Abstract
Epstein-Barr virus (EBV)–DNA was prospectively analyzed in plasma and mononuclear cells (MNCs) from peripheral blood in patients with extranodal natural killer (NK)/T-cell lymphoma, nasal type, to evaluate the clinical significance for diagnosis, monitoring the tumor burden, and prognostication. Thirty-three patients were enrolled, and 32 were evaluable. Pretreatment plasma and MNC EBV-DNA was detectable in 14 (range, 50-71 000 copies/mL) and 6 patients (range, 20-780 copies/μg DNA), respectively, and both were well correlated (r = 0.8741, P < .0001). Detectable plasma EBV-DNA was associated with higher clinical stage (P = .02), presence of B symptoms (P = .02), worse performance status (P = .02), and higher serum soluble IL-2 receptor level (P < .0001). Twenty-two patients attained complete response. Plasma EBV-DNA level was significantly higher in nonresponders than in responders (mean, 16 472 vs 2 645 copies/mL; P = .02). Multivariate analysis showed clinical stage (hazard ratio, 9.0; 95% confidence interval, 1.8%-45.0%) and pretreatment plasma EBV-DNA (hazard ratio, 10.6; 95% confidence interval, 1.3%-87.0%) were significant prognostic factors. Three-year overall survival of plasma EBV-DNA positive and negative patients was 42.9% and 94.4%, respectively (P = .0009). Plasma was a preferable sample for this purpose in NK/T-cell lymphoma, nasal type, and EBV-DNA level was a good indicator for response and overall survival.
Introduction
Extranodal natural killer (NK)/T-cell lymphoma, nasal type (NKTCL) is a distinct subtype of non-Hodgkin lymphoma that mainly occurs in nasal–paranasal area, skin, gastrointestinal tract, or other extranodal sites.1 Histopathologic specimens show diffuse infiltration of lymphoma cells with angiocentricity and necrosis.2 Initial recognition of this lymphoma mandated the angiocentric growth pattern for diagnosis.3 However, later consensus criteria do not require the angiocentricity, mainly because of the sampling error.4 Epidemiologically, extranodal NKTCL predominantly presents in East Asia, Southeast Asia, and Central and South America and is associated with Epstein-Barr virus (EBV).1,5 The presence of EBV can be detected by EBV-encoded RNA (EBER) in situ hybridization in formalin-fixed, paraffin-embedded sections6 and is currently an important diagnostic indicator.
Peripheral blood of patients with an EBV-associated tumor, including extranodal NKTCL, Hodgkin lymphoma, and nasopharyngeal cancer, contains fragmented EBV-DNA. The EBV-DNA can be detected by polymerase chain reaction (PCR) methods and has been described useful for quantification of tumor burden and estimation of residual disease.7 The length of EBV-DNA in peripheral blood is usually < 500 bp. Therefore, the detection of EBV-DNA does not mean the existence of EBV viral particles capable of infection. Another problem for EBV-DNA in peripheral blood is that there are 2 different samples for analysis: a liquid component such as plasma or serum and a cell component, such as mononuclear cells (MNCs) or whole blood. There have been no prospective studies that compared the significance of either sample. Most reports in the literature were retrospective studies and did not compare both samples from the same patients. Therefore, we conducted a prospective study to analyze EBV-DNA using quantitative PCR.
Methods
Patients and study design
This is an observational study to analyze EBV-DNA copy numbers with both plasma and MNCs at the same points in clinical course conducted by the NK-cell Tumor Study Group in Japan. The study was approved by both the Study Committee of NK-cell Tumor Study Group and the institutional review board of each participating institution. The primary objective was to evaluate a prognostic value of EBV-DNA copy number for 2-year overall survival. Secondary objectives were comparison of EBV-DNA copy number and pretreatment characteristics and prognostic capability of EBV-DNA during and after treatments. Inclusion criteria were as follows: (1) Histopathologic diagnosis of extranodal NKTCL or aggressive NK-cell leukemia. (2) Patients without other serious complications and those tolerable for chemotherapy and/or radiotherapy. (3) No prior history of chemotherapy or radiotherapy. (4) Patients with written informed consent in accordance with the Declaration of Helsinki. EBV-DNA was measured at 3 different times for each patient: pretreatment, mid-term of treatment (∼ 3 months), and after completion of the series of treatment (∼ 6 months). The diagnosis of extranodal NKTCL was confirmed by immunophenotyping and EBER in situ hybridization,2 and was verified by central pathologic review.
Patient samples and DNA preparation
A 5-mL patient peripheral blood sample was collected into an EDTA-containing tube, sent to the central laboratory (Hokkaido University, Japan), and centrifuged to isolate MNCs and plasma using a Ficoll-Hypaque gradient method. DNA was extracted from plasma MNCs using a QIAamp blood kit (QIAGEN) as recommended by the manufacturer.
EBV-DNA quantification
The real-time quantification of EBV-DNA was performed in a Prism 7700 sequence detector (Applied Biosystems Japan) as described previously.7,8 The BamHI-W region of EBV was amplified. In brief, DNA sample was mixed with 300nM primers and 100nM TaqMan probe labeled with 5′-6-carboxyfluorescein and 3′-6-carboxytetramethylrhodamine and amplified in a 25-μL volume using the TaqMan PCR core reagents kit (Applied Biosystems Japan). Thermal cycling amplification was initiated with a 2-minute incubation at 50°C, followed by an initial denaturation step of 10 minutes at 95°C and then 40 cycles of 95°C for 15 seconds and 56°C for 1 minute were carried out. The primers used for amplification were 5′-CCCAACACTCCACCACACC-3′ (sense) and 5′-TCTTAGGAGCTGTCCGAGGG-3′ (antisense), and the dual-labeled TaqMqn fluorescent probe was 5′-6-carboxyfluorescein-CACACACTACACACACCCACCCGTCTC-6-carboxytetramethylrhodamine-3′, which produced 76-bp amplicon from 3 duplicated portions. The copy number of EBV-DNA in each sample was calculated with a standard curve. For the external control, the β-globin gene was amplified. The amplification was duplicated for each sample and the mean values were used. A positive control of Namalwa cell line DNA and negative water blanks were included in every analysis.
Statistical analysis
The lactate dehydrogenase (LDH) index was calculated at each of the institutes from a patient's serum LDH level divided by the upper limit of serum LDH. The International Prognostic Index score was calculated as described previously.9 The treatment response was assessed according to standard response criteria.10 Overall survival was measured from the date of diagnosis to the date of death or the last follow-up. Correlations between the 2 groups were examined with the χ2 test, Fisher exact test, and the Mann-Whitney U test. Patient survival data were analyzed with the method of Kaplan and Meier and were compared by the log-rank test. Univariate and multivariate analysis were performed using Cox proportional hazard model. Data were analyzed with STATA Version 10 statistical software (Stata).
Results
Patient characteristics
In total, 33 patients were registered from June 2004 to March 2007. All patients were diagnosed with extranodal NKTCL. No patients with aggressive NK-cell leukemia were registered. One patient refused investigation and treatment after registration and went to another hospital. Accordingly, 32 patients were eligible for the study, with a male/female ratio of 20:12 (Table 1). The median age was 55 years old, ranging from 18 to 81. Nineteen patients were in stage IE, 3 in stage IIE, and 10 in stage IV. Twenty patients had B-symptom, and 13 had elevated serum LDH level. Eastern Cooperative Oncology Group performance status was low (0-2) in 28 patients but was 3 in 2 patients. The International Prognostic Index was high-intermediate/high in 10. In 29 patients, the lymphoma involved nasal and paranasal lesions. Two other patients presented with cutaneous involvement, and another patient had an intestinal lesion. Histopathologic diagnosis of extranodal NKTCL was verified in all patients by central pathologic review. All the specimens showed cytoplasmic CD3-positive, CD20-negative, and cytotoxic molecule-positive immunophenotype. CD56 was positive in 30 but was negative in 2 specimens. EBER in situ hybridization was positive in all specimens, with a median positivity of 56% (range, 21%-96%) of lymphoma cells. Front-line treatment included radiotherapy in 2, concurrent chemoradiotherapy in 16, radiotherapy followed by chemotherapy in 8, and chemotherapy alone in 6 specimens. Among 32 patients evaluable for response, 22 patients (69%) achieved complete response, and 5 with partial response, but 4 patients did not show any change, and 1 patient showed progressive disease. The 3-year overall survival was 69.7%. The median follow-up of patients was 2.9 years. Patient data are shown in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Patient characteristics
| Characteristic . | Value . |
|---|---|
| No. | 32 |
| Median age, y | 55 |
| Range | 18-81 |
| Sex, male/female | 20/12 |
| Stage | |
| IE | 19 |
| IIE | 3 |
| IV | 10 |
| B-symptom (%) | 13 (41) |
| LDH elevation (%) | 12 (38) |
| Performance status > 1 (%) | 4 (13) |
| International Prognostic Index high-intermediate/high (%) | 10 (31) |
| Origin | |
| Nasal | 29 |
| Extranasal | 3 |
| Treatment | |
| Radiotherapy | 2 |
| Concurrent chemoradiotherapy | 16 |
| Radiotherapy followed by chemotherapy | 8 |
| Chemotherapy | 6 |
| Response | |
| Complete response | 22 |
| Partial response | 5 |
| Stable disease | 4 |
| Progressive disease | 1 |
| Characteristic . | Value . |
|---|---|
| No. | 32 |
| Median age, y | 55 |
| Range | 18-81 |
| Sex, male/female | 20/12 |
| Stage | |
| IE | 19 |
| IIE | 3 |
| IV | 10 |
| B-symptom (%) | 13 (41) |
| LDH elevation (%) | 12 (38) |
| Performance status > 1 (%) | 4 (13) |
| International Prognostic Index high-intermediate/high (%) | 10 (31) |
| Origin | |
| Nasal | 29 |
| Extranasal | 3 |
| Treatment | |
| Radiotherapy | 2 |
| Concurrent chemoradiotherapy | 16 |
| Radiotherapy followed by chemotherapy | 8 |
| Chemotherapy | 6 |
| Response | |
| Complete response | 22 |
| Partial response | 5 |
| Stable disease | 4 |
| Progressive disease | 1 |
EBV-DNA in MNCs and plasma
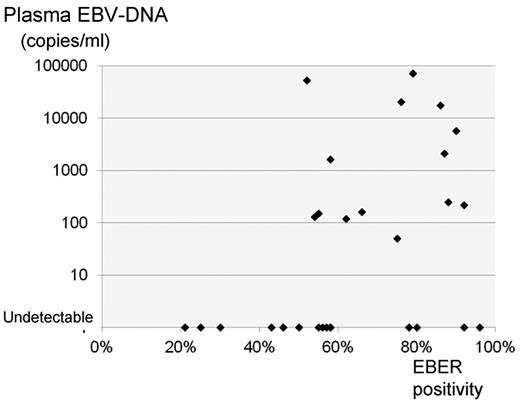
EBV-DNA was detected from MNCs in 6 of 32 patients. The mean copy numbers was 200 copies/μg DNA, ranging from 20 to 780. From plasma, the EBV-DNA was detected in 14 of 32 patients, ranging from 50 to 71 000 copies/mL, and the mean number was 12 354. Significant correlation was observed between MNCs and plasma EBV-DNA copies (r = 0.8741, P < .0001; Figure 1). Plasma EBV-DNA was well correlated with pretreatment clinical stage (P = .02), presence of B-symptom (P = .02), ECOG performance status (P = .02), serum LDH level (P = .05), and soluble IL-2 receptor (P < .0001) but not with regional node involvement (P = .17), nasal versus nonnasal types (P = .16), and serum C-reactive protein (P = .29). A weak but significant correlation between EBER positivity and plasma EBV-DNA level was observed (r = 0.3907, P = .03; Figure 2).
Comparison between plasma and MNC EBV-DNA. Significant correlation was observed between plasma and MNC EBV-DNA copies (r = 0.8741, P < .0001).
Comparison between plasma and MNC EBV-DNA. Significant correlation was observed between plasma and MNC EBV-DNA copies (r = 0.8741, P < .0001).
Correlation of EBER positivity and plasma EBV-DNA level. Patients with < 50% EBER-positive cells did not show detectable plasma EBV-DNA. A weak correlation between EBER positivity and plasma EBV-DNA level was observed (r = 0.3907, P = .03).
Correlation of EBER positivity and plasma EBV-DNA level. Patients with < 50% EBER-positive cells did not show detectable plasma EBV-DNA. A weak correlation between EBER positivity and plasma EBV-DNA level was observed (r = 0.3907, P = .03).
Prognostic value of EBV-DNA
Patients with detectable EBV-DNA showed significantly lower survival than patients without detectable EBV-DNA, either in plasma (P = .0009) or in MNCs (P = .006; supplemental Figure 1). If the copy numbers were divided into low (< 1000 copies/mL plasma or < 100 copies/μg DNA of MNCs) and high copy categories, patients were successfully stratified into 3 prognostic groups (Figure 3).
Survival of patients by pretreatment EBV-DNA. (A) Overall survival was significantly different among the groups of patients with plasma EBV-DNA undetectable, low level (< 1000 copies/mL) and high level (> 1000 copies/mL). (B) Overall survival was significantly different among the groups of patients with MNC EBV-DNA undetectable, low level (< 100 copies/μg DNA) and high level (> 100 copies/μg DNA).
Survival of patients by pretreatment EBV-DNA. (A) Overall survival was significantly different among the groups of patients with plasma EBV-DNA undetectable, low level (< 1000 copies/mL) and high level (> 1000 copies/mL). (B) Overall survival was significantly different among the groups of patients with MNC EBV-DNA undetectable, low level (< 100 copies/μg DNA) and high level (> 100 copies/μg DNA).
Pretreatment plasma and MNC EBV-DNA were significant prognostic factors for overall survival by univariate analysis, as well as clinical stage, performance status, and EBER positivity (Table 2). Multivariate analysis showed clinical stage (hazard ratio [HR], 9.0; 95% confidence interval [CI], 1.8%-45.0%) and pretreatment plasma EBV-DNA (HR, 10.6; 95% CI, 1.3%-87.0%) were significant prognostic factors.
Prognostic factors affecting overall survival
| Variable . | Unfavorable factors . | Univariate . | Multivariate* . | ||
|---|---|---|---|---|---|
| HR (CI) . | P . | HR (CI) . | P . | ||
| Age, y | > 60 | 0.9 (0.2-3.1) | .84 | ||
| Stage | IV | 2.4 (1.4-4.2) | .001 | 9.0 (1.8-45.1) | .007 |
| Performance status | 2-4 | 4.5 (1.3-15.8) | .02 | ||
| Plasma EBV-DNA | Detected | 14.5 (1.8-116) | .01 | 10.6 (1.3-87.0) | .03 |
| MNC EBV-DNA | Detected | 5.4 (1.4-20.3) | .01 | ||
| B symptom | Present | 2.7 (0.8-9.5) | .13 | ||
| EBER | > 75% | 9.3 (2.0-44.0) | .005 | ||
| Variable . | Unfavorable factors . | Univariate . | Multivariate* . | ||
|---|---|---|---|---|---|
| HR (CI) . | P . | HR (CI) . | P . | ||
| Age, y | > 60 | 0.9 (0.2-3.1) | .84 | ||
| Stage | IV | 2.4 (1.4-4.2) | .001 | 9.0 (1.8-45.1) | .007 |
| Performance status | 2-4 | 4.5 (1.3-15.8) | .02 | ||
| Plasma EBV-DNA | Detected | 14.5 (1.8-116) | .01 | 10.6 (1.3-87.0) | .03 |
| MNC EBV-DNA | Detected | 5.4 (1.4-20.3) | .01 | ||
| B symptom | Present | 2.7 (0.8-9.5) | .13 | ||
| EBER | > 75% | 9.3 (2.0-44.0) | .005 | ||
Final model.
Clinical course and change of EBV-DNA
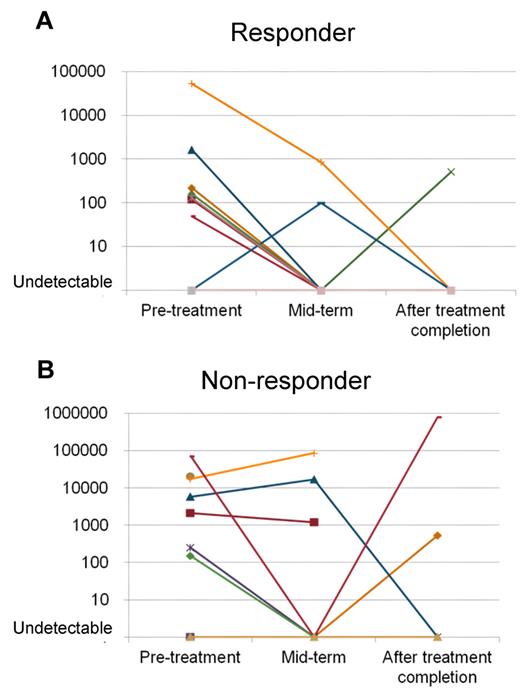
Plasma EBV-DNA copy numbers were well correlated with tumor burden, but not for MNC EBV-DNA. Figure 4 shows the change of plasma EBV-DNA by clinical course. In responders, the EBV-DNA levels went down after the treatment. In contrast, the plasma EBV-DNA was not consistently decreased in nonresponders. It remained in high levels or re-elevated after the initial decrease after treatment. The pretreatment copy number of plasma EBV-DNA was significantly higher in nonresponders compared with responders (mean, 16 472 vs 2645 copies/mL; P = .02).
Change of plasma EBV-DNA during treatment. (A) Plasma EBV-DNA levels of patients who attained complete response (CR) decreased after the treatment and became undetectable. Disease recurrence was observed in 1 patient after ttreatment. (B) For patients who could not attain CR, the plasma EBV-DNA remained at high levels or was re-elevated after the treatment. The pretreatment plasma level was significantly lower for responders (A; mean, 2645 copies/mL) than in nonresponders (B; mean, 16 472 copies/mL).
Change of plasma EBV-DNA during treatment. (A) Plasma EBV-DNA levels of patients who attained complete response (CR) decreased after the treatment and became undetectable. Disease recurrence was observed in 1 patient after ttreatment. (B) For patients who could not attain CR, the plasma EBV-DNA remained at high levels or was re-elevated after the treatment. The pretreatment plasma level was significantly lower for responders (A; mean, 2645 copies/mL) than in nonresponders (B; mean, 16 472 copies/mL).
Discussion
Quantification of EBV-DNA in peripheral blood has been demonstrated as useful for diagnosis and monitoring of EBV-associated diseases. However, the optimal sample type remained uncertain. For patients with EBV primary infection or after transplantation reactivation, it is more common to measure the EBV-DNA in MNCs or whole blood samples.11-13 Comparisons among MNCs, whole blood, and plasma revealed that the sensitivity of plasma was lower than that of MNCs and whole blood.14-17 In contrast, plasma EBV-DNA has been shown to be a valuable diagnostic and prognostic marker for patients with EBV-related tumors.7,18-22 This plasma EBV-DNA was shown to be derived from fragmented DNA from tumor rather than viral particles. In the literature, only the study by Chan et al7 strictly compared plasma and MNC samples from EBV-associated tumors. Although plasma was a preferable sample in the study, it was a retrospective study and included several types of tumors. This prompted us to investigate solely with NK-cell malignancies prospectively, not including nasopharyngeal carcinoma or Hodgkin lymphoma. Here, we simultaneously analyzed the EBV-DNA copy numbers from plasma and MNC samples of extranodal NKTCL patients. Both plasma and MNC EBV-DNA were good indicators for response to treatment and overall survival, but our results indicate that the sensitivity was higher for plasma samples, and the measurement of plasma EBV-DNA was more useful for clinical purpose.
The difference of preferable samples between EBV infection and EBV-associated tumor suggest that the localization of EBV is clearly different. Peripheral blood MNCs are the main target for EBV primary infection or reactivation. In contrast, the EBV virions only reside in tumors, but not in peripheral blood or MNCs. Circulating EBV-DNA for such patients are derived from necrotic or apoptotic tumor cells.23,24 In this context, however, there exists an enigma for the true origin of MNC EBV-DNA. One hypothesis is that it was contaminated and centrifuged from high numbers of plasma-fragmented EBV-DNA. Filtering or other techniques may prevent this contamination, so the result of detected EBV-DNA in MNCs is more accurate. However, this cannot explain the entire results, because one of our patients only showed detectable EBV-DNA in MNCs but not in plasma. This patient presented with stage I disease, and there was no clinical evidence of circulating tumor cells in peripheral blood. Higher numbers of EBV-DNA in MNCs were reported in another study,7 showing the need for further investigation of the EBV residual pattern in patients with extranodal NKTCL.
Hypothetically, the amount of EBV-DNA in plasma reflects the total volume of and replication of tumor. EBER positivity in specimen may reflect the replication of EBV. Therefore, a weak correlation was observed between plasma EBV-DNA and EBER positivity. Here, plasma EBV-DNA was more useful for prognostication, suggesting that it also reflects the tumor burden. If examined in large numbers of patients, the plasma EBV-DNA can be included in prognostic models.25,26 Although the prognosis of extranodal NKTCL is improving using concurrent chemoradiotherapy,27,28 the prognosis is worse than other type of lymphomas. Allogeneic or autologous hematopoietic stem cell transplant is a promising treatment for high-risk patients.29,30 The plasma EBV-DNA also can be used for such stratification.
In conclusion, our study demonstrates pretreatment plasma EBV-DNA copy number is a good indicator for both response to treatment and overall survival in extranodal NKTCL. Measurement of the plasma EBV-DNA is useful for prospective clinical trials and general practice.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the following participants in this study: Asahikawa Medical University, Ehime University, Fujita Health University, Fukushima Medical University, Hiroshima University, Hyogo Cancer Center, Juntendo Shizuoka Hospital, Juntendo University, Komagome Hospital, Kyoto Municipal Hospital, Nagaoka Red Cross Hospital, Nihon Medical University, Mie University, Tokyo Medical and Dental University, University of Tokyo.
This work was supported in part by an unrestricted grant from Kirin Pharma, Tokyo, Japan.
Authorship
Contribution: R.S., M.Y., K.K., and K.O. designed the study, collected data, performed statistical analysis, and prepared manuscript; K.I. and J.S. participated in interpretation and prepared manuscript; G.Y., Y.H., Y.I., H.G., T.K., and M.O. prepared patient data; K.T. conducted viral and molecular analyses; R.H. and S.N. performed central pathologic review; and all authors participated in interpretation of data and approval of final manuscript.
Conflict-of-interest disclosure: K.O. is currently an employee of Eisal Pharmaceutical Co Ltd (Tokyo, Japan). The remaining authors declare no competing financial interests.
Correspondence: Ritsuro Suzuki, Department of HSCT Data Management and Biostatistics, Nagoya University Graduate School of Medicine, 1-1-20 Daiko-Minami, Higashi-ku, Nagoya, 461-0047 Japan; e-mail: r-suzuki@med.nagoya-u.ac.jp.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal