Abstract
Abstract 741
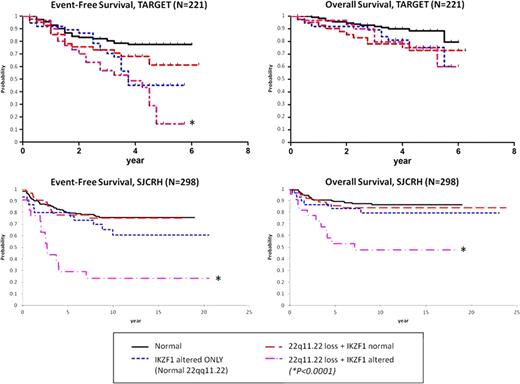
Identifying prognostic biomarkers in childhood acute lymphoblastic leukemia (ALL) is imperative for risk-stratification and intensifying therapy for children at high risk of remission induction failure or relapse. IKZF1 deletions/mutations were recently shown to correlate with poor outcome in ALL, highlighting genetic alterations as prognostic markers (NEJM 360:470, 2009). We recently described focal deletions in VPREB1 located within the lambda variable chain region which are not part of normal light chain rearrangement in precursor B-cell ALL in a local Utah cohort and the TARGET cohort of high-risk ALL patients treated on the Children's Oncology Group P9906 trial. Upon further inspection, we identified a second focal deletion in chromosome 22q11.22, nearly 80 kilobases (Kb) upstream from VPREB1 in the same lambda region. We characterized and correlated this deletion with outcome in childhood ALL using the Utah Cohort (N=60), TARGET P9906 cohort (N=221), and St. Jude Children's Research Hospital cohort (SJCRH, N=298). Microarray data was analyzed (Utah = Molecular Inversion Probe 330K [Affymetrix]; TARGET = SNP 500K & U133A [Affymetrix]; SJCRH = SNP 500K/6.0 & U133A [Affymetrix]) by Nexus Copy Number and Nexus Expression (BioDiscovery, Inc.). Each cohort contains the focal 22q11.22 loss (Hemizygous: Utah = 21%, TARGET = 16%, SJCRH = 18%; and Homozygous: Utah = 7%, TARGET = 16%, SJCRH = 10%). It spans 142 Kb, with the most common recurring region just under 10 Kb in length. The deleted segment encodes no known genes. In the SJCRH cohort, 22q11.22 loss differs by subtype: Down's syndrome 93%, BCR-ABL1 47%, ETV6-RUNX1 46%, Pseudodiploid 38%, Hyperdiploid 30%, Other 33%, MLL 12%, TCF3-PBX1 12%, T-ALL 7%, Hypodiploid 0%. In the TARGET cohort, 22q11.22 loss confers a trend for worse event-free survival (EFS; P=0.17) and overall survival (OS; P=0.06). Compared to normal 22q11.22/IKZF1 (77% EFS) or IKZF1 alternations alone (45% EFS), combined 22q11.22 loss plusIKZF1 alterations drastically reduces EFS to 15% (P<0.0001). This pattern was validated in the larger SJCRH cohort with worse outcome for isolated 22q11.22 loss (EFS; P=0.03, and OS; P=0.02). Once again, compared to normal 22q11.22/IKZF1 (EFS 76%, OS 87%) or IKZF1 alterations alone (EFS 61%, OS 80%), 22q11.22 loss plusIKZF1 alterations reduces EFS to 23% (P<0.0001) and OS to 48% (P<0.0001). In the SJCRH cohort, multiple Cox regression including subtype, age, and WBC reveals that combined 22q11.22/IKZF1 alterations independently predict worse EFS (HR 5.568 [95% CI 2.6–11.7], P<0.0001) and OS (HR 4.989 [95% CI 1.8–13.9], P=0.0021). To understand the basis of this combined worse outcome, we analyzed paired gene expression data for the TARGET (N=198) and SJCRH (N=146) cohorts. We looked for differentially expressed genes in common between both cohorts and included only samples with 22q11.22 homozygous loss to capture the most pronounced effects of the deleted region. We compared: 1) 22q11.22 normal vs. 22q11.22 loss in IKZF1 normal samples, 2) IKZF1 normal vs. IKZF1 alterations in 22q11.22 normal samples, and 3) normal samples vs. combined 22q11.22/IKZF1 altered samples. CCND2 was upregulated 2.1-fold in isolated 22q11.22 loss samples, 1.4-fold in isolated IKZF1 altered samples, and increased to 2.6-fold in combined 22q11.22 loss plusIKZF1 altered samples. Of 23 genes affected by combined 22q11.22/IKZF1 alterations, 9 were associated with resistance to at least one of 4 commonly use antileukemic agents (prednisolone, vincristine, asparaginase, and daunorubicin) in primary ALL based on a previously published dataset (NEJM 351:533,2004; N=177). For example, CCND2 over-expression in diagnostic ALL blasts significantly associated with higher LC50 (resistance) for daunorubicin (P=0.002) and prednisolone (P=0.005). Gene Ontology enrichment for differentially expressed genes in combined 22q11.22/IKZF1 altered samples was significant for anti-apoptosis (P=0.001) and cell growth (P=0.005) biological processes. In summary, we discovered and validated that 22q11.22 loss independently contributes to worse outcome in pediatric ALL in the presence of IKZF1 genetic alterations. These combined alterations may be useful to identify patients with very poor outcomes in childhood ALL. Further work to understand the mechanism of the synergistic effect of combined 22q11.22 loss plusIKZF1 alterations is underway.
Disclosures:
Shams:BioDiscovery, Inc.: Employment.
Author notes
*
Asterisk with author names denotes non-ASH members.
© 2011 by The American Society of Hematology
2011

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal