Abstract
Individual cytokines and groups of cytokines that might represent networks in chronic lymphocytic leukemia (CLL) were analyzed and their prognostic values determined. Serum levels of 23 cytokines were measured in 84 patients and 49 age-matched controls; 17 levels were significantly elevated in patients. Unsupervised hierarchical bicluster analysis identified 3 clusters (CLs) of highly correlated but differentially expressed cytokines: CL1 (CXCL9, CXCL10, CXCL11, CCL3, CCL4, CCL19, IL-5, IL-12, and IFNγ), CL2 (TNFα, IL-6, IL-8, and GM-CSF), and CL3 (IL-1β, IL-2, IL-4, IL-15, IL-17, and IFNα). Combination scores integrating expression of CL1/CL2 or CL1/CL3 strongly correlated (P < .005) with time-tofirst-treatment and overall survival (OS), respectively. Patients with the worst course had high CL1 and low CL2 or CL3 levels. Multivariate analysis revealed that CL1/CL2 combination score and immunoglobulin heavy chain variable region mutation status were independent prognostic indicators for time-to-first-treatment, whereas CL1/CL3 combination score and immunoglobulin heavy chain variable region mutation status were independent markers for OS. Thus, we identified groups of cytokines differentially expressed in CLL that are independent prognostic indicators of aggressive disease and OS. These findings indicate the value of multicytokine analyses for prognosis and suggest therapeutic strategies in CLL aimed at reducing CL1 and increasing CL2/CL3 cytokines.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by a progressive accumulation of monoclonal B lymphocytes whose growth and survival require endogenous and exogenous activation signals.1,2 Considerable progress has been made in understanding this cross talk,3,4 with clinical and translational studies supporting roles for various cytokines and chemokines, together with other soluble factors, surface receptors including adhesion molecules, and antigens in the complex stimulation of leukemic cells within the microenviroment.5,6 However, because many cytokines elevated in different CLL microenvironments are pleiotropic, with overlapping as well as antagonistic actions, determining an integrated profile of coordinately expressed cytokines that may reflect or contribute to CLL disease severity is needed.7
Individual, specific cytokines and chemokines have been reported to be elevated in the sera, plasma, or both of CLL patients and to correlate with clinical course and outcome.8-14 For example, high serum levels of IL-10, a cytokine that regulates inflammation, correlate with shorter survival.10 In addition, plasma levels of CCL3 and CCL4, 2 inflammatory chemokines that regulate cell recruitment and activation, are elevated in CLL and correlate with time-to-first treatment (TTFT)13 ; these chemokines are secreted by nurse-like cells and by CLL cells in response to B-cell receptor (BCR) engagement,15 and their secretion by leukemic cells can be down-regulated by small-molecule inhibitors of BCR signaling,15,16 linking chemokines with another environmental influence on CLL–antigen stimulation.17 Although the list of cytokines and chemokines that correlate with clinical outcomes and prognostic markers in CLL continues to grow, simultaneous analyses of large numbers of cytokines in CLL sera that identify subgroups correlating with pathogenesis, prognosis, or therapeutic responsiveness in CLL are lacking.
Here, we aimed to further elucidate complex direct and indirect CLL cell–microenvironmental interactions by correlating serum levels of immune, inflammatory, and regulatory cytokines and chemokines with clinical outcome variables and existing biologic prognostic factors. We focused on the potential of individual as well as groups of serum cytokines to distinguish CLL patients from healthy subjects and CLL patients with indolent from those with aggressive disease. We found that serum levels of 17 cytokines are significantly higher in CLL patients compared with healthy subjects. In addition, using complementary bioinformatics analyses, we identified 3 distinct clusters (CLs) of highly correlated cytokines that are differentially expressed in CLL patients with indolent and aggressive disease and that serve as independent prognostic indicators.
Methods
Patients and blood collection and processing
Studies were approved by the Institutional Review Board of the North Shore–LIJ Health System. Informed consent was obtained from all subjects in accordance with the Declaration of Helsinki. CLL patients with available data for immunoglobulin heavy chain variable region (IGHV) gene sequences and CD38 expression and age-matched healthy subjects were enrolled. Serum samples were obtained at the first patient visit to our clinic and were stored frozen at −80°C until used. Table 1 summarizes the clinical and laboratory characteristics of CLL and healthy cohorts. CLL patients with clones expressing IGHVs differing by 2% or more from the most similar germ line gene were defined as IGHV-mutated CLL (M-CLL), and those with < 2% difference were defined as IGHV-unmutated CLL (U-CLL).18 Patients with < 30% of the clone expressing CD38 were considered CD38LOW, and those patients with ≥ 30% of the clone expressing CD38 as CD38HIGH.19
Study population clinical and laboratory characteristics
| . | CLL . | Healthy . |
|---|---|---|
| Total no. of subjects* | 84 | 49 |
| Rai stage | ||
| 0-I | 54 | NA |
| II-IV | 21 | NA |
| IGHV mutation status | ||
| Mutated (≥ 2%) | 44 | NA |
| Unmutated (< 2%) | 35 | NA |
| CD38 status | ||
| Low (< 30%) | 45 | NA |
| High (≥ 30%) | 26 | NA |
| Age, y | ||
| Median | 65 | 59 |
| Range | 34-91 | 44-89 |
| . | CLL . | Healthy . |
|---|---|---|
| Total no. of subjects* | 84 | 49 |
| Rai stage | ||
| 0-I | 54 | NA |
| II-IV | 21 | NA |
| IGHV mutation status | ||
| Mutated (≥ 2%) | 44 | NA |
| Unmutated (< 2%) | 35 | NA |
| CD38 status | ||
| Low (< 30%) | 45 | NA |
| High (≥ 30%) | 26 | NA |
| Age, y | ||
| Median | 65 | 59 |
| Range | 34-91 | 44-89 |
NA indicates not applicable.
Clinical data were not available on all 84 CLL patients: Rai stage (75/84), IGHV mutation status (79/84), and CD38 percentages (71/84).
Multiplex cytokine analysis
Levels of IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12, IL-15, IL-17, IFNα, IFNγ, TNFα, GM-CSF, CCL2, CCL3, CCL4, and CCL11 were quantified in sera from 81 CLL patients and 45 healthy age-matched subjects using a multiplex sandwich immunoassay-based protein array system (BioSource International).20,21 Serum levels of CXCL9, CXCL10, CXCL11, CCL17, and CCL19 were quantified in 57 CLL patients and 35 healthy age-matched controls using SearchLight Protein Array, a sandwich immunoassay-based protein array system (Pierce Biotechnology) as per the manufacturer's method. All cytokine determinations were performed in duplicate, and concentrations are reported in picograms per milliliter. For multicytokine analyses (bicluster, mosaic, and discriminant function analysis [DFA]), raw data were logarithmically transformed and standardized across all samples by the z-transform (subtracting mean and dividing by standard deviation). See supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) for a critical discussion of multiplex bead array versus traditional ELISA techniques.
Statistical analysis
Statistical analysis of differences in cytokine levels between groups was performed using a nonparametric Kruskal-Wallis test followed by a Dunn multiple comparisons test. Overall survival (OS) and TTFT were estimated using the Kaplan-Meier product-limit method, and pairwise comparisons were carried out with the log-rank test using a Bonferroni-type correction (P < .01). Cox proportional hazard regression analysis with backward elimination was used to determine multivariable independence (IGHV mutation status, CD38 group, CL1/CL2 combination group, and CL1/CL3 combination group) in predicting OS and TTFT. Statistical analyses were performed using Prism Version 5.00 for Windows (GraphPad Software) and SAS 9.2 (SAS Institute). See supplemental Methods for additional details.
Random forest, hierarchical cluster, and mosaic analyses
Data on all cytokines were used for these analyses. Random Forest analysis (RFA), using R package randomForest (http://cran.r-project.org/web/packages/randomForest/) Version 4.5-34,22 was used to rank cytokine levels in importance in predicting phenotype. Hierarchical clustering was used to identify sets of cytokines whose expression levels correlated among individuals within a population. A heatmap was produced with Bioconductor package made423 Version 1.14 using average linkage on a similarity matrix derived by Pearson moment correlations. Mosaic analysis24 was used to determine the degree to which individual cytokines in sera correlated with one another. See supplemental Methods for more details.
Discriminant function analysis
DFA, a multivariate class distinction algorithm,25 was used to identify linear combinations of cytokines (“roots”) that best discriminate CLL patients with aggressive disease (TTFT < 5 years) from those with mild disease (TTFT ≥ 5 years). Software for DFA was supplied by STATISTICA Version 6 (StatSoft). The overall model significance of the discriminant function was tested by the Wilks λ test.
Results
Serum cytokines and chemokines analyzed
Based on a pilot study analyzing 43 cytokines, a subset of 23 (Table 2) was selected for multiplex serum determinations in CLL and healthy subjects. Using a series of bioinformatic tools, CLL patient cytokine and chemokine levels were analyzed as independent variables for their relation to levels of the same cytokines and chemokines in healthy subjects and for their correlation with disease course (TTFT) and outcome (OS). Comparisons with the latter parameters also were made for groups of cytokines and chemokines developed by clustering and grouping tools.
Serum cytokine levels: CLL versus age-matched healthy subjects
| Cytokine* . | CLL (n = 84) . | Healthy (n = 49) . | P† . | ||
|---|---|---|---|---|---|
| Mean . | Median (range) . | Mean . | Median (range) . | ||
| IL-1β | 38.1 | 6.2 (4-395.9) | 7.9 | 6.2 (4-60.1) | .0038 |
| IL-2 | 32.7 | 3.2 (3.2-560.9) | 6.5 | 3.2 (1.6-69.5) | .0385 |
| IL-4 | 1.4 | 0.3 (0.2-16.6) | 0.9 | 0.3 (0.3-11.5) | .4291 |
| IL-5 | 3.4 | 1.1 (0.2-120.4) | 0.5 | 0.2 (0.1-3.1) | < .0001 |
| IL-6 | 17.3 | 6.9 (0.2-373.7) | 6.1 | 4.4 (0.5-65.7) | .0220 |
| IL-8 | 173.9 | 12.2 (0.8-2465) | 10.2 | 4.5 (1.9-191.9) | .0001 |
| IL-10 | 8.2 | 3.6 (0.3-203.7) | 10.6 | 0.3 (0.2-310.3) | < .0001 |
| IL-12 | 270.3 | 146.5 (5.3-2544.9) | 101.5 | 81.5 (6.7-416) | .0009 |
| IL-15 | 13.8 | 6.4 (1-80.7) | 8.8 | 2.4 (1-47.8) | .1490 |
| IL-17 | 6.9 | 1.3 (0.3-137.7) | 0.9 | 0.3 (0.3-11) | < .0001 |
| TNFα | 7.9 | 1.4 (1.4-117.5) | 1.9 | 1.4 (1.4-6.7) | .0108 |
| IFNα | 15.2 | 2.5 (2.5-136.3) | 5.9 | 2.5 (2.5-49.6) | .0130 |
| IFNγ | 2.0 | 0.9 (0.1-30.7) | 0.8 | 0.3 (0.3-7.7) | .0069 |
| GM-CSF | 15.4 | 1.7 (0.2-412.9) | 20.6 | 1.1 (0.5-783.7) | .1073 |
| CCL2 | 741.6 | 641.8 (95.7-3079.1) | 439.3 | 376.6 (180.7-1468.7) | < .0001 |
| CCL3 | 94.3 | 35.5 (1-2227.3) | 14.0 | 1.0 (1-264.5) | < .0001 |
| CCL4 | 298.3 | 170.6 (1.2-3290.3) | 110.0 | 58.6 (2-1926.1) | < .0001 |
| CCL11 | 159.1 | 146.5 (3-560.3) | 143.4 | 139.4 (30.4-339.2) | .5961 |
| CCL17‡ | 305 | 185 (12.6-1619) | 46.9 | 42 (11.8-84.3) | .0001 |
| CCL19§ | 146.9 | 108.1 (20.3-699.2) | 149.0 | 118.4 (9.4-652.4) | .3566 |
| CXCL9§ | 862.5 | 402.1 (59.3-7302.4) | 257.9 | 197.2 (59.9-1082.4) | .0021 |
| CXCL10§ | 131.6 | 83.1 (13.2-518.6) | 102.9 | 75.5 (14.9-740.5) | .2889 |
| CXCL11§ | 166.5 | 107.2 (13.5-1505.9) | 56.0 | 26.5 (8.8-275) | < .0001 |
| Cytokine* . | CLL (n = 84) . | Healthy (n = 49) . | P† . | ||
|---|---|---|---|---|---|
| Mean . | Median (range) . | Mean . | Median (range) . | ||
| IL-1β | 38.1 | 6.2 (4-395.9) | 7.9 | 6.2 (4-60.1) | .0038 |
| IL-2 | 32.7 | 3.2 (3.2-560.9) | 6.5 | 3.2 (1.6-69.5) | .0385 |
| IL-4 | 1.4 | 0.3 (0.2-16.6) | 0.9 | 0.3 (0.3-11.5) | .4291 |
| IL-5 | 3.4 | 1.1 (0.2-120.4) | 0.5 | 0.2 (0.1-3.1) | < .0001 |
| IL-6 | 17.3 | 6.9 (0.2-373.7) | 6.1 | 4.4 (0.5-65.7) | .0220 |
| IL-8 | 173.9 | 12.2 (0.8-2465) | 10.2 | 4.5 (1.9-191.9) | .0001 |
| IL-10 | 8.2 | 3.6 (0.3-203.7) | 10.6 | 0.3 (0.2-310.3) | < .0001 |
| IL-12 | 270.3 | 146.5 (5.3-2544.9) | 101.5 | 81.5 (6.7-416) | .0009 |
| IL-15 | 13.8 | 6.4 (1-80.7) | 8.8 | 2.4 (1-47.8) | .1490 |
| IL-17 | 6.9 | 1.3 (0.3-137.7) | 0.9 | 0.3 (0.3-11) | < .0001 |
| TNFα | 7.9 | 1.4 (1.4-117.5) | 1.9 | 1.4 (1.4-6.7) | .0108 |
| IFNα | 15.2 | 2.5 (2.5-136.3) | 5.9 | 2.5 (2.5-49.6) | .0130 |
| IFNγ | 2.0 | 0.9 (0.1-30.7) | 0.8 | 0.3 (0.3-7.7) | .0069 |
| GM-CSF | 15.4 | 1.7 (0.2-412.9) | 20.6 | 1.1 (0.5-783.7) | .1073 |
| CCL2 | 741.6 | 641.8 (95.7-3079.1) | 439.3 | 376.6 (180.7-1468.7) | < .0001 |
| CCL3 | 94.3 | 35.5 (1-2227.3) | 14.0 | 1.0 (1-264.5) | < .0001 |
| CCL4 | 298.3 | 170.6 (1.2-3290.3) | 110.0 | 58.6 (2-1926.1) | < .0001 |
| CCL11 | 159.1 | 146.5 (3-560.3) | 143.4 | 139.4 (30.4-339.2) | .5961 |
| CCL17‡ | 305 | 185 (12.6-1619) | 46.9 | 42 (11.8-84.3) | .0001 |
| CCL19§ | 146.9 | 108.1 (20.3-699.2) | 149.0 | 118.4 (9.4-652.4) | .3566 |
| CXCL9§ | 862.5 | 402.1 (59.3-7302.4) | 257.9 | 197.2 (59.9-1082.4) | .0021 |
| CXCL10§ | 131.6 | 83.1 (13.2-518.6) | 102.9 | 75.5 (14.9-740.5) | .2889 |
| CXCL11§ | 166.5 | 107.2 (13.5-1505.9) | 56.0 | 26.5 (8.8-275) | < .0001 |
Cytokine values are reported in picograms per milliliter.
Results of Mann-Whitney nonparametric test comparing median values. Bold values indicate significant difference (P < .05) between CLL and age-matched healthy subjects.
For this cytokine, there were only 36 CLL subjects and 10 controls subjects.
For this cytokine, there were only 63 CLL and 42 healthy subjects.
Analyses of individual cytokines and chemokines
Elevated levels of a discrete cytokines in CLL.
Serum levels for all cytokines tested were equal to or greater in CLL patients than in age-matched healthy subjects (Table 2; Figure 1A). Serum levels of CCL2, CCL3, CCL4, CCL17, CXCL9, CXCL11, IL-1β, IL-2, IL-5, IL-6, IL-8, IL-10, IL-12, IL-17, TNFα, IFNγ, and IFNα were significantly higher (P < .05) in CLL patients.
A subset of cytokines is differentially expressed in the sera of CLL patients compared with healthy age-matched subjects. (A) Levels of cytokines present at significantly higher levels in the sera of CLL patients compared with the sera of healthy subjects are presented as box plots with the bottom and top of the box indicating the 25th and 75th percentiles, respectively. The bar within the box indicates the median value and the ends of the whiskers represent the 10th and 90th percentiles. Outliers are represented by dots. (B) RFA. Cytokines are ranked by their relative importance in discriminating CLL from healthy subjects with CCL3 being the most important predictor of CLL. Only subjects with complete cytokine data (82 total; 31 healthy subjects and 51 CLL patients) were used. Statistical significance was defined as P < .05. For this dataset, the classification error for healthy subjects is 12.9% (4/31) and for CLL subjects 3.9% (2/51). The overall classification error is 7.3% (6/82) keeping the sample size bias or 8.4% [0.5(4/31) + 0.5(2/51)] if we average the 2 error rates. The horizontal axis represents the average decrease in classification accuracy and horizontal bars representing the relative importance of each individual cytokine are sorted by importance along the vertical axis. The horizontal dashed line divides cytokines at the mean value displayed on the horizontal axis and defines the minimum number of cytokines required for maximum classification accuracy.
A subset of cytokines is differentially expressed in the sera of CLL patients compared with healthy age-matched subjects. (A) Levels of cytokines present at significantly higher levels in the sera of CLL patients compared with the sera of healthy subjects are presented as box plots with the bottom and top of the box indicating the 25th and 75th percentiles, respectively. The bar within the box indicates the median value and the ends of the whiskers represent the 10th and 90th percentiles. Outliers are represented by dots. (B) RFA. Cytokines are ranked by their relative importance in discriminating CLL from healthy subjects with CCL3 being the most important predictor of CLL. Only subjects with complete cytokine data (82 total; 31 healthy subjects and 51 CLL patients) were used. Statistical significance was defined as P < .05. For this dataset, the classification error for healthy subjects is 12.9% (4/31) and for CLL subjects 3.9% (2/51). The overall classification error is 7.3% (6/82) keeping the sample size bias or 8.4% [0.5(4/31) + 0.5(2/51)] if we average the 2 error rates. The horizontal axis represents the average decrease in classification accuracy and horizontal bars representing the relative importance of each individual cytokine are sorted by importance along the vertical axis. The horizontal dashed line divides cytokines at the mean value displayed on the horizontal axis and defines the minimum number of cytokines required for maximum classification accuracy.
RFA ranks individual serum cytokines that best discriminate CLL patients from healthy subjects.
RFA indicated that CCL3, IL-5, IL-10, CCL4, CXCL11, IL-8, CCL2, and IL-17 best differentiated between CLL and healthy groups (Figure 1B; supplemental Table 1). RFA was run > 100 times to assess the robustness of cytokine ordering; CCL3, IL-5, IL-10, CCL4, IL-8, and CXCL11 always appeared on top of the list, whereas ordering of cytokines toward the bottom fluctuated, suggesting their contribution was not consistent. Although the full set distinguished CLL from healthy, levels of CCL3 were the dominant discriminator, contributing > 80%, consistent with published data.13
Correlation between individual cytokine levels and prognostic markers.
Individual serum cytokine levels were stratified based on patient's Rai stage, IGHV mutation status, and CD38 expression. Of the 23 cytokines, only IL-12 was correlated (P < .01) with advanced Rai stages (II-IV; Figure 2A), although CCL4 exhibited a strong trend in this regard. CCL3, IL-12, and IL-17 correlated more strongly with U-CLL and IL-8 with M-CLL (Figure 2B; supplemental Table 2). Levels of CXCL9 and IL-12 were elevated in CD38HIGH patients (P < .01; Figure 2C; supplemental Table 3), whereas IL-8, IL-1β, and IL-17 were elevated in CD38LOW patients (P < .001; Figure 2C) compared with healthy subjects. Serum levels of CCL2, CCL4, CCL17, CXCL11, IL-5, and IL-10 were higher in CLL patients (P < .0001), irrespective of IGHV mutation status or CD38 level (supplemental Tables 2-3).
Analysis of serum cytokine levels as a function of Rai stage, IGHV mutation status, CD38 expression, OS, and TTFT. Cytokines for which serum levels are significantly different when patient cohorts are divided by Rai stage (A), M-CLL (good prognosis) and U-CLL (poor prognosis) compared with healthy subjects (B), or CD38HIGH (poor prognosis) and CD38LOW (good prognosis) compared with healthy subjects (C). Data are presented as box plots (see legend to Figure 1 for details; *P < .05, **P < .01, ***P < .001). A result was considered significantly different if P < .05 (A) or P < .017 (B-C) to account for multiple comparisons. (D) Kaplan-Meier analysis of the levels of CXCL10, CXCL11, CCL19, and IL-1β versus survival. CXCL10, CXCL11, and CCL19 independently correlated with shorter survival: CXCL10 (16.3 years vs not reached; P = .012; HR = 3.1; 95% confidence interval [CI], 1.3-7.6), CXCL11 (15.3 years vs not reached; P = .013; HR = 3.1; 95% CI, 1.3-7.7), and CCL19 (16.3 years vs not reached; P = .042; HR = 2.4; 95% CI, 1.0-5.7). IL-1β independently correlated with longer survival (not reached vs 15.3 years; P = .022; HR = 0.4; 95% CI, 0.2-0.9). (E) Kaplan-Meier analysis of the levels of CCL3, CCL4, IL-10, and IL-12 versus TTFT. CCL3, CCL4, IL-10 and IL-12 independently correlated with shorter TTFT: CCL3 (7.6 years vs not reached; P = .031; HR = 2.0; 95% CI, 1.1-3.8), CCL4 (5.4 vs 20.9 years; P = .001; HR = 3.1; 95% CI, 1.6-6.0), IL-10 (7.6 years vs not reached; P = .030; HR = 2.0; 95% CI, 1.1-3.8), and IL-12 (6.8 vs 20.9 years; P = .019; HR = 2.2; 95% CI, 1.1-4.1). Sera were collected and analyzed as described in “Patients and blood collection and processing” and “Multiplex cytokine analysis.”
Analysis of serum cytokine levels as a function of Rai stage, IGHV mutation status, CD38 expression, OS, and TTFT. Cytokines for which serum levels are significantly different when patient cohorts are divided by Rai stage (A), M-CLL (good prognosis) and U-CLL (poor prognosis) compared with healthy subjects (B), or CD38HIGH (poor prognosis) and CD38LOW (good prognosis) compared with healthy subjects (C). Data are presented as box plots (see legend to Figure 1 for details; *P < .05, **P < .01, ***P < .001). A result was considered significantly different if P < .05 (A) or P < .017 (B-C) to account for multiple comparisons. (D) Kaplan-Meier analysis of the levels of CXCL10, CXCL11, CCL19, and IL-1β versus survival. CXCL10, CXCL11, and CCL19 independently correlated with shorter survival: CXCL10 (16.3 years vs not reached; P = .012; HR = 3.1; 95% confidence interval [CI], 1.3-7.6), CXCL11 (15.3 years vs not reached; P = .013; HR = 3.1; 95% CI, 1.3-7.7), and CCL19 (16.3 years vs not reached; P = .042; HR = 2.4; 95% CI, 1.0-5.7). IL-1β independently correlated with longer survival (not reached vs 15.3 years; P = .022; HR = 0.4; 95% CI, 0.2-0.9). (E) Kaplan-Meier analysis of the levels of CCL3, CCL4, IL-10, and IL-12 versus TTFT. CCL3, CCL4, IL-10 and IL-12 independently correlated with shorter TTFT: CCL3 (7.6 years vs not reached; P = .031; HR = 2.0; 95% CI, 1.1-3.8), CCL4 (5.4 vs 20.9 years; P = .001; HR = 3.1; 95% CI, 1.6-6.0), IL-10 (7.6 years vs not reached; P = .030; HR = 2.0; 95% CI, 1.1-3.8), and IL-12 (6.8 vs 20.9 years; P = .019; HR = 2.2; 95% CI, 1.1-4.1). Sera were collected and analyzed as described in “Patients and blood collection and processing” and “Multiplex cytokine analysis.”
Predictive value of individual cytokine levels for OS and TTFT.
To identify specific cytokines associated with clinical outcome, we divided CLL patients into 2 categories for each serum cytokine: those with levels above or below the median. We then used the Kaplan-Meier method and the log-rank test to compare median survival between patients in these categories (Figure 2D; supplemental Table 4). CXCL10, CXCL11, and CCL19 levels independently associated with shorter survival, and IL-1β independently associated with longer survival (Figure 2D), although these associations did not reach the levels of statistical significance required by Bonferroni-like multiple comparison adjustment (P < .01). We also correlated levels of each cytokine with TTFT (Figure 2E); CCL3, CCL4, IL-10, and IL-12 independently associated with shorter TTFT, although the association reached statistical significance (P < .01) only for IL-12.
When the CLL risk factors IGHV mutation status and CD38 expression were fitted with cytokine levels into a Cox proportional hazards regression model with backward elimination, only CXCL10, IL-1β, and IGHV mutation status were significantly associated with OS. When a similar analysis was carried out for TTFT, only CCL4, IL-12, and IGHV mutation status correlated significantly. Multivariable analysis revealed that CXCL10 (hazard ratio [HR] = 4.78; P = .0052), IL-1β (HR = 0.16; P = .0065), and IGHV mutation status (HR = 16.42; P = .0002) were independent prognostic markers for OS, and CCL4 (HR = 2.79; P < .0092), IL-12 (HR = 2.45; P = .0199), and mutation status (HR = 2.54; P = .0171) were independent prognostic markers for TTFT.
Analyses of CLs of cytokines and chemokines
Two-dimensional cluster analysis of serum cytokines defines 3 sets of cytokines relating to TTFT.
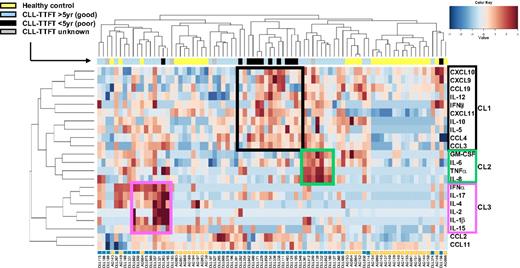
Although analysis of individual cytokines identified 8 associated with clinical outcome parameters, these associations were not robust. Therefore, we performed an unsupervised hierarchical bicluster analysis to identify sets of cytokines that might be coordinately expressed in patients with indolent versus aggressive disease and correlate more strongly with disease outcome. Although CLL patients and healthy subjects clustered separately (Figure 3), CLL patients did not separate into groups based on IGHV mutation status or CD38 expression (supplemental Figure 2). However, patients with shorter TTFT (Figure 3 black bars) generally did group together based on higher levels of a CL of cytokines (CCL3, CCL4, CCL19, CXCL9, CXCL10, CXCL11, IFNγ, IL-5, IL-10, and IL-12) that we termed CL1.
Two-dimensional CL analysis of the cytokine dataset. Expression levels of individual cytokines are represented by shades of blue to red in the central heatmap, with highest values in dark red and the lowest in dark blue. In this unsupervised bicluster analysis, healthy subjects (indicated by yellow boxes directly below heatmap) clustered together in 2 discrete groups, sharply distinct from CLL patients (indicated by turquoise boxes). TTFT status of CLL patients is indicated by blue (≥ 5 years), black (< 5 years), or gray (unknown) coloring in bar above heatmap. CLL patients with short TTFT formed a discrete CL. Three distinct sets of correlated cytokines (CLs) are highlighted by black, green, and pink boxes on heatmap. All available data on the entire panel of cytokines were used for this analysis with the exception of CCL17, because CCL17 values were not available for all patients.
Two-dimensional CL analysis of the cytokine dataset. Expression levels of individual cytokines are represented by shades of blue to red in the central heatmap, with highest values in dark red and the lowest in dark blue. In this unsupervised bicluster analysis, healthy subjects (indicated by yellow boxes directly below heatmap) clustered together in 2 discrete groups, sharply distinct from CLL patients (indicated by turquoise boxes). TTFT status of CLL patients is indicated by blue (≥ 5 years), black (< 5 years), or gray (unknown) coloring in bar above heatmap. CLL patients with short TTFT formed a discrete CL. Three distinct sets of correlated cytokines (CLs) are highlighted by black, green, and pink boxes on heatmap. All available data on the entire panel of cytokines were used for this analysis with the exception of CCL17, because CCL17 values were not available for all patients.
Furthermore, the analysis revealed a group of patients with moderate CL1 levels combined with higher levels of a second CL of correlated cytokines (CL2; GM-CSF, IL-8, TNFα, and IL-6; Figure 3). Notably, the combined group CL1/CL2 had relatively longer TTFT (≥ 5 years; Figure 3 light blue bars) than CL1 alone even though the CL2 group by itself did not exhibit shorter TTFT (see Figure 4A).
Cytokine CLs predict OS and TTFT.
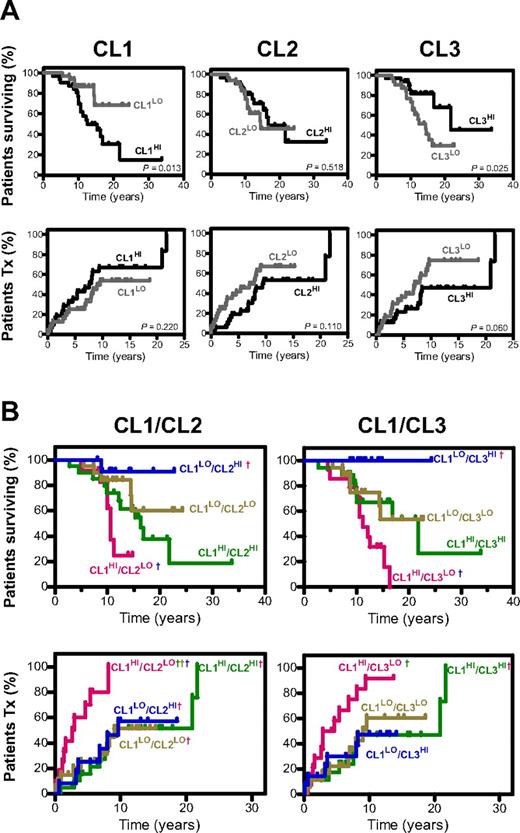
Patients expressing high versus low levels of CL1, CL2, or CL3 cytokines, either individually or in combination, were compared for OS and TTFT (Figure 4; Table 3). Survival analyses using individual CLs revealed that high serum levels of CL1 correlated with shorter OS (CL1HI 12.5 years vs CL1LO not reached; P = .013; Figure 4A top left). In contrast, high levels of CL3 correlated with longer OS (CL3HI 21.8 years vs CL3LO 14.4 years; P = .025; Figure 4A top right) and tended toward longer TTFT (CL3HI 20.9 years vs CL3LO 7.0 years; P = .060; Figure 4A bottom panel). Levels of CL2 did not correlate with OS or TTFT.
Predictive value of cytokine CLs for OS and TTFT. Kaplan-Meier curves depicting correlation between levels of each individual cytokine CL and OS and TTFT (A) and multiple CLs and OS and TTFT (B). In multiple cluster analyses a dagger (†) indicates a statistically significant difference at the P < .01 level (accounting for multiple comparisons) between that combined CL group and a second combined CL group (the identity of which is indicated by the color of the †). Median survival and TTFT (years), HR (95% CI), and P values for combination CL group comparisons are found in supplemental Table 5. The method by which patients were subgrouped by a categorical score based on their integrated cytokine levels within the different cytokine CLs (CL1, CL2, and CL3) is described in supplemental Methods.
Predictive value of cytokine CLs for OS and TTFT. Kaplan-Meier curves depicting correlation between levels of each individual cytokine CL and OS and TTFT (A) and multiple CLs and OS and TTFT (B). In multiple cluster analyses a dagger (†) indicates a statistically significant difference at the P < .01 level (accounting for multiple comparisons) between that combined CL group and a second combined CL group (the identity of which is indicated by the color of the †). Median survival and TTFT (years), HR (95% CI), and P values for combination CL group comparisons are found in supplemental Table 5. The method by which patients were subgrouped by a categorical score based on their integrated cytokine levels within the different cytokine CLs (CL1, CL2, and CL3) is described in supplemental Methods.
OS and TTFT analysis for single and combined cluster groups
| Characteristic . | No. Total . | No. Expired . | No. Treated . | OS . | TTFT . | ||
|---|---|---|---|---|---|---|---|
| Median, y (95% CI) . | P* . | Median, y (95% CI) . | P* . | ||||
| All patients | 84 | 31 | 42 | 16.9 (14.4, —)† | 8.05 (6.8, 21.7) | ||
| CL1 | |||||||
| > Median | 32 | 18 | 21 | 12.5 (10, 21.8) | .013 | 7.6 (3.8, 20.9) | .2196 |
| < Median | 32 | 6 | 15 | — (14.5, —) | 9.6 (7.0, —) | ||
| CL2 | |||||||
| > Median | 32 | 12 | 17 | 16.9 (12.5, —) | .518 | 9.6 (6.8, 21.7) | .110 |
| < Median | 32 | 12 | 19 | 14.5 (10.6, —) | 7.9 (2.8, —) | ||
| CL3 | |||||||
| > Median | 32 | 7 | 15 | 21.8 (16.9, —) | .025 | 20.9 (7.6, 21.7) | .0604 |
| < Median | 32 | 17 | 21 | 14.4 (10.6, 16.3) | 7.0 (2.9, 9.4) | ||
| CL1/CL2 combined group | |||||||
| CL1HI CL2LO | 12 | 7 | 10 | 10.6 (8.6, —) | .0209 | 2.8 (0.6, 5.4) | .0002 |
| CL1HICL2HI | 20 | 11 | 11 | 16.3 (9.8,) | 9.4 (6.8, 21.7) | ||
| CL1LOCL2LO | 20 | 5 | 9 | — (14.4, —) | 9.0 (3.9, —) | ||
| CL1LOCL2HI | 12 | 1 | 6 | — (—) | 9.6 (3.2, —) | ||
| CL1/CL3 combined group | |||||||
| CL1HICL3LO | 14 | 11 | 12 | 11.2 (8.6, 15.3) | .0023 | 3.8 (1.3, 6.8) | .0107 |
| CL1HICL3HI | 18 | 7 | 9 | 21.8 (9.8, —) | 20.9 (5.4, 21.7) | ||
| CL1LOCL3LO | 18 | 6 | 9 | — (8.8, —) | 9.0 (6.8, —) | ||
| CL1LOCL3HI | 14 | 0 | 6 | — — | — (3.2, —) | ||
| CL1HI/CL2LO versus all other groups combined‡ | |||||||
| CL1HICL2LO | 12 | 7 | 10 | 10.6 (8.6, —) | .0102 | 2.8 (0.6, 5.4) | < .0001 |
| all other groups | 52 | 17 | 26 | 21.8 (14.5, —) | 9.4 (7.9, 21.7 | ||
| CL1HI/CL3LO versus all other groups combined§ | |||||||
| CL1HICL3LO | 14 | 11 | 12 | 11.2 (8.6, 15.3) | .007 | 3.8 (1.3, 6.8) | .0009 |
| All other groups | 50 | 13 | 24 | — (16.9, —) | 9.6 (7.9, 21.7) | ||
| Characteristic . | No. Total . | No. Expired . | No. Treated . | OS . | TTFT . | ||
|---|---|---|---|---|---|---|---|
| Median, y (95% CI) . | P* . | Median, y (95% CI) . | P* . | ||||
| All patients | 84 | 31 | 42 | 16.9 (14.4, —)† | 8.05 (6.8, 21.7) | ||
| CL1 | |||||||
| > Median | 32 | 18 | 21 | 12.5 (10, 21.8) | .013 | 7.6 (3.8, 20.9) | .2196 |
| < Median | 32 | 6 | 15 | — (14.5, —) | 9.6 (7.0, —) | ||
| CL2 | |||||||
| > Median | 32 | 12 | 17 | 16.9 (12.5, —) | .518 | 9.6 (6.8, 21.7) | .110 |
| < Median | 32 | 12 | 19 | 14.5 (10.6, —) | 7.9 (2.8, —) | ||
| CL3 | |||||||
| > Median | 32 | 7 | 15 | 21.8 (16.9, —) | .025 | 20.9 (7.6, 21.7) | .0604 |
| < Median | 32 | 17 | 21 | 14.4 (10.6, 16.3) | 7.0 (2.9, 9.4) | ||
| CL1/CL2 combined group | |||||||
| CL1HI CL2LO | 12 | 7 | 10 | 10.6 (8.6, —) | .0209 | 2.8 (0.6, 5.4) | .0002 |
| CL1HICL2HI | 20 | 11 | 11 | 16.3 (9.8,) | 9.4 (6.8, 21.7) | ||
| CL1LOCL2LO | 20 | 5 | 9 | — (14.4, —) | 9.0 (3.9, —) | ||
| CL1LOCL2HI | 12 | 1 | 6 | — (—) | 9.6 (3.2, —) | ||
| CL1/CL3 combined group | |||||||
| CL1HICL3LO | 14 | 11 | 12 | 11.2 (8.6, 15.3) | .0023 | 3.8 (1.3, 6.8) | .0107 |
| CL1HICL3HI | 18 | 7 | 9 | 21.8 (9.8, —) | 20.9 (5.4, 21.7) | ||
| CL1LOCL3LO | 18 | 6 | 9 | — (8.8, —) | 9.0 (6.8, —) | ||
| CL1LOCL3HI | 14 | 0 | 6 | — — | — (3.2, —) | ||
| CL1HI/CL2LO versus all other groups combined‡ | |||||||
| CL1HICL2LO | 12 | 7 | 10 | 10.6 (8.6, —) | .0102 | 2.8 (0.6, 5.4) | < .0001 |
| all other groups | 52 | 17 | 26 | 21.8 (14.5, —) | 9.4 (7.9, 21.7 | ||
| CL1HI/CL3LO versus all other groups combined§ | |||||||
| CL1HICL3LO | 14 | 11 | 12 | 11.2 (8.6, 15.3) | .007 | 3.8 (1.3, 6.8) | .0009 |
| All other groups | 50 | 13 | 24 | — (16.9, —) | 9.6 (7.9, 21.7) | ||
Bold values indicate a significant difference for single cluster comparisons (P < .05) and for pairwise comparisons (P < .01) of CL1/CL2 and CL1/CL3 combined cluster groups.
— indicates not reached.
The phrase “all other groups combined” refers to the combination of the 3 CL1/CL2 cluster groups with better OS (CL1HI/CL2HI, CL1LO/CL2LO, and CL1LO/CL2HI). CL1HI/CL2LO, and “all other groups combined” are the groups used in multivariate analysis.
“All other groups combined” refers to the combination of CL1HI/CL3HI, CL1LO/CL3LO, and CL1LO/CL3HI.
The levels of CL2 and CL3 were variable in CL1HI patients, and in fact, 10 of 12 patients with TTFT < 5 years had low levels of both CL2 and CL3 (Figure 3). Therefore, we next explored whether a categorical score integrating expression of 2 CLs, either CL1/CL2 or CL1/CL3, better correlated with OS or TTFT (Figure 4B; Table 3). Stratification of patients on the basis of various combinations of CL1 and CL2 revealed differences among groups for both OS (P = .0209) and TTFT (P = .0002). Patients who did the worst long-term were CL1HI and CL2LO, whereas those who did the best were CL1LO and CL2HI. Significant differences also were observed when patients were stratified by combining CL1 and CL3 (OS, P = .0023; TTFT, P = .0107), with patients with shortest TTFT and OS being CL1HI and CL3LO and those with the longest being CL1LO and CL3HI. Detailed statistical analyses of single and combination comparisons are reported in supplemental Table 5.
Recursive partitioning analysis testing the prognostic power of CL1, CL2, and CL3 combinations revealed that 43 of 45 CLL patients with TTFT of 5 or more years and 11 of 19 CLL patients with TTFT of < 5 years were correctly identified by CL1LO/CL2HI/CL3HI and CLHI/CL2LO/CL3LO, respectively (with classification success rates of 96% and 58%, respectively; see supplemental Figure 3).
Multivariate analysis of cytokine CL combinations, IGHV mutation status, and CD38 expression as prognostic indicators of OS and TTFT.
CL1/CL2 and CL1/CL3 combination CL groups were fitted into a Cox proportional hazards regression model with backward elimination (Table 4). For multivariate analysis, the 3 CL1/CL2 CL groups with better outcomes (OS and TTFT) were collapsed into a single group that was compared with CL1HI/CL2LO (worst outcome) group. A similar process was performed for CL1/CL3 CL groups.
Multivariate analysis
| Parameters in model . | HR (95% CI) . | P . |
|---|---|---|
| Survival reference (OS)* | ||
| IGHV mutation status (reference: M-CLL) | 6.91 (1.92-24.87) | .003 |
| CL1HICL3LO (ref: all other groups combined) | 5.26 (1.95-14.21) | .001 |
| Treatment (TTFT)† | ||
| IGHV mutation status (reference: M-CLL) | 2.90 (1.36-6.19) | .0059 |
| CL1HICL2LO (reference: all other groups combined) | 6.12 (2.48-15.10) | < .0001 |
| Parameters in model . | HR (95% CI) . | P . |
|---|---|---|
| Survival reference (OS)* | ||
| IGHV mutation status (reference: M-CLL) | 6.91 (1.92-24.87) | .003 |
| CL1HICL3LO (ref: all other groups combined) | 5.26 (1.95-14.21) | .001 |
| Treatment (TTFT)† | ||
| IGHV mutation status (reference: M-CLL) | 2.90 (1.36-6.19) | .0059 |
| CL1HICL2LO (reference: all other groups combined) | 6.12 (2.48-15.10) | < .0001 |
OS indicates overall survival.
TTFT indicates time-to-first treatment.
CL1HI/CL3LO group and IGHV mutation status (but not CL1HI/CL2LO or CD38) retained discriminatory power as independent prognostic factors for OS in multivariate analysis. Subjects in the CL1HI/CL3LO group were 5.3 times more likely to die sooner than those in the other 3 groups (HR = 5.3; P = .001); this was similar to the comparison of U-CLL with M-CLL (HR = 6.91; P = .003). For TTFT, in contrast, the CL1HI/CL2LO group and IGHV mutation status (but not CL1HI/CL3LO or CD38) were independent prognosticators. Subjects in the CL1HI/CL2LO group were 6.1 times more likely to be treated earlier than those in the other groups (HR = 6.1; P < .0001). U-CLL patients were 2.9 times more likely for to be treated earlier than M-CLL (HR = 2.9; P = .006).
Mosaic analysis identifies discrete networks of highly correlated cytokines in better versus worse outcome CLL.
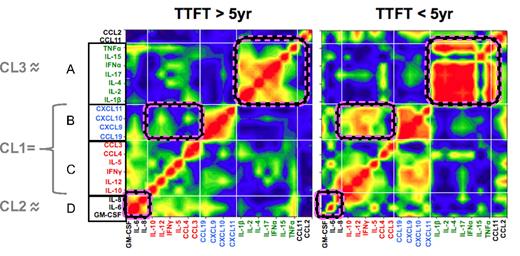
Mosaic analysis was used to evaluate the degree to which serum levels of individual cytokines correlated with one another, and whether these correlations differed between CLL patients with less (TTFT ≥ 5 years) and more (TTFT < 5 years) aggressive disease (Figure 5). Unlike bicluster analysis that uses cytokine levels to develop a color-coded heatmap (Figure 3), the mosaic plot assigns colors based on the strength of correlation between pairs of cytokines, irrespective of serum levels.24 This approach identified 4 broad groups of correlated cytokines (A-D). To a large degree, these groups mirror the CLs identified by bicluster analysis (indicated by gray lines at left of Figure 3); specifically, groups B and C cytokines comprise CL1 and groups A and D are, respectively, CL3 and CL2, except for TNFα, which falls in CL2 and group A. Although correlation profiles were similar in patients with mild and aggressive disease, differences between the 2 sets of patients were apparent (pink-striped boxes in Figure 5); group D cytokines correlated highly in patients with mild disease and groups B and C (which together form CL1) correlated highly in patients with aggressive disease.
Networks of highly correlated cytokines in sera of patients with mild and aggressive CLL. “Connectivity mosaics” representing matrices of correlation coefficients are shown for serum cytokines from patients with TTFT of ≥ 5 years (left) and patients with TTFT of < 5 years (right). Individual cytokines are identically listed on x- and y-axes with A, B, C, and D indicating selected groups of highly correlated cytokines. Color shades indicate the type of correlation, with red = positive, blue = negative, and green = absent correlation. Light white lines indicate the borders of defined CLs of interconnected cytokines. For clarity, we indicate the relationship between groups of highly correlated cytokines determined by mosaic analysis and the 3 cytokine CLs (CL1, CL2, and CL3) identified by unsupervised cluster analysis (indicated in gray at far left). Some areas in the mosaic plot which differ in correlation strength between the 2 TTFT cohorts are delineated by pink-striped edges. All available data on the entire panel of cytokines were used for this analysis with the exception of CCL17, because CCL17 values were not available for all patients.
Networks of highly correlated cytokines in sera of patients with mild and aggressive CLL. “Connectivity mosaics” representing matrices of correlation coefficients are shown for serum cytokines from patients with TTFT of ≥ 5 years (left) and patients with TTFT of < 5 years (right). Individual cytokines are identically listed on x- and y-axes with A, B, C, and D indicating selected groups of highly correlated cytokines. Color shades indicate the type of correlation, with red = positive, blue = negative, and green = absent correlation. Light white lines indicate the borders of defined CLs of interconnected cytokines. For clarity, we indicate the relationship between groups of highly correlated cytokines determined by mosaic analysis and the 3 cytokine CLs (CL1, CL2, and CL3) identified by unsupervised cluster analysis (indicated in gray at far left). Some areas in the mosaic plot which differ in correlation strength between the 2 TTFT cohorts are delineated by pink-striped edges. All available data on the entire panel of cytokines were used for this analysis with the exception of CCL17, because CCL17 values were not available for all patients.
Discriminative patterns identify novel cytokine CLs stratifying CLL patients by disease aggressiveness.
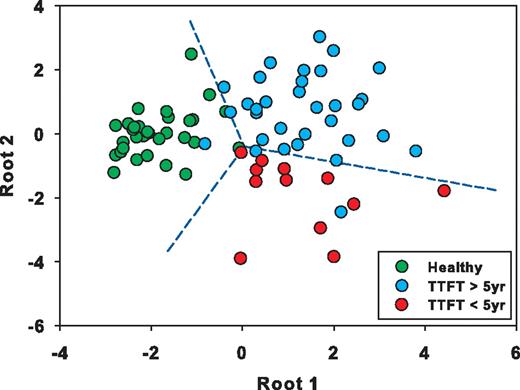
DFA was used to further characterize cytokines that best discriminate CLL patients with less (TTFT ≥ 5 years) versus more (< 5 years) aggressive disease from each other and from healthy subjects. This approach identified CCL3, IL-5, GM-CSF, IL-8, IL-15, CCL2, IL-10, IL-12, IFNα, IFNγ, and IL-2 as the cytokines with the highest discriminatory activity (Figure 6; supplemental Table 5). Using roots comprising this set of cytokines, CLL patients (Figure 6 blue and red dots) are clearly distinguished from healthy subjects (Figure 6 green dots). More importantly, CLL patients with more aggressive disease (red dots) also segregate distinctly from those with milder disease (blue dots).
DFA of TTFT ≥ 5 years versus TTFT < 5 years. Graphic representation of select cytokines determined by DFA analysis to maximally discriminate 3 groups: CLL patients with TTFT ≥ 5 years, CLL patients with TTFT < 5 years, and healthy subjects. DFA results are visualized on a multidimensional plot with roots serving as coordinates and class discrimination power represented by distance between groups. The cytokines comprising root 1 and root 2, sorted by their discriminated capability, are CCL3, IL-5, GM-CSF, IL-8, IL-15, CCL2, IL-10, IL-12, IFNα, IFNγ, and IL-2.
DFA of TTFT ≥ 5 years versus TTFT < 5 years. Graphic representation of select cytokines determined by DFA analysis to maximally discriminate 3 groups: CLL patients with TTFT ≥ 5 years, CLL patients with TTFT < 5 years, and healthy subjects. DFA results are visualized on a multidimensional plot with roots serving as coordinates and class discrimination power represented by distance between groups. The cytokines comprising root 1 and root 2, sorted by their discriminated capability, are CCL3, IL-5, GM-CSF, IL-8, IL-15, CCL2, IL-10, IL-12, IFNα, IFNγ, and IL-2.
Discussion
Growth and survival of CLL cells in situ depend on complex sets of signals received from the microenvironment, including cytokines, engagement of costimulatory molecules, antigen, or a combination.5,17 CLL cell trafficking to sites where these stimulatory influences can be attained depends on chemokines, acting in concert with adhesion molecules.6 Gaining a more complete understanding of the regulation of each of these pathways is essential to better understand CLL. Because the actions of certain cytokines and chemokines can be modulated by the actions of other cytokines and chemokines, identifying groups of molecules that robustly correlate with clinical course and outcome, the focus of the current work, has the potential to improve disease prognostication and may offer crucial insight into networks of signals that could be targeted by therapies.
In this exploratory study, we quantified serum levels of 23 cytokines in CLL patients and healthy subjects and analyzed these findings, based on individual mediators and also on sets of mediators grouped by a variety of analytical tools. For the former approach, we determined that serum levels of 17 cytokines are higher in CLL patients than healthy subjects (Figure 1; Table 2). Of these cytokines, a subset is elevated irrespective of disease severity, whereas others correlate with prognostic markers or clinical outcome measures (Figure 2; supplemental Tables 2-5). This distinction suggests that expression of certain cytokines may be driven by and potentially impact pathways common to CLL disease itself, irrespective of severity (eg, intrinsic and extrinsic signaling cascades critical for homeostatic proliferation and survival of CLL cells), whereas expression of others may be responsible for or reflect pathways specific for more aggressive disease and have greater impact on disease progression (eg, angiogenesis, inflammation, antitumor responses, and CLL and accessory cell recruitment and activation).
This is the first demonstration that CCL17, CXCL11, IL-5, and IL-17 exist at higher levels in CLL sera, suggesting new molecules relevant to CLL biology. CCL17 and CXCL11 are chemokines that modulate immune cell trafficking and adhesion. Although this is the first report of elevated levels of CCL17 protein in CLL sera, CLL cells have been shown to express CCL17 mRNA.26 Because CCL17 recruits activated CD4+CD40L+ T cells expressing CCR4, CLL or accessory cells may recruit CD4+ T cells via CCL17 that, after CD40L/CD40 interactions, could supply survival and growth signals to CLL clones.26 Although this is the first report that CXCL11 is elevated in CLL sera, CXCL9, a closely related chemokine, is elevated in CLL patient sera.27 Because both CXCL9 and CXCL11 are ligands for CXCR3, a receptor highly expressed on CLL cells,28 these chemokines may regulate CLL cell homing to tissues where they receive growth and survival signals. The T-helper (Th) 2 cytokine IL-5 stimulates normal B-cell growth and differentiation and increases immunoglobulin secretion29 ; however, because its effects on CLL cells have not been characterized, there is no framework on which to speculate on the relevance of elevated IL-5 levels in CLL. Finally, IL-17, an inflammatory and angiogenic cytokine produced primarily by CD4+ Th17 cells,30 can mediate both pro- and antitumor properties,31,32 and may influence CLL growth, survival, or both.
Other cytokines found elevated in CLL patient sera in this study (CCL2, CCL3, CCL4, CXCL9, IL-1β, IL-2, IL-6, IL-8, IL-10, IL-12, TNFα, IFNα, and IFNγ) have been reported previously as higher in CLL blood or tissues.8-14,33 Of these cytokines, CCL3, CCL4, IL-1β, IL-6, IL-8, IL-10, and IL-12 correlate with survival10 or TTFT.13 In our study, CCL3 and CCL4 were among the most effective in distinguishing CLL from healthy subjects (Figure 1B) and also correlated with shorter TTFT (Figure 2E), as did IL-10 and IL-12. In contrast, we found IL-1β and IL-8, previously shown to associate with poor prognosis in CLL,10-12 to correlate with good prognostic markers, increased survival, or both (Figures 2–3); this discrepancy could reflect variable expression of these cytokines during the disease or their pleiotropism, exerting both pro-and antitumor actions in different contexts.34 In addition, this apparent discrepancy indicates the downside of correlating the complexities of disease severity with levels of individual cytokines.
Thus, although analyses of individual cytokine levels is informative, a central finding in this study is that identification of CLs of coordinately regulated cytokines has an advantage by directing attention to linked actions and better predicting patient outcome than individual cytokines.
CL1, a CL identified by its association with shorter TTFT in unsupervised bicluster analysis (Figure 3) comprises 10 cytokines expressed in a highly coordinated manner in a subset of CLL patients. The coordinate expression of CL1 cytokines in patients with poor prognosis may reflect underlying differences in disease-related pathways active in aggressive versus indolent disease. Alternatively, CL1 cytokines may directly or indirectly influence, alone or in combination, immune and inflammatory mechanisms impacting survival, growth, and migration of CLL cells. CCL3 and CCL4, for example, are secreted by CLL cells in response to BCR engagement and by nurse-like cells,15,16 and levels may be higher in aggressive disease where microenvironmental stimuli may be more influential. In addition, recruitment and subsequent activation of macrophages by CCL3 and CCL4 are parts of a defined functional loop leading to TNF, VEGF, and CD49 production that could promote survival and growth of CLL cells.35 IL-12, another CL1 member, is proinflammatory and inhibits CLL cell apoptosis,36 and its association with a “poor outcome CL” may reflect this. IL-12 also can induce IFNγ that triggers macrophage production of 3 other CL1 members, CXCL9, CXCL10, and CXCL11, that could contribute migratory and survival signals to CLL cells by signaling via CXCR3.28 Of note, levels of another CL1 member, CCL19, strongly correlate with CXCL9, CXCL10, CXCL11, and IL-12 (supplemental Figure 1). Because CCL19 promotes migration of cells across the vascular endothelium, it could facilitate trafficking of CLL cells to tissue sites where survival and growth signals are encountered.37 Finally, IL-10 and IL-5 are Th2 cytokines that suppress cell-mediated immune responses critical for effective antitumor immunity;38,39 therefore, elevated levels might favor tumor growth. Based on these functions, CLL clones in CL1HI patients would exist in a cytokine milieu favoring their survival and growth.
In contrast to CL1 cytokines, higher levels of CL2 and CL3 cytokines associated with longer TTFT in unsupervised bicluster analysis (Figure 3), suggesting that expression of these cytokines might be induced by pathways active in indolent disease, or alternatively, that their expression might be inhibited by pathways active in aggressive disease. High serum levels of CL3 cytokines (IL-1β, IL-2, IL-4, IL-15, IL-17, and IFNα) correlate with significantly longer OS, irrespective of the levels of CL1. This finding was somewhat unexpected in that at least one CL3 cytokine has been reported previously to independently promote CLL cell growth, survival, or both in vitro40 ; another apparent discrepancy that highlights the differences encountered when analyzing individual versus coordinate CLs of immune molecules. CLL patients are typically immunocompromised with defective T-cell, NK-cell, and accessory cell functions, and this could contribute to impaired antitumor responses.41 CL3 includes the common γ-chain cytokines IL-2, IL-4, and IL-15, potent T-cell survival factors42 that could buttress immune function in CLL. In addition, although IL-15 stimulates CLL cell proliferation directly,40 it also stimulates dendritic cell (DC) maturation and NK- and CD8 T-cell cytotoxicity43 that could enhance antitumor responses and a less permissive milieu for CLL cells. Because mosaic analysis indicated IL-15 levels are not only low in patients with aggressive disease but also correlate inversely with levels of other CL3 cytokines and TNF (Figure 5), the beneficial effects of CL3 cytokines may be best realized in the presence of IL-15. Another CL3 cytokine, IL-1β, promotes differentiation of naive T cells into Th17 cells,30 a T-cell subset that secretes IL-17. Th17 cells exert antitumor effects in certain human cancers, and we have shown that Th17 levels correlate with longer OS in CLL.44 IL-17 is induced by IL-15 in certain settings;45 therefore, a milieu high in CL3 might promote IL-17–mediated antitumor pathways. Based on these functional profiles, CLL clones in CL3HI patients would exist in a cytokine milieu that activates immune cells and favors antitumor responses.
Unlike CL3, CL2 did not independently associate with good prognosis (Figure 4A middle panels) and its 4 cytokines (IL-6, IL-8, GM-CSF, and TNFα) have not previously been associated with good outcome in CLL; in fact, all 4 cytokines have been independently linked to poor outcome.10-12 Therefore, the positive correlation between CL2 and good outcome that we observe seems paradoxical. However, elevated levels of these 4 proinflammatory cytokines are characteristic features of Th1-skewed immune responses, which promote antitumor immunity.38 Because induction of effective antitumor immunity in CLL may be compromised by deficient DC function,41 TNFα, GM-CSF, and IL-6, which promote maturation and activation of DCs, might overcome this deficit. Alternatively, CL2 might associate with good outcome if one or more of the cytokines comprising CL2 interferes with CL1-triggered protumor pathways. Finally, it is possible that the association of high levels of CL2 with better outcomes is not the result of a direct antitumor action or interference but rather is a consequence of its down-regulation by one or more CL1 cytokines. If that were the case, then patients with high levels of CL1 (associated with worse outcome) would have low levels of CL2 and those with low levels of CL1 (associated with better outcome) would have higher levels of CL2, as we observed here. This scenario is in line with the fact that the CL1 cytokine IL-10, an anti-inflammatory cytokine, inhibits expression of the proinflammatory cytokines that comprise CL2. This interpretation is also in line with our finding that CL2 cytokines, unlike CL1 and CL3, do not correlate independently with TTFT or OS.
Furthermore, although CL1/CL2 and CL1/CL3 combinations are both effective at distinguishing “poor” and “good” outcome patients, the effects of CL2 and CL3 seem distinct, based on multivariate analysis indicating that CL1/CL2 CL combinations are discriminatory for TTFT, whereas CL1/CL3 CL combinations are discriminatory for OS. Thus, TTFT may be influenced by cytokine signals that impact initiation and early progression of CLL, whereas OS could depend not only on those pathways but also on the effectiveness of host antitumor responses and patient responsiveness to therapy. If this were the case, CL2 cytokines might ablate CL1 negative effects, resulting in benefit in early years, whereas later CL3 cytokines might directly exert positive benefit, consistent with the independent protective effect of CL3 (Figure 4A).
In summary, this initial correlative study suggests that CLL patients can be divided into discrete subsets based on defined serum cytokine patterns that might reflect underlying immune and inflammatory mechanisms impacting survival, growth, and migration of CLL cells. This conclusion is supported by our finding that multivariate algorithms not only identified outcome-related interrelationships among cytokines but also suggested potential roles for other cytokines not overexpressed in CLL, but whose expression in combination with other cytokines may favor either poor (eg, CCL19 in CL1) or good (eg, IL-15 in CL3) outcomes. Therefore, cytokine CL measurements may be valuable in risk stratification and treatment design and also provide insight into inflammatory and immunologic pathways active in CLL patients.
Our findings also suggest additional questions that deserve investigation. For example, in the long term, refining the membership of individual cytokines within each CL might create a minimal, integrated set with greater prognostic value, because it would not be surprising if some of the members of the CLs we defined are not causes of the observed clinical outcomes but themselves represent consequences of these processes. Forward stepwise DFA of our current data, a first step in accomplishing this, identified 11 cytokines that discriminate CLL patients with indolent from aggressive disease and CLL from healthy subjects (Figure 6). This cytokine pattern overlapped significantly (with the exception of CXCL9 and CXCL11) with those cytokines identified by our unsupervised CL (Figure 3) and mosaic analyses (Figure 5). Based on commonality among these 3 correlative analytic methods, CCL3, IL-5, IL-10, IL-12, IFNγ (CL1); GM-CSF, IL-8 (CL2); and IL-15, IFNα, IL-2 (CL3) may be key cytokines in CLL pathobiology, at least among those we have studied.
Furthermore, functional studies are necessary to elucidate the interrelationships among these cytokines and pathophysiologic and protective mechanisms in which these CLs participate. Indeed, insights into the roles of various cytokines and the CLL tumor microenvironment have revealed new therapeutic targets,46 and currently clinical trials are addressing the efficacy of immunomodulatory agents in CLL.46,47 Because our data reveal heterogeneity among patients with regard to “CL” combinations, these differences might underlie differential responsiveness of patients to various therapies. For example, in patients with refractory or relapsing disease, a higher ratio of CL1 to CL2/CL3 might exist; therefore, screening serum levels for CL composition pretreatment might identify responsive patients and posttreatment might help understand drug effectiveness. A therapeutic regimen that suppresses CL1, but spares or enhances CL2, CL3, or both, may provide considerable benefit. Furthermore, immunomodulatory drugs that seem to act to a large extent by down-regulating prosurvival factors such as VEGF, TNF, IL-8, and IL6,46,48 may act in part by altering cytokine CL composition or relationships. The finding that tumor flare reaction that can occur with lenalidomide treatment predicts clinical response in CLL49 suggests that modulation of inflammation contributes to the effectiveness of this drug; this is consistent with our finding that CLs of proinflammatory cytokines correlate with better outcome (CL2 and CL3).
In addition, cytokine profiles within bone marrow and lymphoid tissues, where CLL cells receive important growth and survival signals, should be analyzed because serum levels may not reflect those at such sites. In addition, analyses of cytokine levels among cohorts homogeneous for point of disease, treatment status, and serially along disease course will be informative because in our study serum cytokines were analyzed at a single time point in a broad cohort of patients that included newly diagnosed patients as well as patients whose disease was diagnosed at an earlier point, prior to obtaining serum samples. Furthermore, extending the breath of cytokines analyzed to others not included in our study could be helpful, including CXCL12, CXCL13, CX3CL1, IL-21, BAFF, and APRIL,6,50 all of which have been identified by others as playing a role in this disease. Finally, combining functional and expression studies is essential to definitively dissect the physiologic extent that these cytokines play (either directly or indirectly) in CLL and whether their levels are elevated as a cause or consequence of other stimuli within the CLL microenvironment; if the latter, they might be prognostic but not necessarily impact CLL biology.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Gloria Telusma for expert technical support and Drs Patricia K. Mongini and Percio Gulko for helpful discussions of the data.
This work was supported in part by the National Center for Research Resources (grant M01RR018535); the Karches Foundation, the Prince Family Foundation; the Marks Foundation; the Jerome Levy Foundation; the Leon Levy Foundation; the Tebil Foundation Inc; and the Joseph Eletto Leukemia Research Fund.
National Institutes of Health
Authorship
Contribution: X.-J.Y., N.C., and B.S. contributed to the overall design of the research; X.-J.Y., M.C., and B.S. carried out laboratory-based studies; I.D. and W.L. carried out statistical modeling studies; X.-J.Y., I.D., W.L., C.S., S.Y., N.C., and B.S. analyzed the results and prepared the figures; X.-J.Y., P.J., S.L.A., J.E.K., K.R.R., N.C., and B.S. wrote the paper; S.L.A., J.E.K., and K.R.R. were involved in the care of the patients and contributed clinical samples and data; and all authors reviewed and contributed to the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Barbara Sherry, The Feinstein Institute for Medical Research, 350 Community Dr, Manhasset, NY 11030; e-mail: bsherry@nshs.edu.

![Figure 1. A subset of cytokines is differentially expressed in the sera of CLL patients compared with healthy age-matched subjects. (A) Levels of cytokines present at significantly higher levels in the sera of CLL patients compared with the sera of healthy subjects are presented as box plots with the bottom and top of the box indicating the 25th and 75th percentiles, respectively. The bar within the box indicates the median value and the ends of the whiskers represent the 10th and 90th percentiles. Outliers are represented by dots. (B) RFA. Cytokines are ranked by their relative importance in discriminating CLL from healthy subjects with CCL3 being the most important predictor of CLL. Only subjects with complete cytokine data (82 total; 31 healthy subjects and 51 CLL patients) were used. Statistical significance was defined as P < .05. For this dataset, the classification error for healthy subjects is 12.9% (4/31) and for CLL subjects 3.9% (2/51). The overall classification error is 7.3% (6/82) keeping the sample size bias or 8.4% [0.5(4/31) + 0.5(2/51)] if we average the 2 error rates. The horizontal axis represents the average decrease in classification accuracy and horizontal bars representing the relative importance of each individual cytokine are sorted by importance along the vertical axis. The horizontal dashed line divides cytokines at the mean value displayed on the horizontal axis and defines the minimum number of cytokines required for maximum classification accuracy.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/19/10.1182_blood-2011-03-342436/4/m_zh89991180940001.jpeg?Expires=1763543879&Signature=1WW9~HSrXOrvld5Th4QumtNFDb0LVterFNiTF2Bf9~3LF8pntPrHZFEYmSoB2Bj9C53UN-0UsGzLO535i8NOJ2ynz37fa5lmo3fecVQe4CO83efqrpVIwkGu2Z~7-OMGeFa-ermeuAgnawNHz4wafN7F-nPQ4sbHcxoVvHzFgbWCoijviP8v9w8fwXBuR-TYv2cblz4E1Eqf-imGtGhJqXhWtq9sMscypjkdG5W6nmk5Xt7qhoHEb51lIIygroIO6-b-HmCfx1QkW~OW5cN0HtlZ8EAPydv~66~Icc3JDt2gL3UpHSPjCQvpuLZr3I9yLADiGquE9Z0TjBj1WmIRSw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Analysis of serum cytokine levels as a function of Rai stage, IGHV mutation status, CD38 expression, OS, and TTFT. Cytokines for which serum levels are significantly different when patient cohorts are divided by Rai stage (A), M-CLL (good prognosis) and U-CLL (poor prognosis) compared with healthy subjects (B), or CD38HIGH (poor prognosis) and CD38LOW (good prognosis) compared with healthy subjects (C). Data are presented as box plots (see legend to Figure 1 for details; *P < .05, **P < .01, ***P < .001). A result was considered significantly different if P < .05 (A) or P < .017 (B-C) to account for multiple comparisons. (D) Kaplan-Meier analysis of the levels of CXCL10, CXCL11, CCL19, and IL-1β versus survival. CXCL10, CXCL11, and CCL19 independently correlated with shorter survival: CXCL10 (16.3 years vs not reached; P = .012; HR = 3.1; 95% confidence interval [CI], 1.3-7.6), CXCL11 (15.3 years vs not reached; P = .013; HR = 3.1; 95% CI, 1.3-7.7), and CCL19 (16.3 years vs not reached; P = .042; HR = 2.4; 95% CI, 1.0-5.7). IL-1β independently correlated with longer survival (not reached vs 15.3 years; P = .022; HR = 0.4; 95% CI, 0.2-0.9). (E) Kaplan-Meier analysis of the levels of CCL3, CCL4, IL-10, and IL-12 versus TTFT. CCL3, CCL4, IL-10 and IL-12 independently correlated with shorter TTFT: CCL3 (7.6 years vs not reached; P = .031; HR = 2.0; 95% CI, 1.1-3.8), CCL4 (5.4 vs 20.9 years; P = .001; HR = 3.1; 95% CI, 1.6-6.0), IL-10 (7.6 years vs not reached; P = .030; HR = 2.0; 95% CI, 1.1-3.8), and IL-12 (6.8 vs 20.9 years; P = .019; HR = 2.2; 95% CI, 1.1-4.1). Sera were collected and analyzed as described in “Patients and blood collection and processing” and “Multiplex cytokine analysis.”](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/19/10.1182_blood-2011-03-342436/4/m_zh89991180940002.jpeg?Expires=1763543879&Signature=rpNVkeQVeF3Lv0xZmXgh6znTdwS8-tvFduYO~hfNQkk6Qafy-e-HgxVDYuoZp0~~qAXZLx7BEsvR74wbMEXG2r4LdhgRWmerAnJLaNNubhcvmlkjX0-DWL-ZKhap16bI1DzJ1qRiUYCrdNuU75JhleUy0oVN52VdJU2qbc7PkZrydDZiLGz9qCMbt~6reALI1OiA3r0iy-7g3ESkBVXqtNWmSdju7mYtsak8E2b5NYfFAqzCbdDdILCIGtoszZsw1-xa~2IPQbJQEuY0KGtNk8zeGZE4FcJ3BGmQ1~LgUClFvm2QCXzJo9bhwmce3jBoj2DSnQOPq~M6CXg-c3bm9Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)