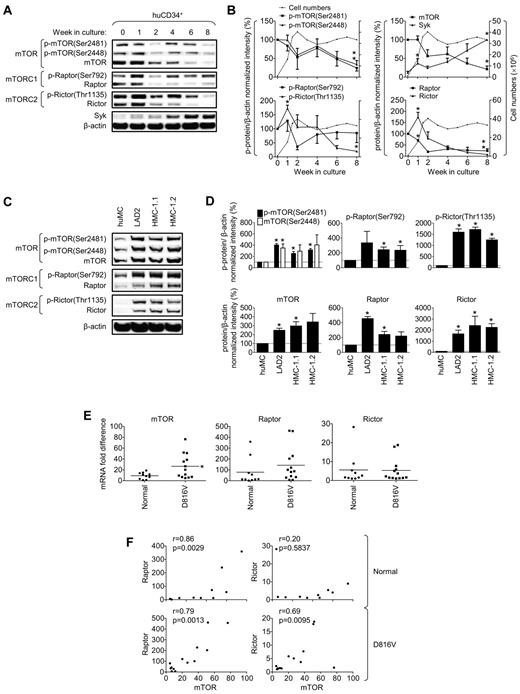
To investigate the relative roles of mTORC1 and mTORC2 in human MC expansion and survival, we initially examined changes in mTOR, raptor, and rictor, the major components of mTORCs, during huMC development. The activity of mTOR was assessed by evaluation of phosphorylation at Ser2448 and autophosphorylation at Ser2481, which reflects the kinase activity of mTOR.25,26 Raptor activity was assessed by phosphorylation of the regulatory residue (Ser792),27 and rictor activity by phosphorylation of the regulatory residue (Thr1135; Figure 2A).28 As described previously,29 the cells rapidly proliferated during the first 2 weeks of culture, after which time the numbers plateaued (Figure 2B). By 7-8 weeks, the cells reached maturity, being terminally differentiated, nondividing, and having the morphological characteristics of mature MCs (ie, well-condensed nuclei and abundant granules; data not shown).
mTORC1 and mTORC2 during development of huMCs from CD34+ progenitors, in neoplastic MC lines, and in cells with D816V-mutated KIT from patients with systemic mastocytosis. (A) Peripheral blood CD34+ progenitors were cultured for 8 weeks. (B) mTORC1 and mTORC2 were then analyzed by immunoblotting and compared with total cell numbers (dashed lines) during the course of 8 weeks. (C-D) mTORC1 and mTORC2 were analyzed in huMCs, LAD2, HMC-1.1, and HMC-1.2 MCs by immunoblotting. (E) Expression of mTOR, raptor, and rictor mRNA in normal cells (healthy volunteers, n = 10) or cells with the KIT D816V mutation (n = 13) of donor's BM mononuclear cell fraction aspirates was analyzed, and differences in mRNA expression of each protein between the group of normal and KIT D816V donors and (F) correlation of mRNA expression of the mTORC components within each group of donors evaluated. In panels B and D, data represent means (cell counts) or means and SEM (phosphorylation and protein levels; n = 3; 3 donors or 3 independent sample preparations). In panel B, differences between phosphorylation or protein expression in 0- and 1-week-old cells, respectively, and 0- and 8-week-old cells are indicated (*P < .05 by t test). In panel D, differences between huMCs and each type of the myeloproliferative MCs are indicated (*P < .05 by t test). In panel E, bars represent mean of values determined in each group and differences between the group of normal and KIT D816V donors are indicated (*P < .05 by t test). In panel F, the correlation was evaluated by Spearman correlation test between normal donors (n = 10) and KIT D816V mutation donors (n = 13).
mTORC1 and mTORC2 during development of huMCs from CD34+ progenitors, in neoplastic MC lines, and in cells with D816V-mutated KIT from patients with systemic mastocytosis. (A) Peripheral blood CD34+ progenitors were cultured for 8 weeks. (B) mTORC1 and mTORC2 were then analyzed by immunoblotting and compared with total cell numbers (dashed lines) during the course of 8 weeks. (C-D) mTORC1 and mTORC2 were analyzed in huMCs, LAD2, HMC-1.1, and HMC-1.2 MCs by immunoblotting. (E) Expression of mTOR, raptor, and rictor mRNA in normal cells (healthy volunteers, n = 10) or cells with the KIT D816V mutation (n = 13) of donor's BM mononuclear cell fraction aspirates was analyzed, and differences in mRNA expression of each protein between the group of normal and KIT D816V donors and (F) correlation of mRNA expression of the mTORC components within each group of donors evaluated. In panels B and D, data represent means (cell counts) or means and SEM (phosphorylation and protein levels; n = 3; 3 donors or 3 independent sample preparations). In panel B, differences between phosphorylation or protein expression in 0- and 1-week-old cells, respectively, and 0- and 8-week-old cells are indicated (*P < .05 by t test). In panel D, differences between huMCs and each type of the myeloproliferative MCs are indicated (*P < .05 by t test). In panel E, bars represent mean of values determined in each group and differences between the group of normal and KIT D816V donors are indicated (*P < .05 by t test). In panel F, the correlation was evaluated by Spearman correlation test between normal donors (n = 10) and KIT D816V mutation donors (n = 13).
The expression of the mTOR, raptor, and rictor proteins was observed to be markedly higher during the expansion stage of culture (ie, the first 2 weeks) than at later time points, and there was a significant increase in rictor expression during the first week of culture (Figure 2A-B). The slow decline in the expression of mTOR, rictor, and raptor during the subsequent weeks mirrored the diminishing expansion rate of the cells. By 8 weeks, when the cells were terminally differentiated, the levels of expression were markedly lower than in dividing cells. In contrast, there was a steady increase in Syk expression during this time, reflecting increased MC expansion and maturity. The degree of phosphorylation of mTOR, raptor, and rictor during expansion and maturation of the cells (Figure 2A-B) largely mirrored the changes in protein content, indicating that there was little developmental change in phosphorylation of these molecules per se. Irrespective of the reduced expression and activation of the mTORC1 and mTORC2 components in mature huMCs, these components could be activated in the huMCs after activation via KIT or antigen after overnight SCF depletion (supplemental Figure 2A-B).
With the exception of raptor, the observed developmental decreases in protein expression were not reflected by similar decreases in mRNA levels (supplemental Figure 2C), indicating that down-regulation of the expression of the mTORC proteins occurred posttranscriptionally. Nevertheless, these data suggest that cell expansion was associated with elevated mTORC1 and mTORC2 signaling and that once the huMCs were terminally differentiated, mTORC1 and mTORC2 levels were significantly down-regulated.
Elevated expression and activation of mTORC1 and mTORC2 in neoplastic human MC lines
The increased mTORC1 and mTORC2 levels and activation associated with developing/expanding MCs led us to next assess whether the expression and activation of mTORC2, in addition to mTORC1, was preferentially elevated in neoplastic, compared with nonneoplastic, MCs. As we reported previously,10 both increased protein expression and increased constitutive phosphorylation of mTOR was enhanced in the human HMC-1.1, HMC-1.2, and LAD2 MC lines compared with the terminally differentiated huMCs (Figure 2C-D). Phosphorylation and protein expression of raptor and rictor were also found to be markedly elevated in these lines compared with huMCs (Figure 2C). However, as observed during huMC development, the increase in phosphorylation of rictor and raptor (Figure 2D top) was largely a consequence of the increased expression of protein (Figure 2D bottom) rather than being a net increase in phosphorylation per se. Furthermore, as observed previously in the huMC progenitors, these changes in protein levels were not correlated with changes in mRNA (supplemental Figure 2D); therefore, as was the case for developing huMCs, the elevated levels of mTOR, raptor, and rictor observed in the cell lines may reflect posttranscriptional regulation. Nevertheless, our data reveal that the activities of both mTORC1 and mTORC2 were markedly elevated in the neoplastic human MC lines compared with the terminally differentiated huMCs.
mTORC1 and mTORC2 expression in systemic mastocytosis
The observation that KIT activates mTOR-mediated pathways10 and that mTOR expression and indices of mTORC activities are constitutively elevated in neoplastic MCs led us to examine whether increased expression of mTOR, raptor, and rictor was also evident in BM aspirates from patients with the D816V KIT mutation–associated MC proliferative disorder mastocytosis compared with normal individuals.30 Because of the limited numbers of MCs present in these samples, expression was determined at the mRNA level in the BM mononuclear cells, a fraction of which are MC precursors. As shown in Figure 2E, mTOR mRNA was significantly elevated in the BM fraction from the mastocytosis patients. Although we did not detect significantly elevated levels of raptor or rictor in the overall patient population, mTOR, rictor, and raptor levels tended to be higher in patients with the aggressive form of the disease (supplemental Figure 3). Furthermore, we observed a significant correlation between expression levels of rictor and mTOR, as well as between raptor and mTOR, in the mastocytosis patients (Figure 2F). In contrast, in healthy donors, although raptor mRNA levels were closely correlated with mTOR RNA levels, those for rictor were not. These and the aforementioned data led us to hypothesize that mTORC1 and/or mTORC2 may play a role in the expansion and/or survival of neoplastic and, potentially, nonneoplastic MCs.
Inhibition of mTORC1 and mTORC2: effect on MC homeostasis
We used 2 approaches to examine how mTORC1 and mTORC2 may differentially affect the expansion and survival of the normal and neoplastic MCs: (1) inhibition of mTORC1 and mTORC2 activities through the use of selective inhibitors in normal and neoplastic huMCs; and (2) inhibition of mTORC activities in these cells through shRNA KD of protein expression of raptor and rictor.
Rapamycin has been used previously to determine the role of mTORC1 in biologic responses.31 However, at least under acute conditions, this compound only poorly inhibits the phosphorylation of 4E-BP1 and, although prolonged exposure can down-regulate mTORC2 activity, short-term exposure is ineffective at blocking this response.22 Therefore, to more effectively explore the potential involvement of mTORC2, we used Torin1, a compound described recently to block both mTORC2 and mTORC1 activities, including 4E-BP1 phosphorylation.22 In agreement with the previous report,22 in SCF-stimulated huMCs, we found that Torin1 inhibited mTORC1 and mTORC2, whereas rapamycin only inhibited mTORC1 and only partly inhibited 4E-BP1 (supplemental Figure 4). Similar results were observed in neoplastic MCs (data not shown). From these studies, effective concentrations of Torin1 (200nM) and rapamycin (200nM) were selected for further analysis.
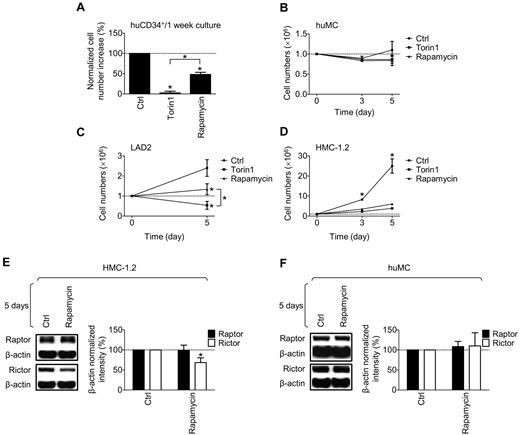
As shown in Figure 3A, Torin1 produced an almost complete ablation of the developing CD34+ progenitor cell population during the rapid expansion stage, whereas rapamycin only partially reduced this expansion. In contrast, in the mature, nondividing huMCs, both Torin1 and rapamycin had a minimal effect on cell numbers when exposed for 5 days or less (Figure 3B). As was the case with rapamycin,10 Torin1 did not affect degranulation in response to antigen or antigen/SCF (supplemental Figure 5A), and only partially inhibited cytokine production in the mature huMCs (supplemental Figure 5B). Furthermore, Torin1 did not reduce the numbers of nondividing MCs in BM aspirates from human subjects (data not shown). As with the expanding huMC progenitors, in the LAD2 MCs, Torin1 was more effective at inhibiting cell expansion than was rapamycin (Figure 3C), whereas both inhibitors greatly diminished expansion of the more rapidly dividing HMC-1.2 MCs (Figure 3D). The differences in the extent of inhibition in response to rapamycin in the HMC-1.2 MCs compared with huMCs may reflect additional inhibition of mTORC2 in the HMC-1.2 MCs, because prolonged exposure of HMC-1.2 MCs (Figure 3E), but not huMCs (Figure 3F), to rapamycin resulted in reduced rictor expression. Nevertheless, from these data, we conclude that mTORC1 and/or mTORC2 play a critical role in the homeostasis of rapidly dividing MCs/MC progenitors but not in terminally differentiated huMCs.
Role of mTORC1 and mTORC2 in the growth of CD34+ cells, mature huMCs, and LAD2 and HMC-1.2 MCs. (A) One-week-old cultures of huCD34+ progenitors were supplemented with vehicle alone (Ctrl), mTORC1/mTORC2 (Torin1, 200nM), or mTORC1 (rapamycin, 200nM) inhibitor, cultured for 72 hours, and the effect of the drugs on cell numbers was evaluated. (B) One million huMCs were incubated as in panel A and the numbers of cells in the cultures were determined and evaluated. (C) One million LAD2 MCs were incubated as in panel A, and the number of cells in the cultures was determined. (D) One million HMC-1.2 MCs were incubated as in panel A, and the number of cells in the cultures was determined. (E-F) HMC-1.2 MCs (E) or huMCs (F) were supplemented with vehicle alone (Ctrl) or rapamycin (200nM), cultured for 5 days, and the effects on raptor and rictor protein levels were evaluated. In panels A through F, the data represent means and SEM (A-B, n = 3, and F, n = 2, where n represents the number of donors; C-D, n = 3, and E, n = 6, where n represents the number of independent sample preparations) and differences among the differentially treated samples (*P < .05; for C-D by 1-way ANOVA and for E-F, by t test) are indicated.
Role of mTORC1 and mTORC2 in the growth of CD34+ cells, mature huMCs, and LAD2 and HMC-1.2 MCs. (A) One-week-old cultures of huCD34+ progenitors were supplemented with vehicle alone (Ctrl), mTORC1/mTORC2 (Torin1, 200nM), or mTORC1 (rapamycin, 200nM) inhibitor, cultured for 72 hours, and the effect of the drugs on cell numbers was evaluated. (B) One million huMCs were incubated as in panel A and the numbers of cells in the cultures were determined and evaluated. (C) One million LAD2 MCs were incubated as in panel A, and the number of cells in the cultures was determined. (D) One million HMC-1.2 MCs were incubated as in panel A, and the number of cells in the cultures was determined. (E-F) HMC-1.2 MCs (E) or huMCs (F) were supplemented with vehicle alone (Ctrl) or rapamycin (200nM), cultured for 5 days, and the effects on raptor and rictor protein levels were evaluated. In panels A through F, the data represent means and SEM (A-B, n = 3, and F, n = 2, where n represents the number of donors; C-D, n = 3, and E, n = 6, where n represents the number of independent sample preparations) and differences among the differentially treated samples (*P < .05; for C-D by 1-way ANOVA and for E-F, by t test) are indicated.
Inhibition of mTORC1 and mTORC2: effects on MC proliferation and survival
Because mTORC inhibition had a more significant outcome on rapidly dividing MCs than on nondividing MCs, it appeared that mTORC1 and/or mTORC2 may play a more critical role in cell growth than in cell survival. Therefore, we next examined how mTORC inhibition affected indices of these parameters. To determine cell viability, we analyzed both caspase cleavage and PS externalization on the cell surface. Treatment of huMCs with Torin1 and rapamycin for 5 days did not lead to caspase cleavage, as shown by the lack of accumulation of cleaved caspases and depletion of their noncleaved counterparts (Figure 4A). The absence of caspase cleavage products was not because of direct inhibition of the apoptotic process, as shown by UV exposure (supplemental Figure 6A). Torin1 and rapamycin also failed to induce externalization of PS on the cell surface, as detected by annexin V staining (Figure 4B). In contrast, conditions known to induce MC apoptosis (UV radiation and imatinib treatment) induced marked caspase cleavage and allowed annexin V staining. The inhibitors had only a moderate impact on the metabolic activity of the huMCs, as determined by the MTT assay (Figure 4C).
Role of mTORC1 and mTORC2 in survival and metabolic activity of mature huMCs and HMC-1.2 MCs. (A) HuMCs were cultured in the presence of vehicle alone, mTORC1/mTORC2 (Torin1, 200nM) or mTORC1 (rapamycin, 200nM) inhibitors and caspases were analyzed by immunoblotting. As a positive control, cell samples from UV-irradiated cells cultured for 8 hours were used. (B) HuMCs were cultured as in panel A for 5 days and cell viability was determined by annexin V assay. As a positive control, imatinib (2μM) was used. (C) HuMCs were cultured as in panel A and metabolic activity of the cells was determined by the MTT assay and evaluated. (D-F) HMC-1.2 MCs were investigated as for huMCs in panels A through C, with the exception that dasatinib treatment in panel E was performed for 3 days. (G-J) HuMCs or HMC-1.2 MCs were cultured as in panel A and the numbers (G and I) and cell viability (H and J) were evaluated. In panels A and D, the blots are representative of 3 donors (A) or 3 independent sample preparations (D). In panels B, C, and E through J, data represent means and SEM (n = 3; 3 donors or 3 independent sample preparations) and differences among the differentially treated samples of each MC type are indicated (*P < .05 by 1-way ANOVA).
Role of mTORC1 and mTORC2 in survival and metabolic activity of mature huMCs and HMC-1.2 MCs. (A) HuMCs were cultured in the presence of vehicle alone, mTORC1/mTORC2 (Torin1, 200nM) or mTORC1 (rapamycin, 200nM) inhibitors and caspases were analyzed by immunoblotting. As a positive control, cell samples from UV-irradiated cells cultured for 8 hours were used. (B) HuMCs were cultured as in panel A for 5 days and cell viability was determined by annexin V assay. As a positive control, imatinib (2μM) was used. (C) HuMCs were cultured as in panel A and metabolic activity of the cells was determined by the MTT assay and evaluated. (D-F) HMC-1.2 MCs were investigated as for huMCs in panels A through C, with the exception that dasatinib treatment in panel E was performed for 3 days. (G-J) HuMCs or HMC-1.2 MCs were cultured as in panel A and the numbers (G and I) and cell viability (H and J) were evaluated. In panels A and D, the blots are representative of 3 donors (A) or 3 independent sample preparations (D). In panels B, C, and E through J, data represent means and SEM (n = 3; 3 donors or 3 independent sample preparations) and differences among the differentially treated samples of each MC type are indicated (*P < .05 by 1-way ANOVA).
In the HMC-1.2 MCs, rapamycin induced no evidence of cleaved caspases (Figure 4D). Nevertheless, in these cells, Torin1 resulted in a moderate accumulation of the cleaved caspases. When we analyzed these cells for annexin V staining (Figure 4E), we found that neither Torin1 nor rapamycin had a major effect on the induction of PS externalization. We also determined that the difference in the sensitivity to Torin1 was not because of the D816V mutation in HMC-1.2, because HMC-1.1 MCs, which lack the KIT D816V mutation, showed comparable responses to the inhibitors as HMC-1.2 MCs (data not shown). However, both rapamycin and Torin1 reduced the metabolic activity of these cells, as determined by the MTT assay, especially compared with the huMCs (compare Figure 4 panel C with panel F).
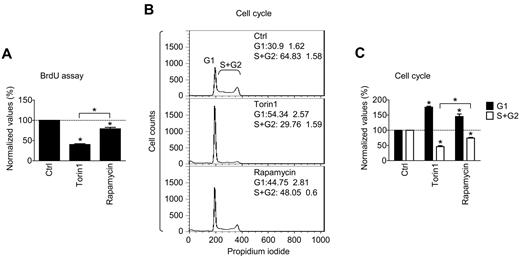
In contrast to these observations, when huMCs and HMC-1.2 MCs were exposed to the inhibitors for an extended period of time (12 days), there was a significant decrease in the number and viability of both huMCs and HMC-1.2 MCs (Figure 4G-J). Nevertheless, our data suggest that although prolonged exposure to rapamycin and Torin1 results in reduced numbers and viability of MCs, the decrease in cell numbers observed in the dividing cells treated with the mTORC inhibitors for a shorter period of time was not a consequence of the induction of apoptosis, but rather may be related to an inhibition of cell division. We therefore determined the potential of Torin1 and rapamycin to inhibit cell proliferation as monitored by the BrdU assay in HMC-1.2 MCs. As shown in Figure 5A, both Torin1 and rapamycin treatment resulted in a decrease in BrdU incorporation in the HMC-1.2 MCs. However, Torin1 showed a significantly more effective inhibition of BrdU incorporation than did rapamycin. These data suggest that the mTORC inhibitors reduced HMC-1.2 MC numbers through arrest of cell division, at least during short-term (5 days) exposure. To further explore this possibility, we examined the effect of the inhibitors on cell cycle progression using flow cytometry. As shown in Figure 5B-C, the ability of Torin1 and rapamycin to induce inhibition of cell proliferation could be explained by cell cycle arrest of MCs at the G1 phase.
Role of mTORC1 and mTORC2 in proliferation and cell cycle of HMC-1.2 MCs. (A) HMC-1.2 MCs were supplemented with vehicle alone (Ctrl), mTORC1/mTORC2 (Torin1, 200nM), or mTORC1 (rapamycin, 200nM) inhibitors and cultured for 72 hours. Incorporation of BrdU was determined with the BrdU assay and evaluated. (B-C) HMC-1.2 MCs were supplemented with vehicle alone and inhibitors as in panel A, cultured for 72 hours, and subjected to cell cycle analysis by flow cytometry (B) and evaluated (C). In panels A and C, data represent means and SEM (n = 3; 3 independent sample preparations) and differences among the differentially treated samples are indicated (*P < .05 by 1-way ANOVA). In panel B, data are means and SEM (n = 3; 3 independent sample preparations).
Role of mTORC1 and mTORC2 in proliferation and cell cycle of HMC-1.2 MCs. (A) HMC-1.2 MCs were supplemented with vehicle alone (Ctrl), mTORC1/mTORC2 (Torin1, 200nM), or mTORC1 (rapamycin, 200nM) inhibitors and cultured for 72 hours. Incorporation of BrdU was determined with the BrdU assay and evaluated. (B-C) HMC-1.2 MCs were supplemented with vehicle alone and inhibitors as in panel A, cultured for 72 hours, and subjected to cell cycle analysis by flow cytometry (B) and evaluated (C). In panels A and C, data represent means and SEM (n = 3; 3 independent sample preparations) and differences among the differentially treated samples are indicated (*P < .05 by 1-way ANOVA). In panel B, data are means and SEM (n = 3; 3 independent sample preparations).
shRNA KD of raptor and rictor: consequences for survival and proliferation in MCs
The mTORC1 and mTORC2 inhibition studies thus far had supported the concept that mTORC1 and/or mTORC2 activity can dictate MC numbers largely through regulation of progression through cell cycle, but that, over the long term, they may also play a role in cell survival. However, the ability of Torin1 to inhibit both mTORC1 and mTORC2, and of rapamycin to inhibit mTORC2 in addition to mTORC1 with prolonged exposure, did not permit us to define the exact roles of these complexes in MC expansion and/or survival. Nevertheless, the more robust inhibition obtained with Torin1 on parameters of MC expansion suggested that mTORC2 may be the predominant mTORC regulating proliferation of these cells. To investigate this possibility and to further define the respective roles of mTORC1 and mTORC2 in MC homeostasis, we attempted to selectively KD raptor or rictor in these cells using targeted shRNA.
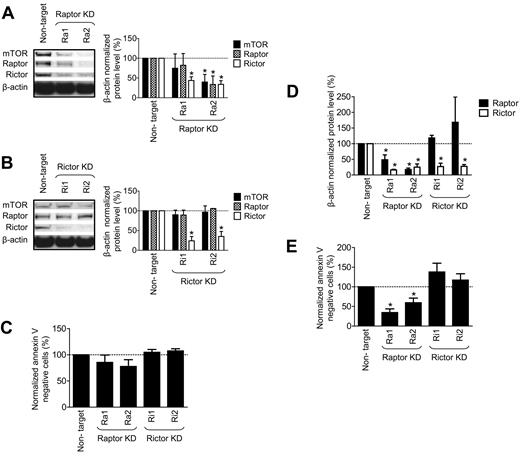
We analyzed the role of raptor and rictor in normal huMCs by transfecting the cells with 2 raptor-targeting or 2 rictor-targeting vectors and compared their performance to the nontarget vector. The nontarget vector minimally affected mTOR, rictor, and raptor expression compared with the nontransfected cells (data not shown). However, as shown in Figure 6A, targeting of raptor led to decreased expression of not only raptor, but also of rictor and mTOR. Regardless, shRNA targeting rictor led to selectively decreased expression of rictor, and had no effect on the expression of raptor or mTOR (Figure 6B). As shown in Figure 6C, the marked KD of rictor produced by both shRNA constructs had no effect on cell viability as determined by annexin V binding. However, both raptor KD shRNAs produced a slight decrease in viability after the 7-day posttransfection time interval. To determine whether this represented the initiation of a progressive decrease in cell viability, we extended the culture time of the transfected KDs for an additional 3 days after transfection. As is apparent from the results shown in Figure 6D, raptor KD continued to maintain decreased expression of both raptor and rictor, whereas rictor KD was still specific for rictor; again, rictor KD had no impact on cell viability (Figure 6E). However, raptor KD now caused a progressive decrease in cell viability. Culturing these KD cells for an additional 2 weeks resulted in the death of all raptor KD cells, but no deterioration of viability of the rictor KD cells (data not shown). These data show that mTORC2 is not required to maintain cell viability of mature huMCs. However, although the raptor-targeted shRNA also knocked down mTOR and rictor, the compromised viability of the huMCs in the absence of a similar response with the rictor-targeted shRNA together suggest a role for the mTORC1 in the long-term survival of huMCs.
mTORC2, but not mTORC1, is dispensable for the survival of huMCs. HuMCs were transfected with luciferase (nontarget) or 2 raptor-targeting shRNA vectors (Ra1 and Ra2; A), or 2 rictor-targeting shRNA vectors (Ri1, Ri2; B), and expression of the mTOR, raptor, and rictor proteins were determined by immunoblotting and evaluated at day 7 after transfection. Cell viability of the transfected cells was determined with the annexin V assay (C). Expression of raptor and rictor (D) and cell viability (E) was determined by, respectively, immunoblotting and annexin V assay at day 10 after transfection. In panels A through E, data represent means and SEM (n = 3; 3 donors) and differences between the cells transfected with control (nontarget) and the targeted shRNAs are indicated (*P < .05 by t test).
mTORC2, but not mTORC1, is dispensable for the survival of huMCs. HuMCs were transfected with luciferase (nontarget) or 2 raptor-targeting shRNA vectors (Ra1 and Ra2; A), or 2 rictor-targeting shRNA vectors (Ri1, Ri2; B), and expression of the mTOR, raptor, and rictor proteins were determined by immunoblotting and evaluated at day 7 after transfection. Cell viability of the transfected cells was determined with the annexin V assay (C). Expression of raptor and rictor (D) and cell viability (E) was determined by, respectively, immunoblotting and annexin V assay at day 10 after transfection. In panels A through E, data represent means and SEM (n = 3; 3 donors) and differences between the cells transfected with control (nontarget) and the targeted shRNAs are indicated (*P < .05 by t test).
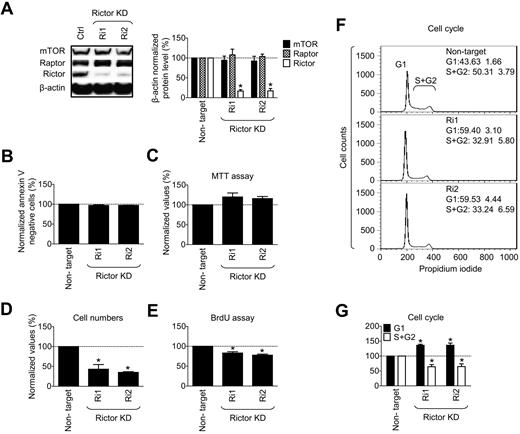
Finally, we sought to provide support for our conclusion that mTORC2 regulates the proliferative potential of expanding MCs. We evaluated the effects of shRNA-mediated rictor KD on proliferation of the HMC-1.2 MCs. As shown in Figure 7A and as observed in the huMCs, the rictor-targeted shRNA constructs resulted in decreased expression of rictor in the absence of effects on expression of raptor or mTOR. Rictor KD did not compromise cell viability (Figure 7B) or metabolic activity (Figure 7C) of the cells. However, it had a significant inhibitory impact on cell proliferation (Figures 7D-E). Similar data were obtained at an earlier time point after transfection and after extended culturing of the transfected cells (supplemental Figure 6B-C). In addition, in analyzing cell cycle parameters, we found that rictor KD caused cell cycle arrest at the G1 phase (Figure 7F-G). Therefore, these data support the conclusion that mTORC2 regulates homeostasis in neoplastic MCs by controlling cell proliferation through progression through the cell cycle.
mTORC2 is not essential for survival but is essential for proliferation of myeloproliferative HMC-1.2 MCs. (A) HMC-1.2 MCs were transfected and analyzed at day 7 after transfection as in Figure 6B. At day 7 after transfection, cell viability of the transfected cells was determined by annexin V assay (B), metabolic activity by MTT assay (C), cell expansion by evaluation of cell number increase after the 24 hours (D), cell proliferation by BrdU assay (E), and cell cycle analysis by flow cytometry (F-G). In panels A-G, data represent means and SEM (n = 3; 3 independent transfection), and differences between the cells transfected with control (nontarget) and each of rictor-targeted shRNAs are indicated (*P < .05 by t test).
mTORC2 is not essential for survival but is essential for proliferation of myeloproliferative HMC-1.2 MCs. (A) HMC-1.2 MCs were transfected and analyzed at day 7 after transfection as in Figure 6B. At day 7 after transfection, cell viability of the transfected cells was determined by annexin V assay (B), metabolic activity by MTT assay (C), cell expansion by evaluation of cell number increase after the 24 hours (D), cell proliferation by BrdU assay (E), and cell cycle analysis by flow cytometry (F-G). In panels A-G, data represent means and SEM (n = 3; 3 independent transfection), and differences between the cells transfected with control (nontarget) and each of rictor-targeted shRNAs are indicated (*P < .05 by t test).
Discussion
In the present study, we sought to determine the respective roles of mTORC1 and mTORC2 in the growth, development, and survival of neoplastic and nonneoplastic MCs. We showed that mTORC1 and mTORC2 expression and/or activities are elevated during huMC development from CD34+ progenitors and in neoplastic huMCs compared with their terminally differentiated, nondividing counterparts. We also showed that, whereas mTORC1 may contribute to the regulation of MC survival, mTORC2 selectively regulates the proliferative capacity of the expanding MC populations.
The studies investigating the respective roles of mTORC1 and mTORC2 in MC expansion and survival were initiated by our previous10 and present observations of elevated mTOR, rictor, and raptor expression in dividing MCs and MC lines compared with nondividing mature MCs, as well as by our present observations of significantly elevated mTOR mRNA in the BM mononuclear cell fraction taken from D816V-positive mastocytosis patients compared with healthy subjects. In the HMC-1.1, HMC-1.2, and LAD2 MC lines and in normal MCs during development, we also observed enhanced expression of mTORC1 and mTORC2 components at the protein level compared with nondividing, mature huMCs. However, the levels of mRNA for these proteins (except for raptor in the developing MCs) were largely unchanged. These observations are consistent with the conclusion that, in addition to transcriptional regulation, protein expression of mTORC components can be significantly up-regulated posttranscriptionally. Although the mechanism for the enhanced levels of mTORC proteins in the dividing MC lines and expanding MC cultures is unclear, it is possible that they may reflect miRNA regulation of mTORC components in these cells. mTOR expression has been reported to be regulated by miRNA in childhood adrenocortical tumors,32 and miRNA can suppress protein expression not only by degradation of the targeted mRNA, but also by repression of its translation.33 However, other regulatory mechanisms, including protein degradation or translation “on demand,”34,35 are also possible.
Regardless of the mechanism, our data clearly show significant up-regulation of mTORC1 and mTORC2 components in developing and in neoplastic MCs. These observations led us to hypothesize that the up-regulation of mTORC1 and/or mTORC2 may selectively confer preferential survival and/or proliferative potential in these cell populations, and that, once MCs are terminally differentiated and nondividing, these complexes might be relegated to a minor role in their homeostasis. This conjecture was supported by our data demonstrating that: (1) mouse MCs defective in mTOR expression failed to expand from their BM progenitors; (2) KI mice had relatively fewer MCs in the peritoneum than WT mice; and (3) the dual mTORC1/mTORC2 inhibitor Torin1, and to a lesser extent, the more selective mTORC1 inhibitor, rapamycin, were able to prevent the expansion of huMCs from their progenitors under conditions where there was no effect of these agents on huMCs once mature and terminally differentiated. The marked attenuation in the numbers of HMC-1.2 MCs cultured in the presence of the mTORC inhibitors compared with those that were left untreated further illustrated the dependency of the proliferative/survival capacity of these MCs on intact mTOR catalytic activity.
These data, together with the expression and activity profile of components of mTORC1 and mTORC2 during MC development and their relatively low levels in the mature MCs, suggested that these complexes play a more critical role in cell proliferation than in cell survival. Certainly, the significant reduction of cell division, as monitored by BrdU incorporation and cell cycle progression after treatment of the cells with the inhibitors, support this conclusion. However, although there was limited evidence of compromised cell survival under identical conditions, more prolonged incubation resulted in increased cell death. From these inhibitor studies, therefore, although we could clearly define a role for the mTORCs in cell expansion and perhaps survival, the ability of Torin1, and to a certain extent rapamycin, to block both mTORC1 and mTORC2 precluded us from defining what the specific roles were based on the actions of these inhibitors. Nevertheless, the higher degree of inhibition of BrdU incorporation obtained with Torin1, which more effectively targeted mTORC2 compared with rapamycin, indicates that mTORC2 and mTORC1 have divergent roles in the regulation of MC proliferation.
Evidence to further support differential roles of mTORC1 and mTORC2 in MC growth and survival was provided by the shRNA KD studies, which revealed that a decrease in rictor expression reduced the numbers and proliferation of expanding HMC-1.2 MCs, but had no adverse effects on the viability of MCs. In contrast to the effects of Torin1 and rapamycin on huMCs, however, there was no evidence of cell death after prolonged KD of mTORC2 in the absence of mTORC1 KD. Together with the data revealing that indices of cell division were similarly reduced in HMC-1.2 MCs after rictor KD in the absence of marked apoptosis, these data support the conclusion that mTORC2 preferentially regulates MC division but not survival. Nevertheless, rictor KD appeared to have a slightly greater effect on cell expansion than on progression through the cell cycle. Although these observations may be explained by the effects on the cell cycle accumulating over time, resulting in an apparently slightly greater effect on cell numbers, it also may indicate that mTORC2 can partially regulate cell numbers independently of effects on cell cycle progression.
The experiments using raptor-targeted shRNA were less unequivocal than those conducted with the rictor-targeted shRNA, in that the raptor-targeted shRNA also resulted in reduction of rictor and mTOR expression in huMCs and HMC-1.2 MCs (data not shown). Although these observations may reflect off-target inhibition by the raptor-targeted shRNA, the possibility also exists that raptor in some way stabilizes mTORC2 and thereby prevents its degradation. This may also provide an explanation as to why long-term rapamycin treatment can inhibit mTORC236 in the absence of similar effects on short-term exposure.37,38 Certainly, this conclusion is supported by our observation that long-term exposure to rapamycin down-regulated rictor, but not raptor, expression in HMC-1.2 MCs but not in huMCs. Regardless of the effects of the raptor-targeted shRNA on mTORC2, the increased apoptosis observed in the huMCs with raptor-targeted, but not rictor-targeted, shRNA suggests that whereas the mTORC2 preferentially regulates MC proliferation, mTORC1 may preferentially play a role in MC survival. These data would explain why long-term exposure to Torin1 and rapamycin, both of which block mTORC1, reduced huMC viability, whereas rictor-shRNA and short-term inhibitor exposure only blocked indices of cell proliferation.
In conclusion, this study provides evidence that mTORC2 may preferentially regulate homeostasis in expanding and neoplastic MCs compared with their terminally differentiated counterparts. These findings may have important implications for approaches aimed at selectively reducing MC burden associated with inflammation, tumorigenesis, and MC proliferative disorders. Following on the reports of the roles of mTOR in cell growth and survival in other systems,39-43 rapamycin has been investigated as a potential approach to block proliferating MCs.44 However, this may in part be because of antisurvival effects through blockade of mTORC1. The results reported in the current study demonstrate that targeting mTORC2 would provide a much more selective approach in which rapidly dividing neoplastic MCs would be preferentially targeted, whereas mature, tissue-resident MCs would be relatively unaffected.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Nathanael S. Gray (Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA) for providing Torin1, Dr Arnold S. Kirshenbaum (NIAID, NIH, Bethesda, MD) for CD34+ cells, Dr Amy Klion (NIAID) for normal BM controls, and the clinical research staff at the NIAID for their medical assistance.
Financial support for this work was provided by the Division of Intramural Research of the NIAID and the National Cancer Institute, NIH.
National Institutes of Health
Authorship
Contribution: D.S., M.-S.K., S.S., O.S., I.M., and T.M.W. conducted the experiments and/or analyzed the data; D.S., M.-S.K., and A.M.G. designed the experiments; T.M.W. and D.D.M. supervised the studies on clinical samples; S.Z., B.A.M., and W.D. generated the KI mice, genotyped and isolated samples, and provided input for the studies on these mice; D.S., T.M.W., D.D.M., and A.M.G. wrote the manuscript; and D.D.M. and A.M.G. supervised the research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alasdair M. Gilfillan, PhD, Laboratory of Allergic Diseases, NIAID, NIH, Bldg 10, Rm 11C206, 10 Center Dr, MSC 1881, Bethesda, MD 20892-1881; e-mail: agilfillan@niaid.nih.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal