Abstract
Monoclonal B-cell lymphocytosis (MBL) is classified as chronic lymphocytic leukemia (CLL)–like, atypical CLL, and CD5− MBL. The number of B cells per microliter divides CLL-like MBL into MBL associated with lymphocytosis (usually detected in a clinical setting) and low-count MBL detected in the general population (usually identified during population screening). After a median follow-up of 34 months we reevaluated 76 low-count MBLs with 5-color flow cytometry: 90% of CLL-like MBL but only 44.4% atypical CLL and 66.7% CD5− MBL persisted over time. Population-screening CLL-like MBL had no relevant cell count change, and none developed an overt leukemia. In 50% of the cases FISH showed CLL-related chromosomal abnormalities, including monoallelic or biallelic 13q deletions (43.8%), trisomy 12 (1 case), and 17p deletions (2 cases). The analysis of the T-cell receptor β (TRBV) chains repertoire showed the presence of monoclonal T-cell clones, especially among CD4highCD8low, CD8highCD4low T cells. TRBV2 and TRBV8 were the most frequently expressed genes. This study indicates that (1) the risk of progression into CLL for low-count population-screening CLL-like MBL is exceedingly rare and definitely lower than that of clinical MBL and (2) chromosomal abnormalities occur early in the natural history and are possibly associated with the appearance of the typical phenotype.
Introduction
The presence of monoclonal B-cell expansions in the peripheral blood (PB) of asymptomatic persons has long been known.1 Multiparameter flow cytometry2,3 recently allowed to recognize a preclinical hematologic condition characterized by small B-cell clones detectable in the PB of otherwise healthy persons and named monoclonal B-cell lymphocytosis (MBL).1,4 Most (75%) MBLs have the phenotype (CD5+, CD23+, CD20low, sIglow) of chronic lymphocytic leukemia (CLL-like MBL). The remaining MBLs are classified as atypical CLL (CD5+, CD20bright) and CD5-negative (CD5−) MBL.1,4 The prevalence of CLL-like MBL strikingly increases with age, reaching a frequency > 20% in persons > 60 years5,6 and being also more frequent among relatives of patients affected by CLL.7-10
Because among the elderly the MBL frequency is 100 times higher than that of CLL, MBL might represent a “normal” aspect of the immune system, especially of the immunosenescence process. Therefore, the central problem of CLL-like MBL is to define how it relates to CLL,11 specifically to understanding which are the molecular features12-14 that make them a potential preleukemic condition12 and which is their clinical effect.
The International Workshop on Chronic Lymphocytic Leukemia guidelines15 indicate that the diagnosis is CLL-like MBL and not yet stage 0 CLL when the number of circulating clonal B cells is below the threshold of 5 × 109 cells/L. This brings in the issue that, within MBL, the number of circulating clonal B cells can be heterogeneous, and CLL-like MBL may be further divided into 2 different general subgroups, clinical MBL and population-screening MBL.4,12 Clinical MBL is usually diagnosed in a clinical setting, is associated with lymphocytosis, and has a concentration of clonal B cells > ∼ 1500/μL.13 On the contrary, population-screening MBL, also defined “low-count MBL,”14 is only detected during screening studies of healthy persons in the general population with the use of highly sensitive technical procedures and is characterized by < ∼ 50 clonal B cells/μL.4 The median absolute count of clonal B cells in population-screening MBL is 1 cell/μL, and in 75% of the cases the count is < 3 cells/μL.12 These numbers strikingly differ from the median concentration of clonal B cells in clinical MBL, which is ∼ 3000/μL, thus representing the majority of lymphocytes. Although the potential risk of progression of clinical MBL (previously defined MBL with lymphocytosis) into clinically overt CLL is ∼ 1.1% per year,13 limited data are available on the outcome of low-count population-screening MBL even if it is the most common form in the general population and the prevalence is ≥ 100-fold than the prevalence of CLL. Conceivably, the progression of population-screening MBL into CLL is uncommon,16 and most might simply be an epiphenomenon of immunosenescence, that is, an intensification of the progressive restriction of Ig and TCR repertoire observed in the elderly.17-20 It becomes therefore important to investigate which molecular and biologic features of low-count population-screening MBL are associated with the risk of progression to focus medical attention onto the rare risky subjects and to refrain from useless and prolonged monitoring most of the otherwise healthy persons.
To this end we took advantage of our cohort of 137 MBL cases, detected during a population screening of 1779 healthy persons > 18 years of age.14 More than one-half of the originally cases of diagnosed MBL were reevaluated after a median follow-up of ∼ 3 years. We here report that 90% of CLL-like MBL clones persisted over time, whereas atypical CLL and CD5− CLL MBL tended to be transient. None of the low-count population-screening MBLs developed an overt leukemia even if they were found to carry 13q deletions with the same frequency as CLL.21 This suggests that 13q deletion occurs early in the natural history of MBL/CLL and is probably associated with the typical CLL phenotype rather than with the progression into an overt disease.
Methods
Study population
The baseline study was conducted between 2005 and 2008 and enrolled 1779 healthy persons from Val Borbera, a rural valley in Northern Italy that is isolated from the surrounding areas by geographic barriers.14 The whole population is currently involved in a study not aimed at elucidating any particular health problem but rather at dissecting potential genetic components of common diseases because it is probable to derive from a limited number of common ancestors (ie, “genetically isolated”).
Among the initial participants we identified 138 cases of MBL; 1 male was affected by a splenic marginal zone lymphoma whose leukemic phase was detected by our analysis and was subsequently excluded from the study (final prevalence, 137 of 1779; 7.7%). Of the 137 originally diagnosed MBL subjects, 76 persons participated to a second visit after a median follow-up of 34 months (range, 11-50 months). Forty-five persons refused to take part in the follow-up visit, and 16 subjects were deceased, for unrelated causes, at the time of the second evaluation. The persons studied were > 18 years of age with a median age of 66 years (range, 25-92 years); 29 were women and 47 men. The study population has been carefully investigated from the clinical and genealogic point of view during the previous study. Clinical records were collected on all 76 persons who underwent routine blood tests, including a complete blood count with differential. No person had a known history of CLL. The follow-up evaluation was performed over 1 year (2009).
The research protocol was approved by the Institutional Ethics Committee at San Raffaele Scientific Institute, Milano, Italy, and all participants gave written informed consent in accordance with the Declaration of Helsinki.
Blood samples, cell preparation, staining, and FACS analysis
EDTA PB samples obtained from all persons enrolled were processed within 24 hours after blood withdrawal as previously described.14 The following antibody mixes were used: FITC-conjugated F(ab)2–anti-κ, PE–anti-λ light chain (Dako), PE-cyanin7 (Cy7)–labeled anti-CD20, PE-cyanin5 (Cy5)–conjugated anti-CD5 and PE-Texas Red–conjugated anti-CD19 (Beckman Coulter). For each sample, up to 500 000 events were acquired on a FC500 equipped with 488 argon ion laser and 635 red HeNe laser and analyzed with the CXP software system (Beckman Coulter) according to the gating strategy previously described.14 MBL has been divided into CLL-like MBL, atypical CLL MBL, and CD5− MBL, based on the immunophenotypic profile.1,4 The latter categories are based on the occurrence of an unbalanced κ/λ ratio (> 3:1 or < 1:3) within CD5+ or CD5− B lymphocytes, respectively. In contrast CLL-like MBL is defined on the basis of the distinct CD5brightCD20dim expression pattern on CD19+ B cells.14 Of the 54 persistent CLL-like MBL clones, 43 cases (79.6%) were clearly monoclonal, expressing κ light chains in 37 cases (68.5%) and λ light chain in 6 cases (11.1%); in 5 cases (9.3%) surface light chain expression was not detectable, this being suggestive of monoclonality,14 whereas the 6 remaining cases (11.1%) were polyclonal.14
T-cell study
To study lymphocytes subpopulations the following antibodies mix has been used: CD25 FITC-labeled, CD4 PE-conjugated, and CD16 PE-Cy5–conjugated mixed with CD56 PE-Cy5–conjugated (Beckman Coulter). For each sample, up to 100 000 events were acquired and analyzed on FC500.
To study the repertoire of the TCR variable genes of the β chain (TRBV) used by T lymphocytes circulating in the blood of persons with CLL-like MBL, PB mononuclear cells have been stained with the “Io test Beta Mark TCR Vβ Repertoire kit” (Immunotech, Beckman Coulter), and a mix of PE-Cy5–conjugated anti-CD4 and PE-Cy7–labeled anti-CD8 (Beckman Coulter). T-cell subsets have been analyzed according to the previously described gating strategy, and a restricted TRBV expression was considered when the expression of a single TRBV chain was higher than the mean value plus 3 SDs (generally > 20%).17
For quality control purposes, we daily used 0.4 mL of Flow-Check Fluorospheres (Beckman Coulter) mixed with 0.2 mL of Flow-Check 770 (Beckman Coulter PC7 [770/488] Setup Kit) to assess flow cytometric optical alignment and fluidics system. In addition, we daily controlled light scatter intensity, fluorescence intensity, and hydrodynamics with the use of 0.4 mL of Flow-Set Fluorospheres (Beckman Coulter) mixed with 0.2 mL of Flow-Set 770 (Beckman Coulter PC7 [770/488] Setup Kit) to assess optimal conditions for quantitative analysis of human leukocytes.
PCR amplification of IGHV-D-J rearrangements and sequence analysis
IGHV-D-J rearrangements have been analyzed in selected samples after DNA extraction, PCR amplification, and nucleotide sequencing as previously described.14 Systematic resequencing of Ig genes has not been performed because of paucity of material, which was mainly used to perform FISH analysis. A monoclonal IGHV-D-J rearrangement was determined in 3 additional cases in addition to those previously published14 : case VB094, expressing IGHV4-59/61 (95.6% of identity to germ line), case VB266, expressing IGHV3-21 (89.1% of identity); and case VB1698 expressing IGHV3-15 (93.0% of identity) genes, respectively.
FACS and FISH
CLL-like MBL cells have been purified on High Speed Sorter MoFLo (Dako). The following antibody mixes were used: PE-Cy7–labeled anti-CD20, PE-Cy5–conjugated anti-CD5 and PE-Texas Red–conjugated anti-CD19 (Beckman Coulter).
Sorted CD5+CD20dim monoclonal B cells underwent interphase FISH analysis after fixation in methanol/acetic acid (3:1), using standard protocols.22,23 The FISH panel included probes for the detection of trisomy 12, deletions of 11q22.3 (ATM), 13q14.3 (D13S319), 13q34 (LSI13q34), and 17p13 (TP53) (Vysis LSI p53/LSI ATM and LSI D13S319/LSI 13q34/CEP 12 Multi-color Probe; Abbott Molecular). Nikon Eclipse 90i microscope (Nikon Instruments) and a Qicam Fast 1394 (Qimaging) camera were used. Acquisition was done with a Genikon (Nikon Instruments S.p.a. Italy) software with the use of a FISH 3D Acquisition Module. When possible, FISH signals for each probe were scored in ≥ 50 interphase nuclei. Cutoffs were set on mean values ± 2 SDs, derived from analysis on healthy donor sorted B cells: Del17p, 6.7% ± 2.2%; Del11q, 3.4% ± 4.6%; Del13q14.3, 4.9% ± 10.8%; Del13q34, 5.4% ± 15.5%; and trisomy 12, 0.7% ± 2.3%.
Statistical methods
Median, mean, SD, maximum, minimum, 25th and 75th percentiles (for continuous variables), and relative frequencies (for categorical variables) were calculated with SPSS software (SPSS 16.0; SPSS Inc). To define statistically significant differences between subgroups, Mann-Whitney test and Fisher exact test were used for continuous and categorical variables, respectively. All P values were 2-sided and regarded as statistically significant only if < .05.
Results
Low-count population-screening CLL-like MBL persists over time
Between 2005 and 2008 we enrolled 1779 healthy persons, and flow cytometry detected the presence of MBL in 137 subjects (128 previously described14 ), 96 CLL-like MBL, 21 atypical-CLL MBL, and 20 CD5− MBL.1
After a median follow-up of 34 months (range, 11-50 months), we reanalyzed the PB of 76 subjects (47 men, 29 women; median age, 66 years; range, 25-92 years). Sixteen subjects with MBL had died of unrelated reasons, and the remaining subjects were not available for the follow-up study. All subjects had a normal absolute white blood cell count (mean value, 7.0 × 109 cells/L; range 2.7-12.8 × 109 cells/L) and a normal absolute lymphocyte count (ALC; mean value, 2.4 × 109 cells/L; range, 0.8-3.9 × 109 cells/L), except for 4 cases whose lymphocyte counts were slightly above the value of 4.0 × 109 cells/L (range, 4.1-5.3 × 109 cells/L). The mean absolute B-cell count was 184.5 × 106 cells/L (range, 18.2-1508.8 × 106 cells/L). Among the 76 reevaluated subjects, 60 originally had a CLL-like MBL, 9 an atypical-CLL MBL, and 9 a CD5− MBL. Two subjects with MBL had a double clone.
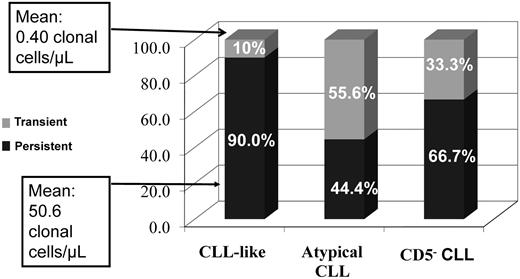
At reevaluation 54 of 60 CLL-like MBL (90.0%), but only 4 of 9 atypical-CLL MBL (44.4%) and 6 of 9 CD5− MBL (66.7%) clones were still detectable in the same original carriers (Figure 1). The 6 transient CLL-like MBL clones were originally present in small numbers (median concentration of aberrant B cells, 0.40/μL) close to the detection limit of the acquisition protocol used. The 2 subjects with double clones were reconfirmed.
CLL-like MBL remains stable over time. Percentages of persistent or transient clones at follow-up analysis in CLL-like, atypical-CLL, and CD5− MBL. At the first evaluation the mean clonal B-cell count of persistent CLL-like MBL was 50.6 cells/μL, whereas transient CLL-like clones had a mean value of 0.40 cells/μL.
CLL-like MBL remains stable over time. Percentages of persistent or transient clones at follow-up analysis in CLL-like, atypical-CLL, and CD5− MBL. At the first evaluation the mean clonal B-cell count of persistent CLL-like MBL was 50.6 cells/μL, whereas transient CLL-like clones had a mean value of 0.40 cells/μL.
Among the persistent MBL, 61 of 64 (95.3%) had a normal ALC (mean value, 2.4 × 109 cells/L; range, 0.8-3.9 × 109 cells/L). In the remaining 3 cases, ALCs were slightly above the 4.0 × 109 cells/L value (4.1, 4.3, and 5.3 × 109 cells/L). The mean absolute B-cell count of the persistent MBL was 190.8 × 106 cells/L (range, 21.6-1508.8 × 106 cells/L), and clonal B cells had a mean value of 62.3 × 106 cells/L (range, 0.2-1249.3 × 106 cells/L).
Population-screening CLL-like MBL remains stable over time
Among the 54 CLL-like persistent cases, 35 were men and 19 were women, with a median age of 66 years (range, 40-92 years). One case (VB094) was excluded from further analysis because of being treated with chemotherapy for a solid tumor.
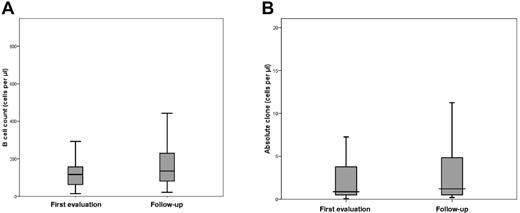
The mean white blood cell count of the 53 CLL-like MBL cases was 7.1 × 109 cells/L (range, 2.7-12.8 × 109 cells/L), the mean ALC was 2.5 × 109 cells/L (range, 0.8-4.1 × 109 cells/L), and the mean absolute B-cell count was 197.0 × 109 cells/L (range, 20.8-1508.8 × 109 cells/L). The clonal CD19+CD5+ B cells were 51.4 × 109 cells/L (range, 0.2-1249.3 × 109 cells/L). All values were not significantly different from the previous assessment after a median follow-up of 34 months (Figure 2A-B). None of the CLL-like clones developed a clinically relevant lymphocytosis; one case only had 4.1 × 109 lymphocytes/L, decreasing from 5.3 × 109 cells/L at the original evaluation.
B-cell count and absolute clone values in persistent CLL-like MBL are not significantly different between first and follow-up evaluations. The box-plots show the distribution of B-cell count (A) and absolute clone (B) values in persistent CLL-like MBL at follow-up evaluation. Notched boxes represent 25th and 75th percentile values, and the line in the middle corresponds to median value. Vertical lines represent the highest and the lowest values that are not outliers or extreme values. The differences between the 2 time points are not statistically significant.
B-cell count and absolute clone values in persistent CLL-like MBL are not significantly different between first and follow-up evaluations. The box-plots show the distribution of B-cell count (A) and absolute clone (B) values in persistent CLL-like MBL at follow-up evaluation. Notched boxes represent 25th and 75th percentile values, and the line in the middle corresponds to median value. Vertical lines represent the highest and the lowest values that are not outliers or extreme values. The differences between the 2 time points are not statistically significant.
When we considered the size of the clone, 48 of 53 clones had a clonal cell count of ≤ 50 aberrant B cells/μL (mean value, 3.6 cells/μL; range, 0.2-50.4 cells/μL) and represented only a small proportion of total B-cell count (mean, 2.8%; range, 0.1%-33.5%). Of the 5 of 53 remaining cases, 4 had < 500 cells/μL (247.1, 319.1, 336.7, 399.3 cells/μL) and 1 had < 1500 cells/μL (1249.3 cells/μL), all below the characteristic limit of clinical MBL.
After a median follow-up of 34 months, none of the CLL-like clones showed relevant changes in the cell count (Figure 2B). In particular, among the 48 cases with < 50 clonal B cells/μL, the mean difference between the first and the follow-up evaluation was < 1 cell/μL (0.93).
Low-count population-screening CLL-like MBL has recurrent CLL-related genomic aberrations
We sorted clonal B cells from 16 low-count population-screening CLL-like MBLs with FACS (median value, 5.3 cells/μL) and performed FISH analysis (Table 1) to evaluate the 4 most frequent aberrations detected in patients with CLL (del13q, trisomy 12, del11q, and del17p).21
CLL-like clones molecular parameters
| ID . | Sex . | Age, y . | Follow-up, mo . | Clone, cells/μL . | Light chain . | FISH . | IGHV gene . | IGHV identity, % . | T-cell clones . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First visit . | Follow-up . | CD4+ . | CD8+ . | CD4highCD8low . | CD4lowCD8high . | ||||||||
| VB0010 | F | 65 | 50 | 5.35 | 6.10 | κ | ND | IGHV3-30 | 97.7 | No clones | No clones | TRBV3 = 22.4% TRBV7.1 = 22.4% TRBV8 = 33.9% TRBV13.6 = 32.1% | No clones |
| VB0094* | M | 73 | 48 | 16.11 | 1.63 | κ | Normal | IGHV4-59/-61 | 95.6 | No clones | No clones | TRBV8 = 23.1% | No clones |
| VB0106 | M | 83 | 48 | 1.03 | 0.35 | λ | ND | IGHV3-30 | 97.8 | ND | ND | ND | ND |
| VB0266 | M | 74 | 47 | 6.56 | 8.16 | κ | ND | IGHV3-21 | 89.1 | No clones | No clones | TRBV2 = 25.3% TRBV8 = 27.4% | No clones |
| VB0274 | F | 67 | 47 | 7.26 | 11.26 | κ | ND | IGHV4-59/-61 | 95.0 | No clones | TRBV9 = 55.0% | No clones | No clones |
| VB0314 | F | 78 | 47 | 5.79 | 4.82 | λ | Del13q14.3† (19%), trisomy 12 (28%) | IGHV4-59/-61 | 94.4 | No clones | No clones | No clones | No clones |
| VB0323 | M | 75 | 47 | 0.51 | 0.61 | κ | ND | IGHV4-34 | 97.7 | ND | ND | ND | ND |
| VB0399 | M | 66 | 46 | 0.74 | 0.22 | κ | ND | IGHV3-48 | 93.9 | No clones | No clones | No clones | TRBV17 = 21.4% |
| VB0400 | M | 65 | 46 | 0.61 | 0.32 | κ | ND | IGHV3-64 | 86.8 | No clones | No clones | No clones | TRBV2 = 22.2% |
| VB0457 | F | 83 | 46 | 0.44 | 2.22 | κ | Normal | IGHV3-23 | 100.0 | No clones | TRBV8 = 28.4% | TRBV2 = 21.1% | TRBV22 = 21.4% |
| VB0537 | M | 74 | 47 | 1.79 | 0.75 | κ | ND | IGHV3-30 | 97.5 | ND | ND | ND | ND |
| VB0588 | F | 65 | 39 | 96.73 | 319.10 | κ | Del13q14.3 (31%) | IGHV4-59 | 93.5 | No clones | No clones | TRBV5.2 = 21.5% | No clones |
| VB0643 | M | 40 | 39 | 0.88 | 0.61 | κ | ND | IGHV3-64 | 93.9 | No clones | No clones | TRBV22 = 50.8% | No clones |
| VB0656 | M | 49 | 39 | 0.88 | 0.46 | κ | ND | IGHV3-7 | 97.0 | No clones | No clones | No clones | No clones |
| VB0667 | M | 65 | 38 | 0.24 | 0.68 | κ | Normal | ND | ND | No clones | No clones | No clones | TRBV22 = 26.1% |
| VB0670 | M | 68 | 37 | 0.24 | 0.21 | κ | ND | ND | ND | ND | ND | ND | ND |
| VB0679 | M | 67 | 38 | 6.34 | 9.25 | λ | ND | IGHV4-59/-61 | 89.4 | No clones | No clones | TRBV7.2 = 39.4% TRBV8 = 30.1% | No clones |
| VB0689 | M | 60 | 38 | 221.33 | 247.09 | κ | Normal | IGHV3-73 | 92.6 | No clones | No clones | TRBV2 = 25.7% | No clones |
| VB0777 | M | 68 | 37 | 0.66 | 0.79 | κ | ND | ND | ND | ND | ND | ND | ND |
| VB0790 | F | 61 | 36 | 1.50 | 13.55 | κ | Normal | IGHV4-30-4 | 94.4 | No clones | No clones | No clones | TRBV13.1 = 48.1% |
| VB0819 | M | 85 | 37 | 0.19 | 0.46 | κ | ND | IGHV3-48 | 88.3 | ND | ND | ND | ND |
| VB0902 | M | 69 | 36 | 0.96 | 6.76 | κ | Normal | IGHV4-39 | 100.0 | No clones | TRBV2 = 35.7% | No clones | No clones |
| VB1002 | F | 63 | 35 | 1.67 | 1.60 | κ | Del17p (80%) | IGHV4-59/-61 | 97.0 | No clones | No clones | TRBV9 = 24.8% TRBV16 = 38.9% | TRBV5.3 = 32.6% TRBV16 = 38.3% |
| VB1091 | F | 46 | 35 | 1.12 | 1.61 | Polyclonal | ND | IGHV3-15 | 93.5 | No clones | TRBV18 = 37.5% | TRBV2 = 30.2% TRBV8 = 19.4% | No clones |
| VB1134 | M | 58 | 34 | 2.36 | 0.46 | κ | Del13q34 (25%) | ND | ND | No clones | No clones | No clones | TRBV13.6 = 22.2% |
| VB1135 | M | 62 | 35 | 0.35 | 0.35 | Negative | ND | ND | ND | ND | ND | ND | ND |
| VB1170 | F | 58 | 34 | 0.85 | 0.33 | κ | ND | ND | ND | No clones | No clones | TRBV2 = 30.2% TRBV17 = 21.1% | TRBV2 = 25.0% TRBV22 = 21.7% |
| VB1189 | M | 80 | 34 | 189.76 | 399.29 | κ | Normal | IGHV3-21 | 88.2 | No clones | TRBV8 = 23.1% | TRVB13.6 = 42.8% | No clones |
| VB1202 | F | 61 | 34 | 4.72 | 18.56 | κ | Del13q14.3 (48%) | ND | ND | No clones | No clones | No clones | No clones |
| VB1231 | F | 64 | 35 | 50.38 | 50.59 | κ | ND | IGHV3-64 | 100.0 | No clones | No clones | No clones | TRBV1 = 22.7% |
| VB1239 | M | 75 | 34 | 0.41 | 0.50 | κ | ND | IGHV3-23 | 92.0 | ND | ND | ND | ND |
| VB1240 | F | 84 | 31 | 1763.90 | 1249.29 | λ | Normal | IGHV3-15 | 98.3 | No clones | No clones | No clones | No clones |
| VB1309 | M | 59 | 24 | 0.50 | 0.56 | Negative | ND | ND | ND | ND | ND | ND | ND |
| VB1320 | F | 61 | 24 | 0.41 | 3.82 | λ | Del13q14.3-q34 (25% and 30%), Del17p (90%) | ND | ND | No clones | No clones | TRBV5.3 = 35.7% TRBV8 = 48.4% TRBV17 = 33.8% | TRBV16 = 32.1% |
| VB1349 | M | 72 | 24 | 0.75 | 1.11 | κ | ND | ND | ND | No clones | TRBV1 = 61.5% | No clones | TRBV17 = 21.4% |
| VB1377 | M | 68 | 24 | 3.78 | 2.42 | κ | Del13q34 (28%) | ND | ND | No clones | No clones | No clones | No clones |
| VB1378 | F | 77 | 25 | 0.54 | 0.42 | κ | ND | ND | ND | ND | ND | ND | ND |
| VB1393 | F | 75 | 25 | 0.55 | 1.84 | κ | ND | ND | ND | No clones | TRBV13.2 = 25.8% | No clones | No clones |
| VB1395 | F | 76 | 25 | 1.53 | 1.22 | Polyclonal | ND | ND | ND | No clones | No clones | No clones | No clones |
| VB1451 | F | 66 | 23 | 0.46 | 2.36 | κ | ND | ND | ND | No clones | No clones | TRBV2 = 45.7% | No clones |
| VB1470 | F | 73 | 23 | 0.94 | 4.02 | κ | Del13q14.3† (37%) | ND | ND | No clones | TRBV13.2 = 38.3% | TRBV16 = 40.3% TRBV21.3 = 42.1% | No clones |
| VB1484 | M | 64 | 24 | 0.25 | 0.73 | Polyclonal | ND | ND | ND | ND | ND | ND | ND |
| VB1487 | M | 84 | 23 | 0.06 | 0.53 | κ | ND | IGHV4-59/-61 | 94.1 | No clones | No clones | No clones | TRBV7.1 = 46.4% TRBV8 = 29.4% TRBV17 = 26.9% TRBV22 = 28.6% |
| VB1518 | M | 53 | 22 | 1.21 | 1.16 | Negative | ND | ND | ND | ND | ND | ND | ND |
| VB1523 | M | 60 | 22 | 0.84 | 1.54 | Polyclonal | ND | ND | ND | No clones | No clones | No clones | No clones |
| VB1531 | M | 61 | 23 | 0.50 | 0.75 | Negative | ND | ND | ND | ND | ND | ND | ND |
| VB1592 | F | 92 | 21 | 0.29 | 0.20 | Polyclonal | ND | IGHV3-64 | 98.1 | ND | ND | ND | ND |
| VB1646 | M | 62 | 22 | 9.03 | 4.84 | Negative | ND | ND | ND | No clones | No clones | No clones | No clones |
| VB1686 | M | 83 | 22 | 0.96 | 1.31 | κ | ND | ND | ND | No clones | No clones | No clones | No clones |
| VB1688 | M | 55 | 21 | 1.22 | 1.07 | Polyclonal | ND | IGHV3-21 | 95.1 | ND | ND | ND | ND |
| VB1698 | M | 48 | 21 | 264.98 | 336.74 | λ | ND | IGHV3-15 | 93.0 | No clones | No clones | TRBV2 = 42.2% | TRBV17 = 42.2% |
| VB1711 | F | 56 | 11 | 0.41 | 0.32 | κ | ND | ND | ND | ND | ND | ND | ND |
| VB1712 | M | 72 | 11 | 0.35 | 1.53 | κ | ND | ND | ND | ND | ND | ND | ND |
| VB1736 | M | 48 | 11 | 0.68 | 0.26 | Kappa | ND | ND | ND | ND | ND | ND | ND |
| ID . | Sex . | Age, y . | Follow-up, mo . | Clone, cells/μL . | Light chain . | FISH . | IGHV gene . | IGHV identity, % . | T-cell clones . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First visit . | Follow-up . | CD4+ . | CD8+ . | CD4highCD8low . | CD4lowCD8high . | ||||||||
| VB0010 | F | 65 | 50 | 5.35 | 6.10 | κ | ND | IGHV3-30 | 97.7 | No clones | No clones | TRBV3 = 22.4% TRBV7.1 = 22.4% TRBV8 = 33.9% TRBV13.6 = 32.1% | No clones |
| VB0094* | M | 73 | 48 | 16.11 | 1.63 | κ | Normal | IGHV4-59/-61 | 95.6 | No clones | No clones | TRBV8 = 23.1% | No clones |
| VB0106 | M | 83 | 48 | 1.03 | 0.35 | λ | ND | IGHV3-30 | 97.8 | ND | ND | ND | ND |
| VB0266 | M | 74 | 47 | 6.56 | 8.16 | κ | ND | IGHV3-21 | 89.1 | No clones | No clones | TRBV2 = 25.3% TRBV8 = 27.4% | No clones |
| VB0274 | F | 67 | 47 | 7.26 | 11.26 | κ | ND | IGHV4-59/-61 | 95.0 | No clones | TRBV9 = 55.0% | No clones | No clones |
| VB0314 | F | 78 | 47 | 5.79 | 4.82 | λ | Del13q14.3† (19%), trisomy 12 (28%) | IGHV4-59/-61 | 94.4 | No clones | No clones | No clones | No clones |
| VB0323 | M | 75 | 47 | 0.51 | 0.61 | κ | ND | IGHV4-34 | 97.7 | ND | ND | ND | ND |
| VB0399 | M | 66 | 46 | 0.74 | 0.22 | κ | ND | IGHV3-48 | 93.9 | No clones | No clones | No clones | TRBV17 = 21.4% |
| VB0400 | M | 65 | 46 | 0.61 | 0.32 | κ | ND | IGHV3-64 | 86.8 | No clones | No clones | No clones | TRBV2 = 22.2% |
| VB0457 | F | 83 | 46 | 0.44 | 2.22 | κ | Normal | IGHV3-23 | 100.0 | No clones | TRBV8 = 28.4% | TRBV2 = 21.1% | TRBV22 = 21.4% |
| VB0537 | M | 74 | 47 | 1.79 | 0.75 | κ | ND | IGHV3-30 | 97.5 | ND | ND | ND | ND |
| VB0588 | F | 65 | 39 | 96.73 | 319.10 | κ | Del13q14.3 (31%) | IGHV4-59 | 93.5 | No clones | No clones | TRBV5.2 = 21.5% | No clones |
| VB0643 | M | 40 | 39 | 0.88 | 0.61 | κ | ND | IGHV3-64 | 93.9 | No clones | No clones | TRBV22 = 50.8% | No clones |
| VB0656 | M | 49 | 39 | 0.88 | 0.46 | κ | ND | IGHV3-7 | 97.0 | No clones | No clones | No clones | No clones |
| VB0667 | M | 65 | 38 | 0.24 | 0.68 | κ | Normal | ND | ND | No clones | No clones | No clones | TRBV22 = 26.1% |
| VB0670 | M | 68 | 37 | 0.24 | 0.21 | κ | ND | ND | ND | ND | ND | ND | ND |
| VB0679 | M | 67 | 38 | 6.34 | 9.25 | λ | ND | IGHV4-59/-61 | 89.4 | No clones | No clones | TRBV7.2 = 39.4% TRBV8 = 30.1% | No clones |
| VB0689 | M | 60 | 38 | 221.33 | 247.09 | κ | Normal | IGHV3-73 | 92.6 | No clones | No clones | TRBV2 = 25.7% | No clones |
| VB0777 | M | 68 | 37 | 0.66 | 0.79 | κ | ND | ND | ND | ND | ND | ND | ND |
| VB0790 | F | 61 | 36 | 1.50 | 13.55 | κ | Normal | IGHV4-30-4 | 94.4 | No clones | No clones | No clones | TRBV13.1 = 48.1% |
| VB0819 | M | 85 | 37 | 0.19 | 0.46 | κ | ND | IGHV3-48 | 88.3 | ND | ND | ND | ND |
| VB0902 | M | 69 | 36 | 0.96 | 6.76 | κ | Normal | IGHV4-39 | 100.0 | No clones | TRBV2 = 35.7% | No clones | No clones |
| VB1002 | F | 63 | 35 | 1.67 | 1.60 | κ | Del17p (80%) | IGHV4-59/-61 | 97.0 | No clones | No clones | TRBV9 = 24.8% TRBV16 = 38.9% | TRBV5.3 = 32.6% TRBV16 = 38.3% |
| VB1091 | F | 46 | 35 | 1.12 | 1.61 | Polyclonal | ND | IGHV3-15 | 93.5 | No clones | TRBV18 = 37.5% | TRBV2 = 30.2% TRBV8 = 19.4% | No clones |
| VB1134 | M | 58 | 34 | 2.36 | 0.46 | κ | Del13q34 (25%) | ND | ND | No clones | No clones | No clones | TRBV13.6 = 22.2% |
| VB1135 | M | 62 | 35 | 0.35 | 0.35 | Negative | ND | ND | ND | ND | ND | ND | ND |
| VB1170 | F | 58 | 34 | 0.85 | 0.33 | κ | ND | ND | ND | No clones | No clones | TRBV2 = 30.2% TRBV17 = 21.1% | TRBV2 = 25.0% TRBV22 = 21.7% |
| VB1189 | M | 80 | 34 | 189.76 | 399.29 | κ | Normal | IGHV3-21 | 88.2 | No clones | TRBV8 = 23.1% | TRVB13.6 = 42.8% | No clones |
| VB1202 | F | 61 | 34 | 4.72 | 18.56 | κ | Del13q14.3 (48%) | ND | ND | No clones | No clones | No clones | No clones |
| VB1231 | F | 64 | 35 | 50.38 | 50.59 | κ | ND | IGHV3-64 | 100.0 | No clones | No clones | No clones | TRBV1 = 22.7% |
| VB1239 | M | 75 | 34 | 0.41 | 0.50 | κ | ND | IGHV3-23 | 92.0 | ND | ND | ND | ND |
| VB1240 | F | 84 | 31 | 1763.90 | 1249.29 | λ | Normal | IGHV3-15 | 98.3 | No clones | No clones | No clones | No clones |
| VB1309 | M | 59 | 24 | 0.50 | 0.56 | Negative | ND | ND | ND | ND | ND | ND | ND |
| VB1320 | F | 61 | 24 | 0.41 | 3.82 | λ | Del13q14.3-q34 (25% and 30%), Del17p (90%) | ND | ND | No clones | No clones | TRBV5.3 = 35.7% TRBV8 = 48.4% TRBV17 = 33.8% | TRBV16 = 32.1% |
| VB1349 | M | 72 | 24 | 0.75 | 1.11 | κ | ND | ND | ND | No clones | TRBV1 = 61.5% | No clones | TRBV17 = 21.4% |
| VB1377 | M | 68 | 24 | 3.78 | 2.42 | κ | Del13q34 (28%) | ND | ND | No clones | No clones | No clones | No clones |
| VB1378 | F | 77 | 25 | 0.54 | 0.42 | κ | ND | ND | ND | ND | ND | ND | ND |
| VB1393 | F | 75 | 25 | 0.55 | 1.84 | κ | ND | ND | ND | No clones | TRBV13.2 = 25.8% | No clones | No clones |
| VB1395 | F | 76 | 25 | 1.53 | 1.22 | Polyclonal | ND | ND | ND | No clones | No clones | No clones | No clones |
| VB1451 | F | 66 | 23 | 0.46 | 2.36 | κ | ND | ND | ND | No clones | No clones | TRBV2 = 45.7% | No clones |
| VB1470 | F | 73 | 23 | 0.94 | 4.02 | κ | Del13q14.3† (37%) | ND | ND | No clones | TRBV13.2 = 38.3% | TRBV16 = 40.3% TRBV21.3 = 42.1% | No clones |
| VB1484 | M | 64 | 24 | 0.25 | 0.73 | Polyclonal | ND | ND | ND | ND | ND | ND | ND |
| VB1487 | M | 84 | 23 | 0.06 | 0.53 | κ | ND | IGHV4-59/-61 | 94.1 | No clones | No clones | No clones | TRBV7.1 = 46.4% TRBV8 = 29.4% TRBV17 = 26.9% TRBV22 = 28.6% |
| VB1518 | M | 53 | 22 | 1.21 | 1.16 | Negative | ND | ND | ND | ND | ND | ND | ND |
| VB1523 | M | 60 | 22 | 0.84 | 1.54 | Polyclonal | ND | ND | ND | No clones | No clones | No clones | No clones |
| VB1531 | M | 61 | 23 | 0.50 | 0.75 | Negative | ND | ND | ND | ND | ND | ND | ND |
| VB1592 | F | 92 | 21 | 0.29 | 0.20 | Polyclonal | ND | IGHV3-64 | 98.1 | ND | ND | ND | ND |
| VB1646 | M | 62 | 22 | 9.03 | 4.84 | Negative | ND | ND | ND | No clones | No clones | No clones | No clones |
| VB1686 | M | 83 | 22 | 0.96 | 1.31 | κ | ND | ND | ND | No clones | No clones | No clones | No clones |
| VB1688 | M | 55 | 21 | 1.22 | 1.07 | Polyclonal | ND | IGHV3-21 | 95.1 | ND | ND | ND | ND |
| VB1698 | M | 48 | 21 | 264.98 | 336.74 | λ | ND | IGHV3-15 | 93.0 | No clones | No clones | TRBV2 = 42.2% | TRBV17 = 42.2% |
| VB1711 | F | 56 | 11 | 0.41 | 0.32 | κ | ND | ND | ND | ND | ND | ND | ND |
| VB1712 | M | 72 | 11 | 0.35 | 1.53 | κ | ND | ND | ND | ND | ND | ND | ND |
| VB1736 | M | 48 | 11 | 0.68 | 0.26 | Kappa | ND | ND | ND | ND | ND | ND | ND |
ND indicates not done.
Case excluded from further analysis because treated with chemotherapy for a solid tumor.
Biallelic Del13q14.3.
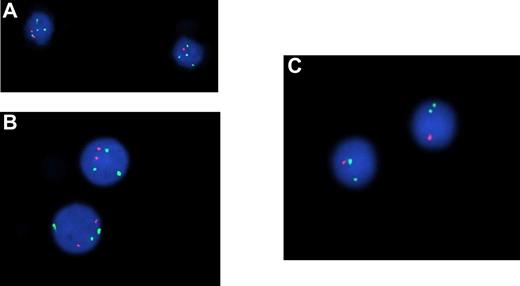
In 8 of 16 subjects analyzed (50.0%) we found genomic abnormalities (Table 1). Aberrations involving the chromosome 13 long arm were the most frequent (7 of 16 cases; 43.8%), 5 cases (31.3%) had a deletion of 13q14.3 that in 2 subjects was biallelic; 2 subjects (12.5%) had only a deletion of 13q34 (Figure 3A), and one had both 13q34 and 13q14.3 regions deleted. The percentage of CLL-like cells carrying these abnormalities was 19%-48%. One case had trisomy 12 in 28% of isolated B cells (Figure 3B) and 2 subjects had a deletion of the 17p region in 80% and 90% of sorted CLL-like cells (Figure 3C), respectively. More than 1 cytogenetic abnormality was identified in 2 cases (12.5%); one presented concomitant trisomy 12 and del13q14.3, the other one showed del13q34/13q14.3 and del17p. Deletion of 11q was never detected. The distribution of the cytogenetic abnormalities was not different between κ and λ clones.
Genomic aberrations are frequently detected by FISH in CLL-like MBL clones. FISH analysis has been performed on CD5+CD20dim purified MBL cells and shows, (A) on the left, a normal nucleus with 2 signals for each probe (Spectrum Aqua, Orange, and Green) corresponding to LSI13q34 locus 13q34, LSI D13S319 locus 13q14.3, and CEP 12 locus 12p11.1-q11, respectively; on the right another nucleus with a heterozygous deletion of the LSI D13S319 locus 13q14.3 (1 Spectrum Orange signal and 2 normal Spectrum Aqua and 2 Spectrum Green signals). (B) Both nuclei show trisomy of CEP 12 locus 12p11.1-q11 (3 Spectrum Green signals) and 2 normal Spectrum Orange signals (LSI D13S319 locus 13q14.3). (C) Both nuclei show the presence of only one signal for LSI p53 locus 17p13.1 (Spectrum Orange) and a normal double signal for LSI ATM locus 11q22.3 (Spectrum Green). Microscope used for these images was Nikon Eclipse 90i with a magnification ×1000 (Objective PLAN APO VC 100X/1:40 oil; 10× ocular; room temperature), and camera used was Qicam Fast 1394 (Qimaging). The imaging medium used was DAPI (Vectashield Mounting Medium with DAPI, vector). Acquisition software was Genikon (Nikon Instruments S.P.A.) and a FISH 3D Acquisition Module was used.
Genomic aberrations are frequently detected by FISH in CLL-like MBL clones. FISH analysis has been performed on CD5+CD20dim purified MBL cells and shows, (A) on the left, a normal nucleus with 2 signals for each probe (Spectrum Aqua, Orange, and Green) corresponding to LSI13q34 locus 13q34, LSI D13S319 locus 13q14.3, and CEP 12 locus 12p11.1-q11, respectively; on the right another nucleus with a heterozygous deletion of the LSI D13S319 locus 13q14.3 (1 Spectrum Orange signal and 2 normal Spectrum Aqua and 2 Spectrum Green signals). (B) Both nuclei show trisomy of CEP 12 locus 12p11.1-q11 (3 Spectrum Green signals) and 2 normal Spectrum Orange signals (LSI D13S319 locus 13q14.3). (C) Both nuclei show the presence of only one signal for LSI p53 locus 17p13.1 (Spectrum Orange) and a normal double signal for LSI ATM locus 11q22.3 (Spectrum Green). Microscope used for these images was Nikon Eclipse 90i with a magnification ×1000 (Objective PLAN APO VC 100X/1:40 oil; 10× ocular; room temperature), and camera used was Qicam Fast 1394 (Qimaging). The imaging medium used was DAPI (Vectashield Mounting Medium with DAPI, vector). Acquisition software was Genikon (Nikon Instruments S.P.A.) and a FISH 3D Acquisition Module was used.
Expansions of monoclonal T-cell populations are frequent in CLL-like MBL
In the hypothesis that CLL-like MBL may coexist with additional abnormalities of the immune system, we investigated also the TRBV with a panel of TRBV antibodies covering 70% of the expressed TCR repertoire17 in 36 cases of MBLs (20 men and 16 women, 14 below and 22 above the age of 65 years; Table 1).
We focused the analysis on CD4+, CD8+, and CD4+/CD8+ double-positive T lymphocytes, the latter being further divided into CD4highCD8low and CD4lowCD8high T cells (Figure 4A,C).17 On the basis of the previously published cutoff value of 20% of the cells expressing any TCR,17 we found ≥ 1 T-cell clone in 27 subjects (27 of 36; 75%) with a total of 53 clones detected.
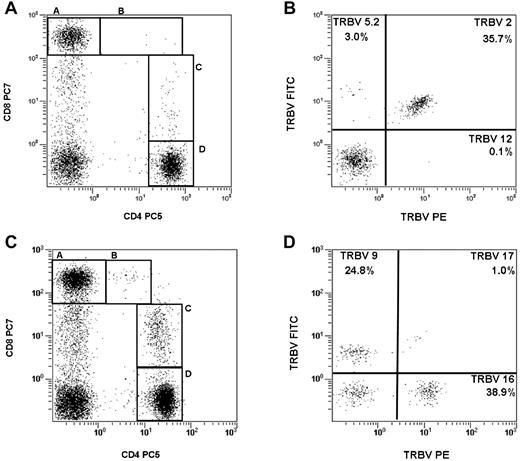
T-cell clones in persons carrying CLL-like MBL. (A) PB T lymphocytes from a representative case (VB0902) are subdivided, based on CD4 and CD8 expression, in 4 subpopulations: CD8+ (gate A), CD4lowCD8high (gate B), CD4highCD8low (gate C), and CD4+ (gate D). (B) CD8+ T lymphocytes (gated through A) of subject VB0902 show a restricted expression of TRBV2 (35.7%, upper right quadrant). (C) T-cell subpopulations in case VB1002. (D) CD4highCD8low T lymphocytes in this second subject (gated through C) show restricted expression for distinct TRBV chain (TRBV9, 24.8%, upper left; and TRBV16, 38.9%, lower right), indicating the presence of 2 distinct clones. In the TCR-Vβ repertoire kit each tube includes a FITC-conjugated mAb, a PE-conjugated mAb, and a third mAb derived from a balanced mixture of a PE- and a FITC-conjugated form. Because of the mutually exclusive expression of each TRBV gene, the “virtual” double-positive population on the upper left quadrant represents a third different TRBV domain.
T-cell clones in persons carrying CLL-like MBL. (A) PB T lymphocytes from a representative case (VB0902) are subdivided, based on CD4 and CD8 expression, in 4 subpopulations: CD8+ (gate A), CD4lowCD8high (gate B), CD4highCD8low (gate C), and CD4+ (gate D). (B) CD8+ T lymphocytes (gated through A) of subject VB0902 show a restricted expression of TRBV2 (35.7%, upper right quadrant). (C) T-cell subpopulations in case VB1002. (D) CD4highCD8low T lymphocytes in this second subject (gated through C) show restricted expression for distinct TRBV chain (TRBV9, 24.8%, upper left; and TRBV16, 38.9%, lower right), indicating the presence of 2 distinct clones. In the TCR-Vβ repertoire kit each tube includes a FITC-conjugated mAb, a PE-conjugated mAb, and a third mAb derived from a balanced mixture of a PE- and a FITC-conjugated form. Because of the mutually exclusive expression of each TRBV gene, the “virtual” double-positive population on the upper left quadrant represents a third different TRBV domain.
No CD4+ clones were found; 8 clones were observed among CD8+ cells (Figure 4B). The double-positive T-cell subsets were present in low percentages with CD4highCD8low cells representing 0.96% of lymphocytes and CD8highCD4low cells 0.24% of lymphocytes. As expected,17 these 2 subsets presented the highest amount of clones with 16 of 36 (44.4%) carrying ≥ 1 clonal population of the CD4highCD8low (Figure 4D) and 13 of 36 (36.1%) of the CD8highCD4low compartment.
Overall, 27 subjects presented ≥ 1 T-cell clonal expansions (range, 1-4 T-cell clonal expansions), of whom 17 of 22 subjects (77.3%) > 65 years of age and 10 of 14 (71.4%) < 65 years. Both frequencies were significantly higher (P = .002) than those observed in the general population.17
TRBV2 (11 of 27; 40.7%) and TRBV8 (9 of 27; 33.3%) were the most frequent genes in the clonal populations. Both genes were more frequent in the MBL population than in the T-cell clones detected in the general population (TRBV2, 18.8%; TRBV8, 7.8%).17
Discussion
Not all MBLs are equal. We have previously found that MBLs identified through population-screening studies,14 with aberrant B cells representing a tiny minority of total B cells, have different biologic and laboratory features compared with clinical MBL associated with lymphocytosis.12 Clinical CLL-like MBL has a risk of progression into overt CLL of ∼ 1.1% per year,13 whereas the progression risk of low-count population-screening MBL is unknown. This is an important issue because the prevalence of population-screening MBL is much higher than that of clinical MBL with a peak of > 75% in persons > 90 years.5 To investigate which features are associated with the risk of progression we have reevaluated, after a median follow-up of 34 months, 76 subjects with MBL previously detected in the PB of a general adult population belonging to a rural community in Northern Italy. We here show that (1) 90.0% of population-screening CLL-like MBL persists over time in contrast to only 44.4% and 66.7% of atypical-CLL and CD5− MBL; (2) most of the population-screening MBLs remain stable, and no progression to clinically overt disease has been recorded; and (3) 50% of population-screening CLL-like MBL carries CLL-related genomic abnormalities, especially monoallelic or biallelic 13q deletions.
The fact that CLL-like MBL clones persist over time strongly implies that a stable modification has occurred within the B lineage and that these clones are not simply related to seasonal or occasional antigenic exposures. Because approximately one-half (44.4% and 66.7%) of atypical-CLL and CD5− MBL became undetectable after 3 years, MBL other than CLL-like may be transient. A plausible possibility is that they are “reactive” monoclonal expansions, conceivably triggered by self-limiting immune stimulations that occur randomly during one's lifetime. Our results differ from those recently published in a series of 12 atypical-CLL and CD5− MBL reevaluated 12 months after the first immunophenotypic analysis.24 All clones were confirmed and even showed a significant increase in the median concentration of clonal B cells. Although this discrepancy may be accounted for both by the original higher median value of the clonal populations and especially by the different length of the follow-up, further studies with larger cohorts are needed to settle the issue.
The persistence of CLL-like MBL closely resembles that of clonal double-positive CD4/CD8 T-cell populations in the elderly, linked to chronic and persistent viral infections (eg, CMV).25,26 Whether persistent stimulation by infectious agents (or self-antigens) may also trigger the development of MBL remains to be elucidated, although the increased frequency of CLL-like MBL in HCV-infected persons27 is in line with this possibility. Clonal T-cell populations, especially double-positive CD4/CD8, were found to be increased in population-screening MBL compared with the general population,17 with TRBV2 and TRBV8 as the most frequently involved genes, possibly suggesting a distinct restriction of T-cell specificity. Finally, more than one-half of the subjects analyzed presented multiple T-cell clones, suggesting a widespread deregulation of the immune system in MBL, another feature potentially related to a prolonged exposure to chronic antigenic stimulation.
After a median follow-up of ∼ 3 years, none of the 76 participants have developed CLL or any other lymphoproliferative disorder, and no one has required medical attention because of the development of clinically relevant lymphocytosis. Most subjects with population-screening MBL remained stable, some even decreased. These results confirm the hypothesis that the natural history of population-screening MBL differs from that of clinical MBL and are in line with the molecular and biologic differences we previously demonstrated.14 Low-count population-screening CLL-like MBL uses an IGHV gene repertoire different from clinical MBL and CLL, shows a distinct gene usage, a higher rate of mutated IGHV, and a rare occurrence of stereotyped receptors.14 On the basis of these evidences, molecular features of the clone could be potentially used to identify those rare subjects at risk of evolution into frank CLL and refrain from useless and prolonged monitoring in most of otherwise healthy persons. To our surprise, in the present work, when we studied by FISH analysis the presence of CLL-related genetic abnormalities, we found that ≤ 50% of cases with low-count population-screening MBL do carry the same CLL-related abnormalities with monoallelic or biallelic 13q deletions in 43.8% of the cases. We also observed trisomy 12 in 1 case and 17p deletion in 2 cases (in 1 case as a sole abnormality). The frequency of 13q deletions is similar to that observed in newly diagnosed CLL and in subjects with clinical MBL5,13,21 and strongly indicates that these lesions occur rather early during the development of MBL and appear to be associated with the acquisition of the typical immunophenotypic profile rather than with the fully fledged leukemic state.16 This finding is in keeping with the observation that the mouse model carrying deletions of the DLEU2/mir15-a/16-1 cluster located at 13q14 region develops a spectrum of lymphoproliferative disorders that range from MBL to CLL to diffuse large B-cell lymphoma.28
Of interest we also detected 17p deletion in 2 cases of population-screening MBL with low numbers of clonal B cells (1.6 and 3.8 cells/μL, respectively); del17p was observed in 80% and 90% of clonal B cells, and in case VB1320 it was coexisting with an additional genetic alteration involving chromosome 13q, but the increase of aberrant B cells was minor at reevaluation (from 0.4 to 3.8 cells/μL, respectively). The detection of 17p deletion per se may not be detrimental because patients with CLL with this abnormality can have a stable disease especially if they also have mutated IGHV genes.29,30 Given the small size of the clone, we were able to sequence the IGHV gene in only 1 of the 2 MBL cases and in particular that one with the 17p deletion as sole abnormality (VB1002) that showed indeed a 97% identity with the germ line IGHV sequence.
In conclusion, our study shows that low-count population-screening CLL-like MBL, as opposed to atypical CLL and CD5− MBL, tends to persist over time. The potential risk of progression into overt CLL is exceedingly rare and definitely less than that of clinical MBL even if they carry CLL genomic aberrations. This suggests that these abnormalities occur early during the natural history of MBL/CLL and are probably associated with the acquisition of the typical phenotype rather than with the progression into a fully fledged leukemic disease.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC; Investigator Grant and Special Program Molecular Clinical Oncology, 5 per mille no. 9965); “Fondazione Piera, Pietro e Giovanni Ferrero”; Fondazione CARIPLO; “CLLGRF–US/European Alliance for the Therapy of CLL,” FIRB and PRIN–Ministero Istruzione, Università e Ricerca (MIUR), Roma, Progetti Integrati Oncologia (PIO)–Ministero della Salute, Roma, Compagnia di San Paolo e Progetto Finalizzato Sanità 2007. C. Scielzo is supported by the EHA Fellowship Program (2009/18), and A.J. is supported by a EHA Partner fellowship program.
Authorship
Contribution: C.F. and L.S. performed the experiments, analyzed the data, and wrote the manuscript; L.P. and A.T. performed FISH experiments; F.C. performed the experiments and analyzed the data; A.D and A.J. designed and performed immunoglobulin studies; C. Scielzo performed the experiments; C. Sala and D.T. updated clinical and genealogic data; F.C.-C. supervised analysis and wrote the paper; and P.G. designed the study, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Federico Caligaris-Cappio, Department of Onco-Hematology, Via Olgettina 60, 20132, Milano, Italy; e-mail: caligaris.federico@hsr.it
References
Author notes
C.F. and L.S. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal