Abstract
Fragments from the extracellular matrix proteins laminin and osteopontin and a sequence from VEGF have potent proangiogenic activity despite their small size (< 10 residues). However, these linear peptides have limited potential as drug candidates for therapeutic angiogenesis because of their poor stability. In the present study, we show that the therapeutic potential of these peptides can be significantly improved by “grafting” them into cyclic peptide scaffolds. Momordica cochinchinensis trypsin inhibitor-II (MCoTI-II) and sunflower trypsin inhibitor-1 (SFTI-1), naturally occurring, plant-derived cyclic peptides of 34 and 14 residues, respectively, were used as scaffolds in this study. Using this approach, we have designed a peptide that, in contrast to the small peptide fragments, is stable in human serum and at nanomolar concentration induces angiogenesis in vivo. This is the first report of using these scaffolds to improve the activity and stability of angiogenic peptide sequences and is a promising approach for promoting angiogenesis for therapeutic uses.
Introduction
Several small peptides have been reported to promote potent angiogenic activity, including peptides derived from the extracellular matrix proteins laminin and osteopontin. A hexapeptide (LAM) from the laminin α1 chain (residues 2105-2110)1 promotes angiogenesis by inducing endothelial cell adhesion, migration, and invasion,2 whereas a heptapeptide (OPN) from osteopontin (residues 162-168)3,4 interacts with α9β1 and α4β1 integrins to regulate hemopoietic stem cells and progenitor cells to promote the recovery of the damaged BM.5,6 OPN also induced angiogenesis in a mouse dorsal air sac assay.7 Mimics of VEGF have also been shown to promote angiogenesis. For example, a peptide that mimics the VEGF helix region (residues 17-25), referred to as the QK peptide, promotes proliferation of endothelial cells derived from bovine aorta and induces capillary formation in Matrigel in vitro.8,9
Pro-angiogenic agents have significant potential in the treatment of a range of conditions, including wound healing, cardiac ischemia, diabetic retinopathy, and rheumatoid arthritis.10,11 Despite this potential, clinical trials carried out with protein therapy and plasmid DNA gene therapy have not produced significant results, primarily because of stability issues.12 Peptides such as LAM, OPN, and QK offer an alternative approach for the design of novel drug leads if the intrinsic instability associated with small, unconstrained peptides can be overcome. An exciting approach to improving the stability of peptides is to take advantage of the stability of disulfide-rich cyclic peptides.13 This approach has recently been exemplified by backbone cyclization of a cone snail peptide to produce an orally active peptide with analgesic activity.14 In addition to cyclizing naturally occurring peptides, grafting small, bioactive sequences into scaffolds that are highly stable has the potential to improve stability and bioavailability.15,16 For example, the cyclotide kalata B1 has recently been used as a scaffold to stabilize a 6-residue peptide sequence with antiangiogenic activity.17 Cyclotides are a large family of plant-derived peptides that contain approximately 30 amino acids and are characterized by a head-to-tail cyclic backbone and cystine knot arrangement of 3 disulfide bonds.18 This structural motif is referred to as the cyclic cystine knot (CCK) and provides exceptional stability to cyclotides.19,20
The small size of the LAM, OPN, and QK peptides makes them ideal candidates for grafting into disulfide-rich cyclic peptide scaffolds. In the present study, we used the plant-derived peptides Momordica cochinchinensis trypsin inhibitor-II (MCoTI-II) and sunflower trypsin inhibitor-1 (SFTI-1) as stable scaffolds. Both MCoTI-II and SFTI-1 are potent trypsin inhibitors, but are structurally quite different. MCoTI-II is a 34–amino acid peptide and is a member of the cyclotide family by virtue of its CCK structural motif,21 but has no sequence similarity to kalata B1 apart from the 6 cysteine residues.22 SFTI-1 contains only a single disulfide bond and 14 amino acids.23 The structures of these frameworks and the targeted grafting sequences are shown in Figure 1. Grafting the bioactive peptide sequences into the cyclic peptide scaffolds resulted in stable and potent angiogenic agents with promising potential in the design of drug leads for therapeutic angiogenesis.
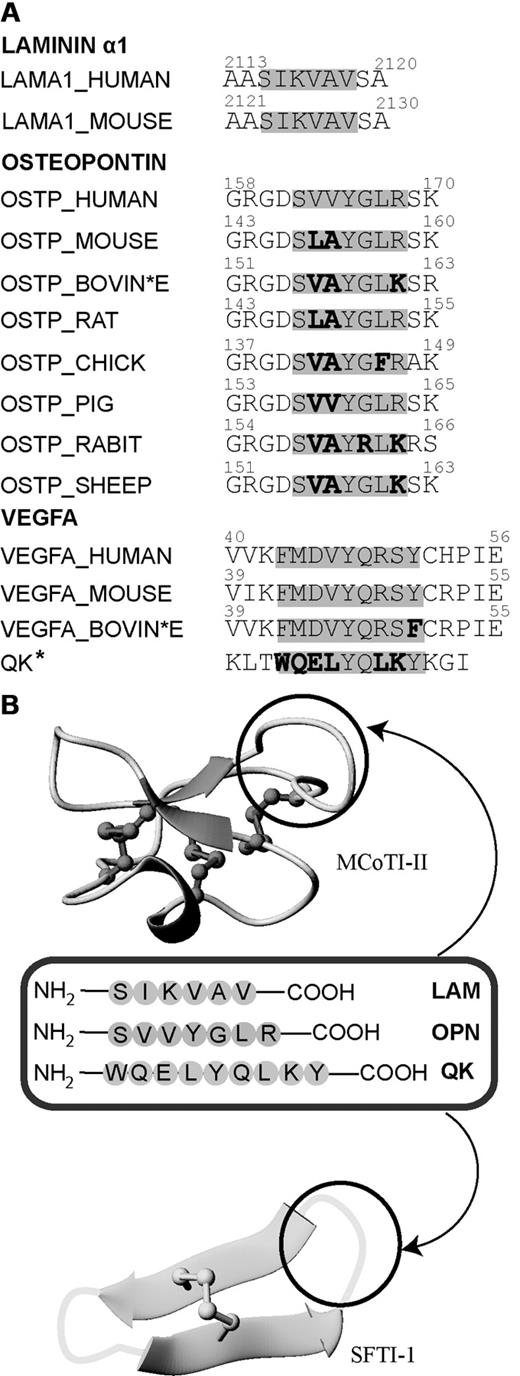
Sequence comparison of laminin α1, osteopontin, and VEGFA in various species and overview diagram of the loops where angiogenic peptides were grafted into MCoTI-II and SFTI-1 scaffolds. (A) Sequence alignment of the laminin α1, osteopontin, and QK sequences from different organisms. Active angiogenesis sequences are highlighted in gray. Nonconserved residues in the active region are bolded. The asterisk indicates that the QK peptide is a chemically designed peptide that mimics the sequence of VEGFA. (B) MCoTI-II (PDB ID: 1HA9) and SFTI-1 (PDB ID: 1JBL) used as scaffolds for grafting 3 major angiogenic sequences. Loop 6 of MCoTI-II and the trypsin-inhibitor loop of SFTI-1 are the regions for introducing pro-angiogenic sequences. These loops are circled in black.
Sequence comparison of laminin α1, osteopontin, and VEGFA in various species and overview diagram of the loops where angiogenic peptides were grafted into MCoTI-II and SFTI-1 scaffolds. (A) Sequence alignment of the laminin α1, osteopontin, and QK sequences from different organisms. Active angiogenesis sequences are highlighted in gray. Nonconserved residues in the active region are bolded. The asterisk indicates that the QK peptide is a chemically designed peptide that mimics the sequence of VEGFA. (B) MCoTI-II (PDB ID: 1HA9) and SFTI-1 (PDB ID: 1JBL) used as scaffolds for grafting 3 major angiogenic sequences. Loop 6 of MCoTI-II and the trypsin-inhibitor loop of SFTI-1 are the regions for introducing pro-angiogenic sequences. These loops are circled in black.
Methods
Peptide synthesis and isolation of MCoTI-II from M cochinchinensis seeds
All grafted peptides (MCo-LAM, MCo-OPN, SFTI-LAM, SFTI-OPN, SFTI-1, and SFTI-QK) were synthesized using manual solid-phase peptide synthesis with Boc chemistry. Peptides were constructed on PAM-Gly-Boc resin with S-tritylmercaptopropionic acid as a linker using 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate to activate the amino acids. Grafted peptides and linear peptides were synthesized on a 0.5-mmol scale. Linear peptides (linear LAM, linear OPN, and linear QK) were synthesized using Fmoc chemistry on a microwave synthesizer using chlorotrityl chloride resin. Peptides made using Boc chemistry were cleaved from the resin using hydrogen fluoride with p-cresol as a scavenger at −5 to 0°C for 1 hour. Fmoc-synthesized peptides were cleaved using a mixture of 95% trifluoroacetyl acid (TFA)/2.5% triisopropylsilyl/2.5% H2O. The TFA was removed by rotary evaporation and the residue partitioned between 50% acetonitrile in water containing 0.1% TFA and cold diethyl ether. The aqueous layer was lyophilized and the resulting crude peptides purified using reverse-phase HPLC (RP-HPLC). Native MCoTI-II peptide was isolated from a M cochinchinensis seed extract as described by Chan et al.24
Purification of peptides
Crude peptides obtained from either plant extracts or chemical synthesis were purified using a series of Phenomenex C18 columns on RP-HPLC. Gradients of 1%/min of 0%-80% solvent B (90% MeCN in 0.045% TFA in H2O) and solvent A (aqueous 0.05% TFA in H2O) were used and the eluant was monitored at 215 and 280 nm. The purity of the peptides was examined by analytical RP-HPLC on a Phenomenex Jupiter 5 μ C18 300 Å 150-mm × 2.0-mm column, and masses were determined by electrospray mass spectrometry.
Cyclization and oxidation conditions
Grafted and native SFTI-1 peptides were folded in solution at 0.1 mg/mL using a range of buffer conditions. Synthetic native SFTI-1 peptide was cyclized and oxidized in a 2-step process. Cyclization was achieved in the presence of 0.1M tris(2-carboxyethyl)phosphine, incubated for 24 hours, and the cyclic product was then purified by RP-HPLC. The cyclic reduced peptide was oxidized in 0.1M ammonium bicarbonate (pH 8.0). In contrast, cyclization and oxidation of MCoTI-II–grafted peptides were done in a single-step reaction with 0.1M ammonium bicarbonate (pH 8.5) for 24 hours. SFTI-1–grafted peptides were oxidized in 0.1M ammonium bicarbonate (pH 8.5).
NMR characterization
Peptides were dissolved at a concentration of approximately 1mM in 90% H2O/10% D2O (vol/vol). D2O (99.9%) was obtained from Cambridge Isotope Laboratories for 1H-nuclear magnetic resonance (1H-NMR) measurements. Spectra were recorded at 290 K for linear and SFTI-1–grafted peptides, whereas spectra for MCoTI-II–grafted peptides were recorded at 298 K. Spectra were recorded on Bruker Avance 500- or 600-MHz spectrometers. NOESY mixing times for linear and SFTI-1–grafted peptides were 300 milliseconds, whereas spectra recorded on MCoTI-II–grafted peptides had mixing times of 200 milliseconds. Two-dimensional spectra included TOCSY, NOESY, COSY, and ECOSY. The latter 2 spectra were only recorded for SFTI-OPN to facilitate structure determination. In addition, SFTI-OPN was dissolved in 100% D2O to determine slowly exchanging amide protons. Spectra were analyzed using SPARKY.25 The sequential assignment procedure pioneered by Wüthrich26 was used to assign the amino acids sequence specifically using TOCSY and NOESY spectra. The 3-dimensional structure of SFTI-OPN was calculated by deriving distance and angle restraints from the NOESY and COSY spectra, respectively. A family of structures that are consistent with the experimental restraints was calculated using the programs CYANA27 and CNS.28 A set of 100 structures was calculated and the 20 lowest-energy structures selected for further analysis. Structures were analyzed using the programs PROCHECK_NMR29 and PROMOTIF30 to generate statistical analyses and a Ramachandran plot. MolMol31 and PyMol32 were used to display the structural ensembles and surfaces of the peptides, respectively.
Serum-stability assay
Human serum from male AB plasma (Sigma-Aldrich) was centrifuged at 17 000g for 10 minutes for the removal of the lipid component. All peptides were tested at a final concentration of 6.6μM after dilution from a stock solution at 200μM. Supernatant was taken out and incubated for 15 minutes at 37°C before the assay. Each test peptide was diluted 1:10 with human serum except controls, which were diluted in PBS. The incubation time points were 0, 3, 8, and 24 hours at 37°C. Controls and test peptides were incubated in parallel at each time point. Serum proteins were denatured by quenching with 40 μL of 6M urea (10 minutes at 4°C), followed by the precipitation of proteins with the addition of 40 μL of 20% trichloroacetic acid (10 minutes, 4°C). These solutions were then centrifuged at 17 000g for 10 minutes. Next, 100 μL of supernatant was taken out in triplicate from the serum-treated and PBS control peptides at each time point and run on the RP-HPLC on a 0.3 mL/min Phenomenex column using a linear 1%/min gradient of 0%-50% solvent B. The elution time for each peptide was determined by the PBS control at the 0 time point. The stability at each time point was calculated as the area of the serum-treated peptide peak on RP-HPLC at 215 nm as percentage of the area of the 0-hour PBS-treated control peptides. Each experiment was done in triplicate.
Enzyme stability assay
Thrombin from human plasma (25 μg/mL of a 0.7μM concentration; Sigma-Aldrich) was prepared in 100mM sodium citrate buffer (pH 6.5). Peptide stock solutions were prepared in the same buffer at a concentration of 200μM. An equal amount of enzyme to peptide was added in an Eppendorf tube and incubated at 37°C in a water bath. Next, 5-μL aliquots were taken at 0, 1, 5, 8, and 24 hours. To each aliquot, 95 μL of 5% formic acid was added to quench the reaction. A blank was prepared by placing 95 μL of 5% formic acid in an analytical vial with 2.5 μL of enzyme added, followed by 2.5 μL of peptide. A control was also prepared by placing 95 μL of 5% formic acid in an analytical vial with only 5 μL of enzyme added. Samples were run for RP-HPLC on a 0.3 mL/min Phenomenex column using a linear 1%/min gradient of 0%-50% solvent B. The elution time for each peptide was determined by the blank at the zero time point. The stability at each time point was calculated as the area of the test peptide peak on RP-HPLC at 215 nm as a percentage of the area of the 0-hour blank. Each experiment was done in duplicate. The same method was applied to metalloproteinase-9 (MMP-9) except using 50mM Tris in 10mM CaCl2 and 150mM NaCl (pH 7.5) as the buffer.
Hemolytic assay
Human RBCs were used to assess the toxicity of the peptides against human cells. Peptide samples were prepared by 2 serial dilutions with PBS, as described previously.24
Cell culture
Rat aortic endothelial cells (RAECs) were seeded at a moderate density (2-3 × 104 cells/cm2) in RPMI medium (GIBCO-BRL) containing 10% FCS (Trace Biosciences), 25 μg/mL of endothelial cell growth promoter (Starrate), and 25 μg/mL of heparin in T75 flasks, and maintained at 37°C in an atmosphere of 5% CO2 in air. Cells were allowed to reach approximately 80% confluence before passaging. Cells were used between passages 1 and 5.
Angiogenesis sprouting assay
RAECs were allowed to form 3-dimensional spheroids in 0.5% methylcellulose (Sigma-Aldrich) in RPMI with 10% FCS within hanging drop wells overnight (1500-2000 cells per well in a 60-well plate). The base gel (1.35 mg/mL of rat-tail collagen and 0.25% methylcellulose in RPMI) was then prepared by adding 500 μL into a 12-well plate, and was allowed to set at 37°C for 30 minutes. Spheroids were collected and washed in serum-free medium, then centrifuged at 42g for 5 minutes. Another 500 μL of a second collagen gel was then added to the spheroids (with or without 10% FCS or the variable of interest), and then the gel containing the spheroids was layered on top of the base gel and allowed to set at 37°C for 30 minutes. Spheroid images were taken using an Olympus BX60 light microscope with a digital camera for baseline measurements under 100× magnification. After 24 hours, cell sprouting was observed in the cell culture and images were taken systematically for every single spheroid. Only independent spheroids were imaged. Cell sprouting was calculated by measuring 30 sprouts at evenly spaced intervals per spheroid using ImageJ 1.42g software. Results are represented as mean sprout length ± SD, and statistical comparisons were made with a Kruskal-Wallis test using Analyze-it 2.20 software. P ≤ .05 was considered significant.
CAM assay using quail egg
The chorioallantoic membrane (CAM) assay was established based on previous studies on chicken and quail eggs as the model for testing angiogenic compounds.33-37 Fertilized quail (Cortunix cortunix) eggs from a local farm in Adelaide, Australia were incubated horizontally at 37°C for 3.5 days in a humidified incubator. On day 5, eggs were sterilized with 70% EtOH/H2O spray and air-dried before opening a 0.5-cm × 0.5-cm window at the center of the egg shell. The embryo dropped by approximately 0.5 cm and the CAM with visible blood vessels were gently pulled down using tweezers to a visible area. The window was sealed with invisible tape, followed by overnight incubation at 37°C. On day 6, embryos that had survived were recorded by observing the color and different vasculature of the blood vessels. Only embryos that survived were implanted with a filter paper presoaked in a test solution. All peptides were initially tested at 50 μg/mL, and those that gave the highest activity were subsequently tested at a range of lower concentrations. VEGF solution (10 ng/mL) and RPMI 1640 medium were used as positive and negative controls, respectively. The window was resealed after the grafting procedure and further incubated at 37°C for another 24 hours. On the next day, eggs were collected from the incubator and chilled on ice for 30 minutes, followed by fixation with 25% glutaraldehyde on the implanted area for 2 hours. Finally, the implanted filter papers were trimmed off and washed with PBS in a 12-well tissue-culture plate. Quantification of blood vessels was on an Olympus SZX12 dissecting microscope with a light box under 32× magnification for blood vessel count using a hand tally counter, and images were taken under 16× magnification. DP controller Version 02.02 software (Olympus) was used to view real-time images and to capture all images. One-way ANOVA and unpaired t test were applied for the statistical comparison analysis using Prism Version 5.0 software (GraphPad). Each test peptide was compared with the negative control and results were represented as mean blood vessel count ± SD. P ≤ .05 was considered significant.
SPR
The interaction between the grafted peptides, MCo-OPN and SFTI-OPN, and the integrin α9β1 was evaluated using a Biacore 3000 surface plasmon resonance (SPR) system. The binding of the scaffolds MCoTI-II and SFTI-1 for the integrin was also evaluated for comparison. The integrin α9β1 was immobilized on the CM5 chip surface by the amine coupling method. Briefly, the dextran surface was activated by injection of a 1:1 (vol/vol) mixture of 0.4M N-ethyl-N'-(3-dimethylaminopropyl)carbodiimide and 0.1M N-hydroxysuccinimide (flow rate 5μL/min for 10 minutes). Next, 50 μg/mL of integrin α9β1 (recombinant human integrin α9β1; R&D Systems), solubilized in 10mM acetate buffer, pH 4.5, was injected over the activated surface (flow rate 5μL/min for 20 minutes). Unreacted carboxyl groups were blocked by 2 35-μL injections of 1M ethanolamine, pH 8.5. A reference channel underwent the same treatment without the integrin step. The integrin immobilization led to a signal of approximately 21 500 response units. The antibody anti-integrin α9β1 (mouse mAb clone Y9A2; Chemicon), which identifies the α9 subunit as a heterodimer in association with β1, was injected over the immobilized α9β1 integrin to evaluate whether the protein was correctly folded (100 μg/mL of anti-integrin α9β1 dialyzed and solubilized in running buffer). Anti-integrin α9β1 bound to immobilized integrin, confirming that at least a portion of the immobilized α9β1 integrin had a correct conformation. The binding of linear OPN, MCo-OPN, SFTI-OPN, MCoTI-II, and SFTI-1 was followed by peptide injection (25, 50, 100, and 200μM) for 3 minutes, followed by 600 seconds of dissociation. The sensor surface was regenerated with a short pulse of 10mM HCl, pH 2, injected for 60 seconds at 5 μL/min. All measurements were performed at 25°C with a flow rate of 5 μL/min. HEPES (10mM) with 150mM NaCl, pH 7.4, was used as running buffer and to prepare all the solutions. All solutions were freshly prepared and filtered (0.22-μm pore size). Response signals were referenced by subtracting the reference flow channel.
Results
Synthesis and characterization of grafted peptides
The sequences of the grafted peptides are shown in Figure 2, labeled with the scaffold name first followed by the code for the linear sequence (eg, SFTI-OPN). The linear peptides were synthesized by Fmoc chemistry and the cyclic peptides by Boc chemistry using a thioester-based chemistry for backbone cyclization.18,38 For the cyclic peptides, the cyclization and oxidation were generally carried out in a single step. Peptides were purified by preparative RP-HPLC and the purity determined by analytical RP-HPLC and mass spectrometry.
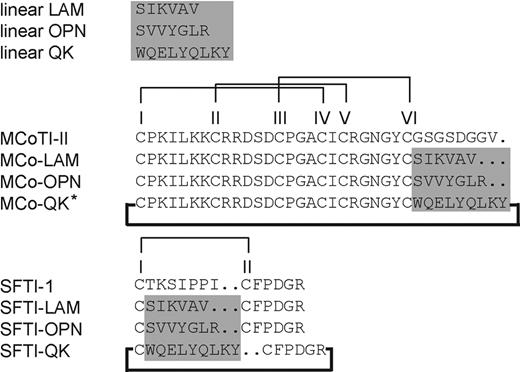
Sequences of the grafted peptides with their corresponding native cyclic peptide scaffolds. The highlighted region is the position where angiogenic sequences were grafted. All grafted peptides were successfully folded except the one labeled with an asterisk. Disulfide bond connectivities are I-IV, II-V, III-VI, and I-II for MCoTI-II- and SFTI-1–grafted peptides, respectively. The bold line indicates that all grafted and native peptides were cyclized.
Sequences of the grafted peptides with their corresponding native cyclic peptide scaffolds. The highlighted region is the position where angiogenic sequences were grafted. All grafted peptides were successfully folded except the one labeled with an asterisk. Disulfide bond connectivities are I-IV, II-V, III-VI, and I-II for MCoTI-II- and SFTI-1–grafted peptides, respectively. The bold line indicates that all grafted and native peptides were cyclized.
Because the SFTI-1–grafted peptides only contain a single disulfide bond, a single isomer was observed after cyclization and oxidation. However, there are 15 possible disulfide connectivities for the MCoTI-II–grafted peptides deriving from the 6 cysteine residues. Despite this potential heterogeneity, MCo-LAM and MCo-OPN folded into one major isomer. Analysis of the peptides with NMR spectroscopy indicated that the native fold was present for all purified peptides (supplemental Figures 1 and 2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
In contrast to the other grafted peptides, the MCo-QK peptide was poorly soluble in the buffer used for the oxidation and cyclization of MCo-LAM and MCo-OPN. A range of different solution conditions was used for MCo-QK, including varying the pH and adding organic solvents, but these solution conditions did not result in cyclization and oxidation into the native-like conformation. Therefore, this peptide was not tested in subsequent assays. However, the peptide with the QK grafted into the SFTI-1 scaffold was highly soluble and was efficiently cyclized and oxidized.
Sprouting activity of grafted peptides on rat and human endothelial cells
Gel-embedded endothelial-cell spheroids were used to measure the neovascular response of the linear sequences and grafted peptides. RAECs were used and spheroid growth induced by the grafted peptides was monitored for 24 hours. In vitro RAEC sprouting assays showed that all RAECs treated with grafted peptides displayed significant sprouting compared with the negative control and corresponding scaffolds (P ≤ .05; Figure 3A-C), except for SFTI-QK, which did not display significant sprouting compared with the negative control. Although SFTI-OPN showed a slightly lower activity compared with its linear sequence, it had a comparable activity to SFTI-LAM and with an improved activity compared with its corresponding scaffold, linear LAM and the negative control. Details of the sprouting assay results are provided in supplemental Figures 3 and 4.
RAEC assay results. (A) Grafted analogs of SFTI-1 with linear LAM and linear OPN. (B) Grafted analogs of SFTI-1 with linear QK. (C) Grafted analogs of MCoTI-II with linear LAM and linear OPN. The after peptide concentrations were: SFTI-1, linear LAM, and SFTI-LAM: 161μM; linear OPN, SFTI-OPN, and MCo-OPN: 5μM; linear QK and SFTI-QK: 0.1μM; FCS only; and negative control (medium only). Two asterisks indicate a significant change relative to the native cyclic scaffold; one asterisk indicates significance relative to negative control with P ≤ .05 and n ≥ 50. Error bars indicate SEM.
RAEC assay results. (A) Grafted analogs of SFTI-1 with linear LAM and linear OPN. (B) Grafted analogs of SFTI-1 with linear QK. (C) Grafted analogs of MCoTI-II with linear LAM and linear OPN. The after peptide concentrations were: SFTI-1, linear LAM, and SFTI-LAM: 161μM; linear OPN, SFTI-OPN, and MCo-OPN: 5μM; linear QK and SFTI-QK: 0.1μM; FCS only; and negative control (medium only). Two asterisks indicate a significant change relative to the native cyclic scaffold; one asterisk indicates significance relative to negative control with P ≤ .05 and n ≥ 50. Error bars indicate SEM.
Angiogenic activity of grafted peptides in an in vivo CAM assay
The linear sequences and grafted peptides were tested in an in vivo CAM assay using C cortunix eggs. The extent of angiogenic activity was determined by quantifying the vessels that grew toward the implant area using a dissecting microscope. Peptides were initially tested at 50 μg/mL, with VEGF used as a positive control. Among the linear sequences, linear OPN had the highest angiogenic effect, followed by linear LAM and linear QK (Figure 4A-B). SFTI-1 had no angiogenic effect in the CAM assay, but, interestingly, MCoTI-II had a significantly higher effect than the negative control, indicating intrinsic angiogenic activity for this scaffold. Grafting the LAM sequence into MCoTI-II resulted in a similar level of activity to the native peptide, but grafting the OPN sequence significantly enhanced the angiogenic activity. The SFTI-OPN graft also had the highest activity for the SFTI-1–grafted peptides (Figure 4B). A 10-fold lower concentration of SFTI-LAM and SFTI-OPN was also used in this assay. Although the blood vessel count for SFTI-LAM was significantly reduced at the lower concentration, the SFTI-OPN blood vessel count was only slightly reduced compared with its activity at the higher concentration. A further 10-fold lower concentration of SFTI-OPN (320nM) also resulted in significant angiogenic activity. A comparison of SFTI-OPN and the linear OPN sequence at the same concentration indicated a significant increase in activity upon grafting into the cyclic peptide scaffold (Figure 4C). Overall, the OPN sequence was the most effective sequence studied, with SFTI-OPN having the highest and MCo-OPN the second highest angiogenic effect. Further details of the CAM assays are given in supplemental Figures 5 and 6.
CAM assay results. (A) Comparison of blood vessel count of the linear and MCoTI-II–grafted peptides. (B) Comparison of blood vessel count of the linear and SFTI-1–grafted peptides. (C) Comparison of blood vessel count with a range of concentrations of linear OPN and SFTI-OPN. Comparison of blood vessel count of the linear OPN and SFTI-OPN. Each group of test and control peptides are n = 12 except VEGF, which is n = 9, and SFTI-OPN 0.5 μg/mL, which is n = 6. Linear angiogenic sequences were grouped as “a” and all grafted peptides were grouped as “b.” Different batches of C cortunix eggs were used for this assay. The following peptide concentrations were tested: VEGF: 0.26nM; linear LAM: 81.2μM; linear OPN: 63.1μM; linear OPN: 32μM; linear OPN 3.2μM; linear QK: 39.4μM; MCoTI-II: 14.5μM; MCo-LAM: 14.6μM; MCo-OPN: 13.9μM; SFTI-1: 33μM; SFTI-LAM: 36.4μM; SFTI-LAM: 3.6μM; SFTI-OPN: 32μM; SFTI-OPN: 3.2μM; SFTI-OPN: 320nM; and SFTI-QK: 24.6μM. All means are significantly different. *P ≤ .05 versus control (medium). Error bars indicate SEM. A red line in each of the bar graph indicates the baseline for a peptide to be considered as having angiogenic activity.
CAM assay results. (A) Comparison of blood vessel count of the linear and MCoTI-II–grafted peptides. (B) Comparison of blood vessel count of the linear and SFTI-1–grafted peptides. (C) Comparison of blood vessel count with a range of concentrations of linear OPN and SFTI-OPN. Comparison of blood vessel count of the linear OPN and SFTI-OPN. Each group of test and control peptides are n = 12 except VEGF, which is n = 9, and SFTI-OPN 0.5 μg/mL, which is n = 6. Linear angiogenic sequences were grouped as “a” and all grafted peptides were grouped as “b.” Different batches of C cortunix eggs were used for this assay. The following peptide concentrations were tested: VEGF: 0.26nM; linear LAM: 81.2μM; linear OPN: 63.1μM; linear OPN: 32μM; linear OPN 3.2μM; linear QK: 39.4μM; MCoTI-II: 14.5μM; MCo-LAM: 14.6μM; MCo-OPN: 13.9μM; SFTI-1: 33μM; SFTI-LAM: 36.4μM; SFTI-LAM: 3.6μM; SFTI-OPN: 32μM; SFTI-OPN: 3.2μM; SFTI-OPN: 320nM; and SFTI-QK: 24.6μM. All means are significantly different. *P ≤ .05 versus control (medium). Error bars indicate SEM. A red line in each of the bar graph indicates the baseline for a peptide to be considered as having angiogenic activity.
Stability of native, linear, and grafted peptides
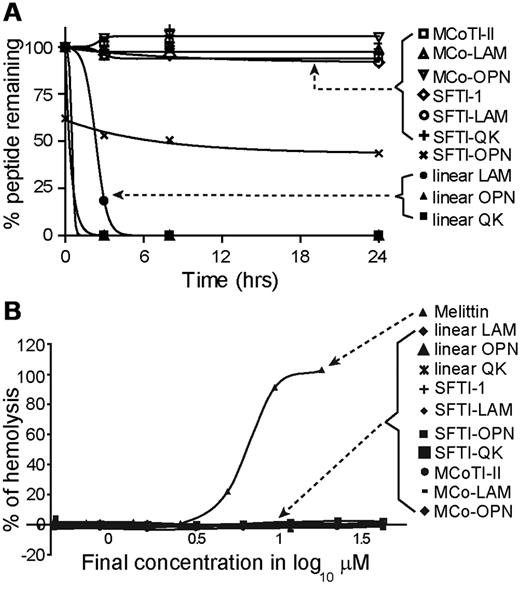
The native, linear, and grafted peptides were incubated in human serum to investigate their stability. All grafted peptides were stable in human serum, with 100% peptide remaining after 24 hours, except for SFTI-OPN, which degraded to approximately 40% of the initial peptide concentration within the first 4 hours and then remained unchanged for the rest of the assay (Figure 5A). The results observed for SFTI-OPN might be related to binding to serum proteins, resulting in an apparent decrease in peptide recovery. In contrast, all linear peptides were significantly degraded within the first 3 hours. Although SFTI-OPN was not as stable as the other grafted peptides, it is more stable than its linear counterpart. Given that the SFTI-OPN has the highest angiogenic activity among the grafted peptides, the stability of this peptide was further tested against thrombin and MMP-9, enzymes involved in angiogenesis. MCoTI-II was also tested in these enzyme-stability assays. It was found that SFTI-1, SFTI-OPN, and linear OPN were stable to thrombin over 24 hours whereas N-benzoyl-Phe-Val-Arg-p-nitroanilide, a known substrate for thrombin, was cleaved. However, in the MMP-9–stability assay, linear OPN was gradually cleaved by the enzyme, whereas SFTI-1, SFTI-OPN, and MCoTI-II were found to be stable over 24 hours (supplemental Figure 7).
Hemolytic and serum-stability assay results. (A) Percentage of peptide remaining in human serum after 24 hours. (B) Percentage of hemolysis of peptides tested in this assay. Melittin was used as a positive control. The data are shown as means ± SD.
Hemolytic and serum-stability assay results. (A) Percentage of peptide remaining in human serum after 24 hours. (B) Percentage of hemolysis of peptides tested in this assay. Melittin was used as a positive control. The data are shown as means ± SD.
Hemolytic activity of all native, linear analogs, and grafted peptides
The hemolytic activity of all peptides was tested in human blood. PBS and Triton-X 100 were used in the assay as positive and negative controls, respectively. Melittin, a highly hemolytic honey bee venom peptide,39 was used as a positive control. None of the grafted peptides exhibited hemolytic activity, as shown in Figure 5B.
Solution structural analysis of SFTI-OPN
To gain an insight into the structure-activity relationships of the grafted angiogenic peptides, the 3-dimensional structure of SFTI-OPN, the most potent grafted peptide was determined. Two conformations were observed for this peptide, most likely as a result of cis or trans isomerization of Pro 13. The αH secondary shifts of the 2 conformations are given in the supporting information (supplemental Figure 1). The αH chemical shift of the trans isomer of SFTI-OPN indicated that the nongrafted region has a similar structure to the native peptide. The ratio of the trans and cis conformations was 70:30 in the NMR spectra and the structure was determined for the predominant trans conformation of SFTI-OPN.
A set of 100 structures was calculated using 98 distance restraints comprising 39 intra-residue, 43 sequential, 8 medium-range, 8 long-range, and 5 dihedral restraints, which were derived from the NMR data (Table 1). The 15 lowest-energy structures, shown in Figure 6A, are in good agreement with the experimental data, show no dihedral angle violations exceeding 3° and no distance violations more than 0.3 Å (Table 1). The root-mean-square deviation over the well-defined region, residues 1-3 and 11-14, is 0.55 ± 0.33 Å for the backbone atoms. The secondary structure of SFTI-OPN was analyzed by PROMOTIF,30 and revealed that a β-hairpin is the major element of secondary structure. The residues comprising the 2 strands are 2-5 and 8-12. A type IV β-turn connects the 2 strands. These structural findings are consistent with the αH secondary shifts analysis, which suggested that the grafted region between Ser4 and Arg10 was disordered (Figure 6A). D2O slow-exchange experiments indicated that there were no slowly exchanging amide protons and therefore no hydrogen bonds to stabilize the structure. A comparison of SFTI-1 with SFTI-OPN is given in Figure 6A and B, indicating that the overall fold is similar between the 2 peptides. A surface representation highlights the distribution of hydrophobic residues in SFTI-OPN and native SFTI-1. SFTI-OPN has a larger hydrophobic surface area in the grafted region. The structural data for SFTI-OPN have been deposited in the Biologic Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu/) with the accession number 17426.
NMR data and refinement statistics for SFTI-OPN
| NMR constraints . | Values . |
|---|---|
| Distance restraints | 98 |
| Long range | 8 |
| Medium range | 8 |
| Sequential | 43 |
| Intra residual restraints | 39 |
| Dihedral restraints | 5 |
| NOE violations > 0.3 Å | 0 |
| Dihedral violations > 3.0° | 0 |
| Energies (kJ mol−1) | |
| Overall | −379.24 ± 12.79 |
| Bonds | 3.40 ± 0.56 |
| Angles | 20.47 ± 3.76 |
| Improper | 2.32 ± 0.57 |
| van der Waals | −7.75 ± 7.34 |
| NOE | 7.28 ± 4.76 |
| cDIH | 0.17 ± 0.13 |
| Dihedral | 57.09 ± 9.47 |
| Electrostatic | −462.23 ± 20.13 |
| r.m.s.d. | |
| Bonds (Å) | 0.0039 ± 0.000 32 |
| Angles (°) | 0.58 ± 0.059 |
| Improper (°) | 0.35 ± 0.044 |
| NOE (Å) | 0.037 ± 0.011 |
| cDIH (°) | 0.78 ± 0.29 |
| Ramachandran analysis (%; residue 1-3, 11-14)† | |
| Most favoured | 80.0 |
| Additionally allowed | 20.0 |
| Generously allowed | 0 |
| Disallowed | 0 |
| Average pairwise r.m.s.d. (Å)* | |
| Backbone | 1.59 ± 0.79 |
| Heavy atoms | 2.93 ± 0.99 |
| Backbone (residue 1-3, 11-14) | 0.55 ± 0.33 |
| Heavy atoms (residues 1-3, 11-14) | 1.44 ± 0.44 |
| NMR constraints . | Values . |
|---|---|
| Distance restraints | 98 |
| Long range | 8 |
| Medium range | 8 |
| Sequential | 43 |
| Intra residual restraints | 39 |
| Dihedral restraints | 5 |
| NOE violations > 0.3 Å | 0 |
| Dihedral violations > 3.0° | 0 |
| Energies (kJ mol−1) | |
| Overall | −379.24 ± 12.79 |
| Bonds | 3.40 ± 0.56 |
| Angles | 20.47 ± 3.76 |
| Improper | 2.32 ± 0.57 |
| van der Waals | −7.75 ± 7.34 |
| NOE | 7.28 ± 4.76 |
| cDIH | 0.17 ± 0.13 |
| Dihedral | 57.09 ± 9.47 |
| Electrostatic | −462.23 ± 20.13 |
| r.m.s.d. | |
| Bonds (Å) | 0.0039 ± 0.000 32 |
| Angles (°) | 0.58 ± 0.059 |
| Improper (°) | 0.35 ± 0.044 |
| NOE (Å) | 0.037 ± 0.011 |
| cDIH (°) | 0.78 ± 0.29 |
| Ramachandran analysis (%; residue 1-3, 11-14)† | |
| Most favoured | 80.0 |
| Additionally allowed | 20.0 |
| Generously allowed | 0 |
| Disallowed | 0 |
| Average pairwise r.m.s.d. (Å)* | |
| Backbone | 1.59 ± 0.79 |
| Heavy atoms | 2.93 ± 0.99 |
| Backbone (residue 1-3, 11-14) | 0.55 ± 0.33 |
| Heavy atoms (residues 1-3, 11-14) | 1.44 ± 0.44 |
Average pairwise r.m.s.d. was calculated among 15 refined structures for mean global backbone and mean global heavy atoms.
Well-defined residues were chosen for analysis.
Structure analysis of SFTI-1 and SFTI-OPN. (A) The surface diagram, ribbon representation, and 15 lowest-energy structures of SFTI-OPN. (B) The surface diagram, ribbon representation, and 20 lowest-energy structures of native SFTI-1. Hydrophobic residues are labeled and highlighted in dark gray. These diagrams were created in MolMol.31
Structure analysis of SFTI-1 and SFTI-OPN. (A) The surface diagram, ribbon representation, and 15 lowest-energy structures of SFTI-OPN. (B) The surface diagram, ribbon representation, and 20 lowest-energy structures of native SFTI-1. Hydrophobic residues are labeled and highlighted in dark gray. These diagrams were created in MolMol.31
Grafted peptides bind to integrin α9β1
The integrin α9β1 has been shown to recognize the angiogenic heptapeptide OPN, and this binding appears to be correlated with OPN-induced angiogenesis.3 SPR studies were performed to evaluate whether the activity of the new pro-angiogenic peptides grafted with the OPN sequence could be recognized by α9β1. The binding of SFTI-OPN and MCo-OPN, and their corresponding scaffolds, to immobilize integrin α9β1 was compared. Injection of SFTI-OPN (Figure 7A) or MCo-OPN (Figure 7B) over α9β1 clearly showed binding of the grafted peptides with the immobilized integrin, whereas the corresponding scaffolds displayed no binding in the concentration range tested (Figure 7A-B). The linear OPN also appeared to show no binding under the conditions tested.
Interaction of cyclic scaffolds versus OPN-grafted peptides with immobilized α9β1 integrin. (A) SPR sensorgrams of SFTI-1 and SFTI-OPN. (B) SPR sensorgrams of MCoTI-II and MCo-OPN. SFTI-OPN and MCo-OPN bound to the integrin, in contrast to SFTI-1 and MCoTI-II.
Interaction of cyclic scaffolds versus OPN-grafted peptides with immobilized α9β1 integrin. (A) SPR sensorgrams of SFTI-1 and SFTI-OPN. (B) SPR sensorgrams of MCoTI-II and MCo-OPN. SFTI-OPN and MCo-OPN bound to the integrin, in contrast to SFTI-1 and MCoTI-II.
Discussion
Stimulation of blood vessel regeneration after cardiac ischemia is a promising therapeutic strategy, and clinical studies have started to assess the potential of angiogenic agents.11 Although small peptide sequences have been shown to have potent angiogenic activity, these peptides are generally easily degraded in vivo. To facilitate the delivery of bioactive peptides to specific target sites, one approach is the use of cyclic peptide scaffolds. This approach is based on the intrinsic stability of these scaffolds,13,40 and we have shown in the current study that grafting angiogenic sequences into them significantly enhances stability. In the case of the OPN peptide, this led to a grafted peptide with nanomolar angiogenic activity.
The cyclic peptide scaffolds used in this study, MCoTI-II and SFTI-1, have significantly different structures despite both having a cyclic backbone. The loops in the cyclic peptide scaffolds chosen for grafting were based primarily on structural considerations. Loop 6 of MCoTI-II had been shown previously to be disordered in solution,21,41 and therefore it was hypothesized that it would be amenable to changes in sequence. The trypsin-inhibitory loop of SFTI-1 is larger than the secondary loop and therefore was more likely to accommodate a larger bioactive sequence. Despite the differences between the scaffolds, all of the grafted peptides except MCo-QK folded with a high yield of the native isomer.
Characterization of the grafted peptides with NMR spectroscopy was valuable for determining whether the native fold had been maintained, particularly for the MCoTI-II–grafted peptides, which have more complex structures than the SFTI-1–grafted peptides. Given that both MCo-LAM and MCo-OPN were able to maintain the native conformation, it is clear that loop 6 is not critical in maintaining the structure of MCoTI-II and therefore is a useful site for introducing foreign sequences into this cyclic peptide scaffold.
Analysis of the angiogenic activity of peptides using a sprouting assay with RAECs provided insights into the angiogenic responses of native, linear, and grafted peptides in vitro. From the RAEC assay, most of the grafted analogs were shown to have significant sprouting compared with the native scaffolds and negative controls. This result shows that once the linear bioactive sequences are grafted into their corresponding cyclic scaffolds, the sequences are able to adopt a conformation in the newly constrained structure that can interact with their receptors on endothelial cells to provoke cell proliferation and movement toward the extracellular matrix. SFTI-QK was found to have a lower neovascular response than its linear counterpart and no significant difference to the negative control, which might be because of distortion of the native helical conformation of linear QK after constraining into the SFTI-1 scaffold. Furthermore, a truncated version of the original full-length sequence of QK peptide was grafted into the SFTI-1 scaffold, which might account for the lower angiogenic activity than previously reported for the QK peptide alone.8
Because in vitro assays are only indicative, an in vivo CAM assay was carried out to further characterize the angiogenic potential of the grafted peptides. A clearer differentiation of the angiogenic effect was observed between the linear sequences and grafted peptides in the CAM assay compared with the sprouting assays. All grafted peptides showed a significant increase in activity compared with the linear sequences, with SFTI-OPN exhibiting the highest angiogenic activity, followed by MCo-OPN, SFTI-LAM, MCo-LAM, and SFTI-QK. MCoTI-II showed a slight increase in angiogenesis activity relative to the negative control, indicating weak intrinsic angiogenic activity of the scaffold. There is no sequence similarity between MCoTI-II and the linear sequences to account for this activity, and detailed structure-activity studies would be required to dissect out the features responsible for this activity. However, the intrinsic activity was weak, and grafting the OPN sequence into the MCoTI-II framework significantly enhanced the angiogenic effect. Overall, both the SFTI-1- and MCoTI-II–grafted peptides were able to maintain or increase the angiogenic activity of all linear sequences. Interestingly, SFTI-OPN has a disordered loop and it is possible that this disorder is related to the enhanced potency of this peptide compared with the other peptides. A similar result was observed when an antiangiogenic peptide sequence was grafted into kalata B1. The peptide with the greatest bioactivity had significantly more structural disorder than the other grafted peptides.17
Analysis of the binding of SFTI-OPN and MCo-OPN to integrin α9β1 using SPR confirmed that the grafted cyclic peptides bind to this receptor. Linear OPN has been shown previously to interact with integrin α9β1 in cell-based assays,3 and the OPN protein was shown to interact via SPR.3 Therefore, it appears that the mechanism of action is maintained in the grafted peptides. Interestingly, in the present study, we found that linear OPN peptide does not bind to the integrin α9β1 under the experimental conditions. The response in SPR is molecular mass dependent, but, given that the linear OPN and SFTI-OPN only differ by a factor of 2 in molecular mass, the lack of binding observed for linear OPN relative to the strong response observed for SFTI-OPN suggests that the binding is significantly enhanced for the grafted cyclic peptide. MCo-OPN displays a slower dissociation rate than SFTI-OPN, indicating stronger binding and therefore suggesting that it might be a better candidate for drug development.
In addition to potent activity, biologic and chemical stability are important in the design of novel drug leads. Plasma stability is an essential factor that affects the half-life of peptides, and therefore all of the grafted peptides were tested in a serum-stability assay. All of the linear sequences showed improvement in stability after grafting into the cyclic scaffolds. In addition, MCo-OPN and SFTI-OPN were stable against the enzymes thrombin and MMP-9. Therefore, it is possible that the enhancement in angiogenic activity observed in the CAM assays might be related to this increase in stability. However, enhanced binding of SFTI-OPN and MCo-OPN to integrin α9β1 relative to linear OPN might also contribute to the enhanced potency.
Previous studies have shown that MCoTI-II and SFTI-1 can be used to design peptides with nonnative bioactivities, although until now, these have been restricted to protease activities. For example, Thongyoo et al42 showed that the active site (loop 1) of MCoTI-II is capable of accommodating a bioactive sequence that leads to the inhibition of a protease from the foot and mouth disease virus (FMDV). In addition, SFTI-1 has been modified to design a peptide that selectively inhibits human kallikrein-related peptidase 4 (KLK4) with potential applications in the treatment of prostate cancer.43 Overall, these peptide scaffolds show significant promise in pharmaceutical applications, and the present study has expanded their potential from simply being involved in protease interactions. Furthermore, we have shown that not only are these scaffolds useful for stabilizing bioactive sequences, but that bioactivity can be enhanced as a result of being constrained in the cyclic peptide scaffolds, making this approach extremely valuable for drug design.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a grant from the National Health and Medical Research Council (NHMRC; grant ID 401600). S.T.H. is a Marie Curie International Outgoing Fellow (PIOF-GA-2008-220318). D.J.C. is an NHMRC Professorial Research Fellow. R.J.C. is an Australian Research Council Future Fellow. N.L.D. is a Queensland Smart State Fellow.
Authorship
Contribution: L.Y.C., D.J.C., J.H.C., and N.L.D. designed the research; L.Y.C., S.G., N.F.W., S.-J.L., R.J.C., and S.T.H. performed the research; L.Y.C., S.G., N.L.D., J.H.C., and S.T.H. analyzed the data; and L.Y.C., D.J.C., and N.L.D. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Norelle L. Daly, The University of Queensland, Institute for Molecular Bioscience, Brisbane, QLD 4072, Australia; e-mail: n.daly@uq.edu.au.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal