Abstract
Identifying the normal cell from which a tumor originates is crucial to understanding the etiology of that cancer. However, retrospective identification of the cell of origin in cancer is challenging because of the accumulation of genetic and epigenetic changes in tumor cells. The biologic state of the cell of origin likely influences the genetic events that drive transformation. We directly tested this hypothesis by performing a Sleeping Beauty transposon mutagenesis screen in which common insertion sites were identified in tumors that were produced by mutagenesis of cells at varying time points throughout the T lineage. Mutation and gene expression data derived from these tumors were then compared with data obtained from a panel of 84 human T-cell acute lymphoblastic leukemia samples, including copy number alterations and gene expression profiles. This revealed that altering the cell of origin produces tumors that model distinct subtypes of human T-cell acute lymphoblastic leukemia, suggesting that even subtle changes in the cell of origin dramatically affect genetic selection in tumors. These findings have broad implications for the genetic analysis of human cancers as well as the production of mouse models of cancer.
Introduction
Scientists have speculated on the origins of cancer cells since the earliest histologic studies of tumors were performed > 150 years ago. However, a definitive cell of origin has not been identified for many tumor types, despite recent technological advances that allow detailed analysis of cancer cells. Despite the challenges of identifying the cell of origin, this goal has persisted because a comparison of tumor cells and the cell of origin may be necessary for identifying the key genetic events that distinguish the normal and early malignant cellular states.
Recent efforts have tried to determine how subtle differences in the cell of origin affect tumor progression. This work used mouse models that were generated by introducing mutations to the same cell lineage within a single tissue at different developmental time points. For example, various gene mutations in earlier stem or progenitor cell populations have been shown to produce tumors with decreased latency in 3 disparate mouse models of cancer: medulloblastoma, intestinal cancer, and leukemia.1-3 These experiments provide a strong indication that the cell of origin influences the efficiency of transformation, each suggesting that transformation potential is lost as cells differentiate. However, human cancers are not thought to exclusively emerge from stem or progenitor cell populations within the body. Thus, it is important to understand how the tumor cell of origin contributes to cancer progression, besides its general influence on tumor susceptibility.
We recently described a Sleeping Beauty (SB)–based transposon system in which mutagenesis in mice can be initiated in a tissue-specific manner using a lox-stop-lox strategy, thus making expression of the SB transposase (SBase) Cre-dependent.4 We devised a forward genetic screen in which transposon mutagenesis is initiated at different developmental time points along the T-cell lineage. Tumors will result from the accumulation of transposon-induced mutations in all cases. However, restricting transposon mutagenesis to later stages of T-cell differentiation will shift the cell of origin to a more differentiated cell type. Comparing the common insertion sites—the functional equivalent of driver mutations in SB-induced tumors—from each tumor model would then provide insight into how the cell of origin affects genetic selection within tumors from the same cell lineage. Additional comparisons between these mouse models and human T-cell acute lymphoblastic leukemia (T-ALL) samples using gene expression and copy number changes may provide correlative evidence for the effects of cell or origin in human cancer (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Methods
Mouse strains
The Cre-inducible RosaSBase-LSL and T2/Onc2 mouse strains were previously described.4,5 The Vav-iCre mice were provided by Dimitris Kioussis (National Institute for Medical Research, Medical Research Council, London, United Kingdom).6 Lck-Cre and CD4-Cre mice were purchased from Taconic Farms and were originally described by Lee et al.7 All procedures using mice were approved and monitored by the Institutional Animal Care and Use Committee at the University of Iowa.
SNP microarray and copy number analysis
Collection and processing of diagnostic and remission BM and peripheral blood samples for Affymetrix single-nucleotide polymorphism (SNP) arrays (using Affymetrix 50K Hind 240, 50K Xba 240, 250k Sty, 250k Nsp, and SNP 6.0 arrays) was performed as previously described in detail.8-10 Primary SNP array data are available from the authors on request (http://hospital.stjude.org/forms/genome-download/request/). All microarray data are available on the Gene Expression Omnibus (GEO) under accession number GSE29849.
Collection and processing of diagnostic and remission BM and peripheral blood samples for Affymetrix SNP arrays has been previously described in detail.8,9 Briefly, SNP array data were processed from CEL files to extract raw signal intensity values using dChip PM-only model-based expression analysis.10 Data were then normalized using a reference normalization algorithm that uses only markers from chromosome known or predicted to be diploid to guide array normalization. For each marker in each array, a normalized log2 ratio analyzed using the circular binary segmentation algorithm implemented in the DNAcopy package from Bioconductor to identify copy number alterations for each tumor sample. We applied the GISTIC method described by Venkatraman et al to the curated segmentation results.11
Microarray analysis
Microarray hybridizations were performed at the University of Iowa DNA Facility. Tumor cDNA was labeled and hybridized to Affymetrix Mouse Gene 1.0 ST arrays. Raw data files were analyzed using the Partek Genomics Suite software to produce a gene summary for each sample. These data were analyzed using Cluster 3 as described, and heatmap images were generated using TreeView.
Results
Altering the cell of origin in mice
To test the impact of the cell of origin in SB-induced T-cell malignancies, we generated 3 independent cohorts of triple-transgenic mice carrying: the Cre-inducible SBase allele (RosaSBaseLsL),4 the mutagenic T2/Onc2 transposon (TG6070 or TG6113),5 and 1 of 3 different Cre transgenes. In these models, the activation of transposon mutagenesis is Cre dependent, but mutagenesis will continue in all daughter cells because the SBase is expressed from the ubiquitous ROSA26 promoter (Figure 1A).4 We generated 3 cohorts of mice in which Cre-induced transposon mutagenesis was initiated in HSCs (Vav-iCre),6 immature thymocytes that lack surface expression of CD4 and CD8 (Lck-Cre),7 or late-stage CD4/CD8 double-positive thymocytes (CD4-Cre)7 (Figure 1B). The triple-transgenic mice were aged and monitored for signs of disease. Mice in the Vav-iCre (Vav-SB) cohort began to develop signs of disease beginning at 10 weeks of age, and all animals were euthanized by 20 weeks of age (Figure 1C). In contrast, their counterparts in the Lck-SB and CD4-SB cohorts had an average survival of 44.6 and 48.9 weeks, respectively. Survival was not affected in double-transgenic mice (RosaSBaseLsL/+; T2/Onc2Tg/+); these animals did not inherit a Cre transgene, and thus did not express the SBase.
Sleeping Beauty (SB)–induced models of T-cell lymphoma that initiate at distinct developmental time points. (A) An overview of the SB mutagenesis screen to identify driver mutations is shown. (B) Three different Cre-transgenic strains were used to induce SB transposase expression, and thus transposon mutagenesis, at distinct stages of differentiation. (C) A Mantel-Cox log-rank test indicated showed that tumor latency varied significantly between all 3 models (Vav-SB vs Lck-SB [P < .0001], Vav-SB vs CD4-SV [P < .001], Lck-SB vs CD4-SB [P = .026]). Tumors developed rapidly in the Vav-SB model (avg = 11 weeks), but much more slowly in the Lck-SB (avg = 45 weeks) and CD4-SB (avg = 49 weeks) models. DN indicates CD4/CD8 double-negative; ISP, immature single positive; and DP = CD4/CD8 double positive.
Sleeping Beauty (SB)–induced models of T-cell lymphoma that initiate at distinct developmental time points. (A) An overview of the SB mutagenesis screen to identify driver mutations is shown. (B) Three different Cre-transgenic strains were used to induce SB transposase expression, and thus transposon mutagenesis, at distinct stages of differentiation. (C) A Mantel-Cox log-rank test indicated showed that tumor latency varied significantly between all 3 models (Vav-SB vs Lck-SB [P < .0001], Vav-SB vs CD4-SV [P < .001], Lck-SB vs CD4-SB [P = .026]). Tumors developed rapidly in the Vav-SB model (avg = 11 weeks), but much more slowly in the Lck-SB (avg = 45 weeks) and CD4-SB (avg = 49 weeks) models. DN indicates CD4/CD8 double-negative; ISP, immature single positive; and DP = CD4/CD8 double positive.
At the time of necropsy, most moribund mice presented with enlarged thymus, spleen, lymph nodes, and liver. Histopathology examination of isolated tissues showed that the triple-transgenic mice developed lymphoid tumors. Further analysis of tumors from the Lck-SB and CD4-SB cohorts indicated that the tumors in these mice were T-cell lymphomas. This finding was supported by the surface immunophenotype and the presence of clonal TCRβ rearrangements in the tumors (supplemental Table 1). A similar analysis showed that ∼ 86% of tumors from the Vav-SB cohort were likewise T-cell lymphomas. The remaining tumors were of B-cell origin (ie, CD19+) and were excluded from this study. No myeloid, erythroid, or megakaryocyte lineage tumors were observed in the Vav-SB cohort, despite that transposon mutagenesis occurs throughout the hematopoietic system in these mice. However, the predominance of T-cell lymphoma in the Vav-SB cohort is not entirely unexpected given that lymphoma is a frequent side effect of increased mutagenesis in mice induced by p53 loss, whole-body irradiation, or chemical carcinogenesis.12-14
Identifying driver mutations
Genomic DNA was prepared from 101 thymic lymphomas isolated from mice at the time of necropsy. To identify transposon/genomic DNA junctions, we subjected each tumor DNA sample to linker-mediated PCR followed by deep sequencing. Each sequence was then trimmed and mapped to the mouse reference genome. Next we applied several criteria to identify clonal transposon insertion events in each sample (supplemental Methods). This produced a final set of 10 203 nonredundant transposon insertion sites found in 101 tumor samples (supplemental Table 2).
We analyzed the final transposon insertion datasets to find regions of the mouse genome that have undergone multiple transposon insertions in independent tumors. These regions are referred to as common insertions sites (CISs). Previous work with the SB system has shown that CISs frequently lie within or near cancer associated genes, and thus represent candidate driver mutations in SB-induced tumors.5,15,16 Two complementary methods were used to identify CISs in our datasets. The underlying assumption of both methods is that transposon insertion occurs in a near random pattern throughout the genome, as this has been demonstrated.5,15-17 The first CIS identification method uses an unbiased Monte Carlo simulation to approximate random transposon integration into TA dinucleotide sites (ie, SB target insertion sites) across the entire mouse genome within each tumor, as previously described.4 This analysis identified 22 CISs in Vav-SB, 30 CISs in Lck-SB, and 46 in CD4-SB tumors (supplemental Table 3).
The T2/Onc2 transposon is designed to modify gene expression only if the transposon integrates within, or near, a gene.18 Therefore, we developed a gene-centric approach to identify CISs (supplemental Methods). This analysis identified 22 candidate driver mutations (gCISs) in Vav-SB, 31 in Lck-SB, and 55 in CD4-SB (supplemental Table 4) tumors. The majority of genes (63%) were identified in both CIS and gCIS gene sets. A nonredundant set of 94 genes was then generated by combining the CIS and gCIS gene sets. We defined these genes as the driver mutations in our SB-induced T-cell lymphoma models. Although ∼ 75% of these genes have additional supporting evidence indicating a role in cancer, 23 genes are novel having not yet been implicated in cancer (supplemental Tables 3-4). In addition, we verified that transposon insertion led to elevated gene expression in the majority of cases (supplemental Figure 2).
Comparing driver mutations
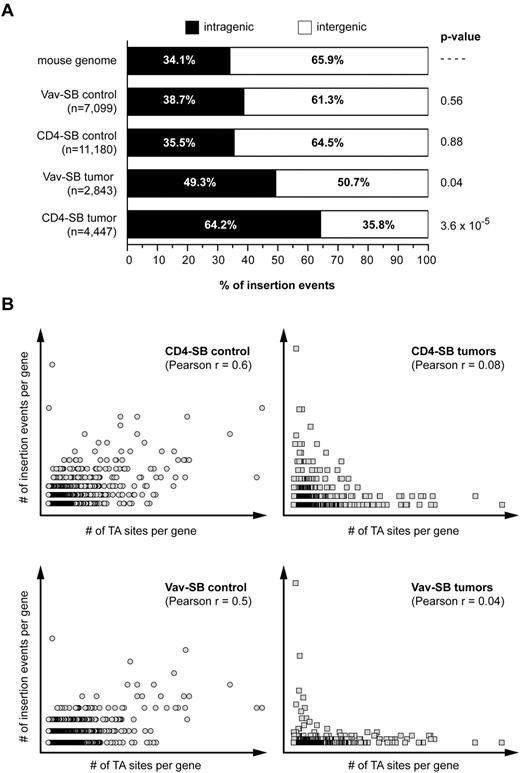
The ultimate goal of this experiment is to determine whether altering the cell of origin affects the process of genetic selection within SB-induced tumors. Implicit in this goal is the assumption that the activity of the SB system does not vary significantly in any of the tumor models. This assumption is based on published results showing that the SB system has little integration site bias in a variety of cell types.5,15-17 However, we sought to ensure that this assumption held true in our tumor models. Therefore, we mapped 7099 and 11 180 transposon insertions in premalignant thymocytes from Vav-SB and CD4-SB mice, respectively. Analysis of these datasets showed that there is not significant insertion site bias with respect to transcribed regions (Figure 2A). In addition, consistent with a random transposon distribution, the number of insertion events within genes in premalignant thymocytes was positively correlated with gene size (Figure 2B). By contrast, this correlation was not present within tumors, suggesting that the selection of transposon insertion sites within tumor cells is nonrandom. Finally, we also considered the possibility that CIS within each model were the result of insertion site bias. However, a χ2 test did not reveal a bias for insertion within the CIS regions identified in either the Vav-SB or CD4-SB tumors (Table 1). Taken together, these results show that the characteristics of SB transposition are not significantly different in the Vav-SB and CD4-SB models.
Transposition within the T-cell lineage does not show significant integration site bias. (A) The distribution of insertion sites within nonmalignant thymocytes and tumors from the Vav-SB and CD4-SB models is shown. A Fisher exact test was used to compare the distribution in each sample to the expected distribution observed in the mouse genome. (B) A Pearson correlation revealed a positive correlation between the number of insertion events within each RefSeq gene and the number of TA sites (ie, SB target sites) found within the gene. However, this correlation is present in nonmalignant control samples (circles) but not in tumor samples (squares).
Transposition within the T-cell lineage does not show significant integration site bias. (A) The distribution of insertion sites within nonmalignant thymocytes and tumors from the Vav-SB and CD4-SB models is shown. A Fisher exact test was used to compare the distribution in each sample to the expected distribution observed in the mouse genome. (B) A Pearson correlation revealed a positive correlation between the number of insertion events within each RefSeq gene and the number of TA sites (ie, SB target sites) found within the gene. However, this correlation is present in nonmalignant control samples (circles) but not in tumor samples (squares).
Comparison of transposon integration sites in normal and malignant T cells
| . | Obs/Exp* . | χ2 . | P . | Obs/Exp* . | χ2 . | P . |
|---|---|---|---|---|---|---|
| Vav-SB control data (7099 events) | CD4-SB control data (11 180 events) | |||||
| Vav-SB CIS regions (81 832 TA sites) | 10/6 | 0.563 | ns | 8/6 | 0.071 | ns |
| CD4-SB CIS regions (191 922 TA sites) | 19/15 | 0.265 | ns | 14/15 | 0.035 | ns |
| Vav-SB tumor data (2843 events) | CD4-SB tumor data (4447 events) | |||||
| VavSB CIS regions (81 832 TA sites) | 249/1 | 255.3 | < .0001 | 91/2 | 84.15 | < .0001 |
| CD4-SB CIS regions (191 922 TA sites) | 64/3 | 60.26 | < .0001 | 296/5 | 289.2 | < .0001 |
| . | Obs/Exp* . | χ2 . | P . | Obs/Exp* . | χ2 . | P . |
|---|---|---|---|---|---|---|
| Vav-SB control data (7099 events) | CD4-SB control data (11 180 events) | |||||
| Vav-SB CIS regions (81 832 TA sites) | 10/6 | 0.563 | ns | 8/6 | 0.071 | ns |
| CD4-SB CIS regions (191 922 TA sites) | 19/15 | 0.265 | ns | 14/15 | 0.035 | ns |
| Vav-SB tumor data (2843 events) | CD4-SB tumor data (4447 events) | |||||
| VavSB CIS regions (81 832 TA sites) | 249/1 | 255.3 | < .0001 | 91/2 | 84.15 | < .0001 |
| CD4-SB CIS regions (191 922 TA sites) | 64/3 | 60.26 | < .0001 | 296/5 | 289.2 | < .0001 |
Obs/Exp indicates no. of observed and expected insertion events; CIS, common insertion site; TA, potential SB insertion sites; and ns, not significant.
Calculation of expected insertion number is based on a total of 162 687 182 TA sites within the mouse reference genome.
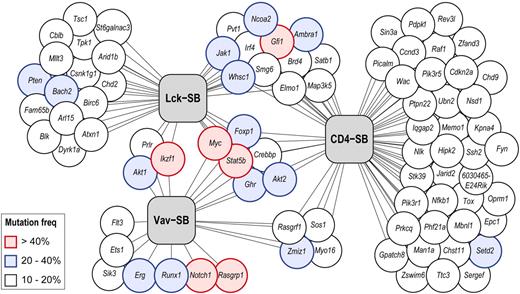
Based on this outcome, we compared the driver mutations identified in the 3 models to determine whether the progression of genetic changes varied within tumors derived from different cells of origin. Figure 3 shows the overlap among the driver mutations that are mutated in ≥ 10% of tumors in at least one model. Only 6 genes meeting these criteria were shared among all 3 models: Myc, Stat5b, Ghr, Foxp1, Akt2, Crebbp (Table 2).
Distribution of driver mutations in 3 independent SB models of T-ALL. Each gene identified as a driver mutation in one or more models is represented as a circle. The connecting lines indicate each gene's association with one or more T-ALL models (represented by gray boxes). The frequency of transposon-induced mutation in each gene is indicated by its color (white, 10%-20%; blue, 20%-40%; red, > 40%). In cases where a gene was identified in multiple models, the highest mutation frequency is indicated.
Distribution of driver mutations in 3 independent SB models of T-ALL. Each gene identified as a driver mutation in one or more models is represented as a circle. The connecting lines indicate each gene's association with one or more T-ALL models (represented by gray boxes). The frequency of transposon-induced mutation in each gene is indicated by its color (white, 10%-20%; blue, 20%-40%; red, > 40%). In cases where a gene was identified in multiple models, the highest mutation frequency is indicated.
Driver mutations identified in SB-induced models of T-ALL (> 15% mutation frequency)
| . | Gene . | Tumor model, % . | COSMIC and CGC . | Tumorscape* . | |||
|---|---|---|---|---|---|---|---|
| Vav-SB . | Lck-SB . | CD4-SB . | Amplification . | Deletion . | |||
| 1 | Akt2 | 33 | 15 | 32 | Ovarian, pancreatic | ||
| 2 | Crebbp | 17 | 15 | 13 | Renal cell, AML, ovarian | Ovarian | |
| 3 | Ghr | 17 | 22 | 18 | MPS | ||
| 4 | Foxp1 | 14 | 22 | 13 | ALL | Lung, prostate, liver, MPS | |
| 5 | Myc | 14 | 30 | 50 | Multiple | Multiple | Ovarian, breast, ALL, MPS |
| 6 | Stat5b | 11 | 59 | 47 | Breast | ||
| 7 | Notch1 | 72 | ns | ns | T-ALL | MPS | |
| 8 | Rasgrp1 | 31 | ns | ns | Lung, melanoma | ||
| 9 | Erg | 28 | ns | – | Ewing sarcoma, prostate, AML | Prostate | |
| 10 | Runx1 | 22 | ns | ns | AML, pre B-ALL, T-ALL | ||
| 11 | Ets1 | 17 | ns | ns | Hematopoietic | Lung, breast, medulloblastoma | |
| 12 | Ikzf1 | 47 | 19 | ns | ALL | Brain | ALL, hematopoietic |
| 13 | Prlr | 19 | 19 | ns | Lung | ||
| 14 | Akt1 | 14 | 22 | ns | Breast, colorectal, ovarian, lung | Lung, brain | |
| 15 | Zmiz1 | 33 | ns | 13 | Breast, prostate | ||
| 16 | Rasgrf1 | 17 | ns | 16 | |||
| 17 | Sos1 | 14 | ns | 16 | |||
| 18 | Bach2 | ns | 26 | ns | ALL, prostate, hematopoietic | ||
| 19 | Pten | – | 22 | ns | Multiple | Multiple | |
| 20 | Arl15 | – | 19 | ns | Lung | ||
| 21 | Atxn1 | – | 19 | ns | Breast, ovarian | Hematopoietic, ovarian | |
| 22 | Blk | – | 19 | ns | |||
| 23 | Csnk1g1 | – | 19 | ns | |||
| 24 | Arid1b | – | 15 | ns | Breast, ovarian | ||
| 25 | Birc6 | ns | 15 | ns | ALL | ||
| 26 | Cblb | – | 15 | – | AML | ||
| 27 | Chd2 | – | 15 | – | Lung | ||
| 28 | Dyrk1a | ns | 15 | ns | |||
| 29 | Fam65b | – | 15 | – | |||
| 30 | Mllt3 | – | 15 | ns | ALL | ||
| 31 | St6galnac3 | – | 15 | – | |||
| 32 | Tpk1 | – | 15 | ns | |||
| 33 | Tsc1 | – | 15 | ns | Hamartoma, renal cell | ||
| 34 | Whsc1 | ns | 33 | 26 | MM | ||
| 35 | Jak1 | ns | 30 | 26 | ALL | ||
| 36 | Gfi1 | ns | 26 | 42 | Lung, brain | ||
| 37 | Brd4 | – | 19 | 16 | Lethal midline carcinoma | ||
| 38 | Ncoa2 | ns | 19 | 21 | AML | Breast | |
| 39 | Pvt1 | – | 19 | 11 | |||
| 40 | Satb1 | ns | 19 | 11 | |||
| 41 | Smg6 | – | 19 | 11 | |||
| 42 | Ambra1 | – | 15 | 21 | |||
| 43 | Elmo1 | – | 15 | 13 | |||
| 44 | Irf4 | – | 15 | 13 | MM | Breast, lung, hematopoietic | |
| 45 | Map3k5 | – | 15 | 11 | Colorectal | ||
| 46 | Setd2 | ns | ns | 21 | Clear cell renal carcinoma | ALL | |
| 47 | Ccnd3 | – | ns | 18 | MM | Colorectal | |
| 48 | Cdkn2a | ns | – | 16 | Multiple | Multiple | |
| 49 | Mbnl1 | ns | ns | 16 | |||
| 50 | Pdpk1 | ns | ns | 16 | Ovarian | ||
| 51 | Wac | ns | ns | 16 | |||
| . | Gene . | Tumor model, % . | COSMIC and CGC . | Tumorscape* . | |||
|---|---|---|---|---|---|---|---|
| Vav-SB . | Lck-SB . | CD4-SB . | Amplification . | Deletion . | |||
| 1 | Akt2 | 33 | 15 | 32 | Ovarian, pancreatic | ||
| 2 | Crebbp | 17 | 15 | 13 | Renal cell, AML, ovarian | Ovarian | |
| 3 | Ghr | 17 | 22 | 18 | MPS | ||
| 4 | Foxp1 | 14 | 22 | 13 | ALL | Lung, prostate, liver, MPS | |
| 5 | Myc | 14 | 30 | 50 | Multiple | Multiple | Ovarian, breast, ALL, MPS |
| 6 | Stat5b | 11 | 59 | 47 | Breast | ||
| 7 | Notch1 | 72 | ns | ns | T-ALL | MPS | |
| 8 | Rasgrp1 | 31 | ns | ns | Lung, melanoma | ||
| 9 | Erg | 28 | ns | – | Ewing sarcoma, prostate, AML | Prostate | |
| 10 | Runx1 | 22 | ns | ns | AML, pre B-ALL, T-ALL | ||
| 11 | Ets1 | 17 | ns | ns | Hematopoietic | Lung, breast, medulloblastoma | |
| 12 | Ikzf1 | 47 | 19 | ns | ALL | Brain | ALL, hematopoietic |
| 13 | Prlr | 19 | 19 | ns | Lung | ||
| 14 | Akt1 | 14 | 22 | ns | Breast, colorectal, ovarian, lung | Lung, brain | |
| 15 | Zmiz1 | 33 | ns | 13 | Breast, prostate | ||
| 16 | Rasgrf1 | 17 | ns | 16 | |||
| 17 | Sos1 | 14 | ns | 16 | |||
| 18 | Bach2 | ns | 26 | ns | ALL, prostate, hematopoietic | ||
| 19 | Pten | – | 22 | ns | Multiple | Multiple | |
| 20 | Arl15 | – | 19 | ns | Lung | ||
| 21 | Atxn1 | – | 19 | ns | Breast, ovarian | Hematopoietic, ovarian | |
| 22 | Blk | – | 19 | ns | |||
| 23 | Csnk1g1 | – | 19 | ns | |||
| 24 | Arid1b | – | 15 | ns | Breast, ovarian | ||
| 25 | Birc6 | ns | 15 | ns | ALL | ||
| 26 | Cblb | – | 15 | – | AML | ||
| 27 | Chd2 | – | 15 | – | Lung | ||
| 28 | Dyrk1a | ns | 15 | ns | |||
| 29 | Fam65b | – | 15 | – | |||
| 30 | Mllt3 | – | 15 | ns | ALL | ||
| 31 | St6galnac3 | – | 15 | – | |||
| 32 | Tpk1 | – | 15 | ns | |||
| 33 | Tsc1 | – | 15 | ns | Hamartoma, renal cell | ||
| 34 | Whsc1 | ns | 33 | 26 | MM | ||
| 35 | Jak1 | ns | 30 | 26 | ALL | ||
| 36 | Gfi1 | ns | 26 | 42 | Lung, brain | ||
| 37 | Brd4 | – | 19 | 16 | Lethal midline carcinoma | ||
| 38 | Ncoa2 | ns | 19 | 21 | AML | Breast | |
| 39 | Pvt1 | – | 19 | 11 | |||
| 40 | Satb1 | ns | 19 | 11 | |||
| 41 | Smg6 | – | 19 | 11 | |||
| 42 | Ambra1 | – | 15 | 21 | |||
| 43 | Elmo1 | – | 15 | 13 | |||
| 44 | Irf4 | – | 15 | 13 | MM | Breast, lung, hematopoietic | |
| 45 | Map3k5 | – | 15 | 11 | Colorectal | ||
| 46 | Setd2 | ns | ns | 21 | Clear cell renal carcinoma | ALL | |
| 47 | Ccnd3 | – | ns | 18 | MM | Colorectal | |
| 48 | Cdkn2a | ns | – | 16 | Multiple | Multiple | |
| 49 | Mbnl1 | ns | ns | 16 | |||
| 50 | Pdpk1 | ns | ns | 16 | Ovarian | ||
| 51 | Wac | ns | ns | 16 | |||
ns indicates not significant (although insertions within the indicated were present in some tumors); and –, no insertions detected.
Tumor types showing amplification and/or deletion at a significant rate (q ≤ 0.25) http://www.broadinstitute.org/tumorscape.
The Vav-SB models showed the greatest difference in mutation profiles compared with either the Lck-SB or CD4-SB models. Interestingly, transposon-induced mutations in Notch1 were common in Vav-SB lymphomas (72%)—a frequency similar to the observed rate of NOTCH1 point mutation in human T-ALL.19 By contrast, Notch1 mutations were rare in Lck-SB (11%) and CD4-SB (5%) lymphomas. Furthermore, we sequenced the Notch1 transcript in all Vav-SB tumors that lacked transposon insertion in the Notch1 locus as well as 15 CD4-SB tumors. We did not detect any intragenic Notch1 mutations similar to those which have been reported in mouse models of spontaneous T-cell lymphoma.20 Transposon-induced mutations in Rasgrp1, Runx1, and Erg were also common in Vav-SB tumors, but these genes were not mutated a significant rate in the remaining models. Instead, Lck-SB and CD4-SB lymphomas frequently had mutations in Whsc1, Jak1, Gfi1, Myc, and Stat5b (Table 2). Although mutations in Myc and Stat5b were found to be significant in all 3 models, the mutation frequencies for these genes were higher in Lck-SB and CD4-SB tumors (Table 2). The significant disparity in mutation profiles for these models suggests that the biologic differences in the cell of origin greatly affect genetic selection during tumor development.
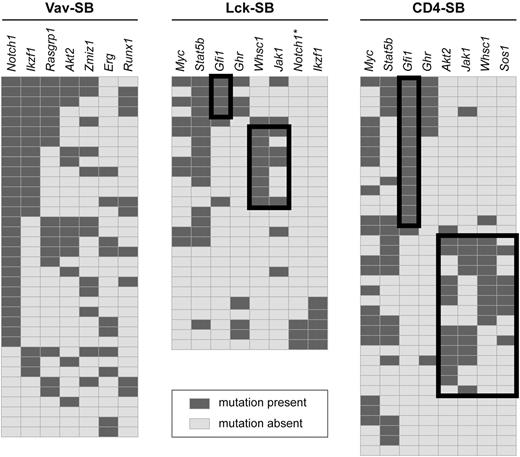
Next, we examined the pattern of gene mutations within each model to determine whether any distinct genetic signatures were evident within each model. The Vav-SB model produced tumors that often involved the same genes in recurring combinations (Figure 4). As a consequence, unique tumor subsets could not be easily distinguished in the Vav-SB model, and thus this model appeared to be homogenous at the genetic level. For example, tumors with a Notch1 insertion also showed frequent mutations in Ikzf1, Rasgrp1, Akt2, Zmiz1, Erg, and Runx1 suggesting that mutations in these genes contribute to Notch1-induced lymphomas. Although mutations in both Notch1 and Ikzf1 have already been shown to cooperate several mouse models of T-cell lymphoma,21-23 the remaining genes have not yet been shown to specifically cooperate with Notch1 mutation to produce T-cell lymphoma. However, Rasgrp1, Akt2, Erg, and Runx1 have all been implicated as oncogenes in mouse models of T-cell lymphoma.24-27
Identification of genetic signatures within each T-ALL model. Genetic signatures were identified within each model by identifying mutations that show significant negative correlation. For each panel, genes are shown as columns as indicated across the top, and individual tumors are shown as independent rows. A dark gray box indicates the presence of a transposon insertion within the indicated gene while a light gray box implies the absence of mutation. Negative correlation between pairs of genes were assessed by Fisher exact test. Black-lined boxes indicate subsets of tumors defined by a negative correlation. For example, mutations in Gfi1 were mutually exclusive with mutations in Whsc1 (P = .018), Akt2 (P = .004), Jak1 (P = .018), and Sos1 (P = .027) in CD4-SB tumors. *Insertion events detected, but below threshold for significance.
Identification of genetic signatures within each T-ALL model. Genetic signatures were identified within each model by identifying mutations that show significant negative correlation. For each panel, genes are shown as columns as indicated across the top, and individual tumors are shown as independent rows. A dark gray box indicates the presence of a transposon insertion within the indicated gene while a light gray box implies the absence of mutation. Negative correlation between pairs of genes were assessed by Fisher exact test. Black-lined boxes indicate subsets of tumors defined by a negative correlation. For example, mutations in Gfi1 were mutually exclusive with mutations in Whsc1 (P = .018), Akt2 (P = .004), Jak1 (P = .018), and Sos1 (P = .027) in CD4-SB tumors. *Insertion events detected, but below threshold for significance.
The interactions among the driver mutations identified in CD4-SB tumors were more complex. First, mutations in Gfi1 were negatively correlated with mutations in Whsc1 (P = .018), Akt2 (P = .004), Jak1 (P = .018), and Sos1 (P = .027). This pattern of mutations clearly defined 2 distinct genetic signatures within the CD4-SB model (Figure 4). The remaining driver mutations identified in the CD4-SB model were equally distributed between these 2 signatures, including commonly mutated genes such as Myc and Stat5b. The biologic mechanism that produced these genetic signatures is not immediately apparent. However, both Gfi1 and Whsc1 are involved in regulating gene expression—Gfi1 as a transcriptional repressor28 and Whsc1 as a H3K36 histone methyltransferase.29 It is possible that overexpression of these proteins affects a common set of genes involved in lymphomagenesis.
Interestingly, the distribution of the driver mutations within tumors from the Lck-SB model again shows features similar to both the Vav-SB and CD4-SB models (Figure 4). Three distinct tumor subtypes were noted in the Lck-SB model. Two of these subtypes are defined by mutations in Gfi1 or Whsc1, as was seen in the CD4-SB model. However, a third subtype is also present that is defined by mutations in Notch1 and Ikzf1, as was seen frequently in Vav-SB tumors (Figure 4). Thus, tumors within the Lck-SB model tend to harbor mutations that are similar to those found in either the Vav-SB or CD4-SB models, with no individual tumor exhibiting features of both models. The biologic origin of this dichotomy in the Lck-SB model is not clear. However, the timing of Cre recombination induced by the Lck-Cre transgene has been shown to be variable, occurring either before or after β selection in thymocytes.30 As a result, some Lck-SB tumors may been initiated just before TCR gene rearrangement—a critical point in T-cell differentiation that likely leads to extensive chromatin remodeling, although the details of this process are poorly understood.31 Thus, it is possible that TCR gene rearrangement represents a transition in the Lck-SB model. For instance, it may be that tumors initiated in cells before TCR rearrangement resemble the Vav-SB model whereas those initiated in cells thereafter resemble CD4-SB tumors.
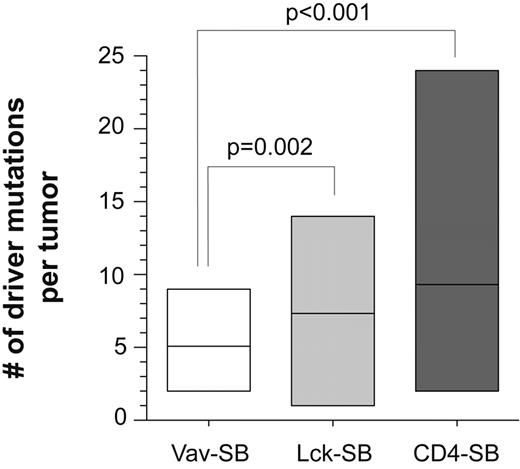
Among the many differences we observed between the 3 lymphoma models, one of the most striking was the significant enrichment in the number of driver mutations identified in the Lck-SB and CD4-SB models versus their Vav-SB counterpart, in which Notch1 mutations predominated. We hypothesized that this difference was a consequence of the high degree of genetic heterogeneity in these tumors, and predicted that the average number of driver mutations per tumor would not differ significantly across models. However, we observed a statistically significant correlation between the average number of driver mutations per tumor and the differentiation state of the cell of origin (Figure 5). Both methods used to identify driver mutations (ie, CIS/gCIS) take into account the sample size and the number of insertion events identified in each sample. Thus, increasing the size of the tumor cohort or the number of transposon insertion sites in individual tumors would result in a more stringent definition of a driver mutation. Finally, we also determined that most CD4-SB animals harbor a single dominant tumor signature by sampling the tumor in different sites throughout the body (supplemental Figure 3). This suggests that the genetic complexity seen in this model is not likely produced by the emergence of multiple independent tumors within each animal.
The number of driver mutations in each tumor is indicated for each of the 3 tumor models. The average number of driver mutations differed significantly across the models. A t test was performed to compare the differences between each pair of models. No significant difference was observed between Lck-SB and CD4-SB tumors.
The number of driver mutations in each tumor is indicated for each of the 3 tumor models. The average number of driver mutations differed significantly across the models. A t test was performed to compare the differences between each pair of models. No significant difference was observed between Lck-SB and CD4-SB tumors.
We next determined whether the increase in the number of driver mutations seen in the CD4-SB model was the result of positive selection, as expected. We anticipated that if this were true, the genes implicated as driver mutations in the CD4-SB model would show a greater overlap with known cancer genes than would be expected by chance. A comparison of the driver mutations from each model to the COSMIC (Catalog Of Somatic Mutations In Cancer) and CGC (Cancer Gene Consensus) databases showed that the gene sets from all SB-induced models overlap significantly with genes mutated in these databases (Table 3). This result suggests that the increase in the number of driver mutations seen in the Lck-SB and CD4-SB tumors has a biologic cause and supports the hypothesis that more mutations are required to transform differentiated cells than are required to transform stem cells.
Comparison of driver mutations to COSMIC and CGC databases
| Model . | No. of driver genes also mutated in COSMIC or CGC* . | P . |
|---|---|---|
| Vav-SB | 11 of 22 (Notch1, Ikzf1, Akt2, Erg, Runx1, Crebbp, Foxp1, Akt1, Flt3, Myc, Sik3) | 2.7 × 10−10 |
| Lck-SB | 17 of 36 (Whsc1, Jak1, Myc, Akt1, Foxp1, Pten, Brd4, Ikzf1, Ncoa2, Smg6, Akt2, Birc6, Cblb, Crebbp, Irf4, Mllt3, Tsc1) | 1.07 × 10−14 |
| CD4-SB | 21 of 37 (Myc, Akt2, Jak1, Whsc1, Ncoa2, Setd2, Ccnd3, Brd4, Cdkn2a, Chd9, Crebbp, Foxp1, Hipk2, Irf4, Picalm, Fyn, Nsd1, Pik3r1, Ptpn22, Raf1, Smg6) | 4.76 × 10−20 |
| Model . | No. of driver genes also mutated in COSMIC or CGC* . | P . |
|---|---|---|
| Vav-SB | 11 of 22 (Notch1, Ikzf1, Akt2, Erg, Runx1, Crebbp, Foxp1, Akt1, Flt3, Myc, Sik3) | 2.7 × 10−10 |
| Lck-SB | 17 of 36 (Whsc1, Jak1, Myc, Akt1, Foxp1, Pten, Brd4, Ikzf1, Ncoa2, Smg6, Akt2, Birc6, Cblb, Crebbp, Irf4, Mllt3, Tsc1) | 1.07 × 10−14 |
| CD4-SB | 21 of 37 (Myc, Akt2, Jak1, Whsc1, Ncoa2, Setd2, Ccnd3, Brd4, Cdkn2a, Chd9, Crebbp, Foxp1, Hipk2, Irf4, Picalm, Fyn, Nsd1, Pik3r1, Ptpn22, Raf1, Smg6) | 4.76 × 10−20 |
The COSMIC and CGC databases currently have data on the mutation frequencies for 18 661 nonredundant genes. Of these, 15 631 have identified mouse orthologues that have also been evaluated by both Monte Carlo and gCIS approaches. We determined the number of driver mutations from each tumor model identified by these methods that have 4 or more independent mutations in the COSMIC database or are present in the CGC database. A Fisher exact test was then performed to determine the significance of the observed overlap.
Comparison of SB models to human T-ALL
Cross-species comparisons of data from insertional mutagenesis screens in mouse models of cancer to gene copy number alterations observed in human tumors are one manner in which cancer-associated genes can be identified among a larger field of candidate genes.15,32 We performed genome-wide copy number analysis on 84 independent human pediatric T-ALL samples using high-density SNP arrays. Recurrent copy number alterations (CNAs) were then identified using GISTIC analysis.33 This identified 31 recurrent copy number alterations that scored as significant (q < 0.25, supplemental Table 5).
We then compared the driver mutations identified in SB-induced lymphomas to the list of genes affected by copy number gain or loss in human T-ALL. First, mouse orthologs were identified for 4314 genes affected by copy number alteration in human T-ALL. Of these genes, 29 that were present in amplicons in human T-ALL were also identified as candidate oncogenes (eg, Jak1, Sos1, Ccnd3, Notch1, Akt2) in SB-induced lymphomas (supplemental Table 5). Similarly, 4 genes which were recurrently deleted in human tumors were also identified as candidate tumor suppressors (Ikzf1, Cdkn2a, Pten, Picalm) in SB-induced lymphomas (supplemental Table 5). In the case of Ikzf1, transposon insertion drives expression of a dominant-negative allele (supplemental Figure 6).
Given the large number of genes evaluated in both mouse and human tumors (n = 15 631), the degree of overlap observed between them is statistically significant (P = .04). This suggests that SB mutagenesis can drive transformation using similar genetic mechanisms that are produced by spontaneous mutations in human tumors. Thus, such comparative analyses will provide additional data to identify which genes, among the large number affected by copy number alteration, are likely to play a direct role in transformation.
Although the comparative genomic analysis between SB-induced lymphomas and human T-ALL produced a significant result, it did not provide insight into how each SB-induced model compared with human T-ALL. This is not surprising because only ∼ 4 CNAs were observed in each sample—a relatively low number compared with other forms of cancer. Therefore, we generated gene expression profiles of SB-induced lymphomas to compare directly to expression profiles of human T-ALL.
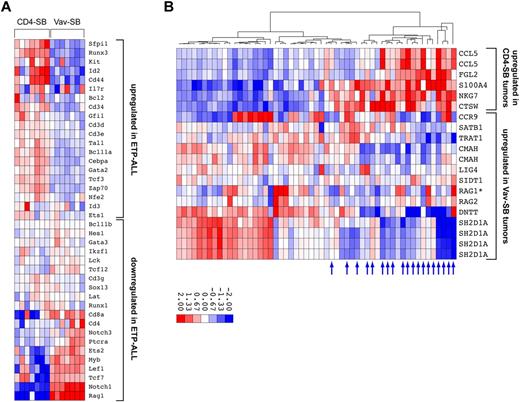
Recently, a new subtype of T-ALL was defined by a distinct immunophenotype and gene expression pattern that is similar to those of early T-cell precursors (ETPs)—and thus is referred to as ETP-ALL.34,35 Gene expression analysis performed by Coustan-Smith and colleagues identified a gene set that distinguishes ETP-ALL from typical T-ALL. We compared the expression patterns in the SB-induced lymphomas using the ETP-ALL gene set.34,36 This revealed a clear correlation between the CD4-SB model and human ETP-ALL, and another between Vav-SB and the more typical T-ALL (Figure 6A, supplemental Figure 4). This result was unexpected because transposon mutagenesis in the CD4-SB model is initiated in the more mature CD4/CD8 double-positive T cells (Figure 1B), yet the tumors clearly exhibit a gene expression pattern characteristic of early T-cell precursors (Figure 6A).
Comparative oncogenomics of SB-induced T-cell lymphomas. (A) Global gene expression was analyzed in a set of 7 Vav-SB and 7 CD4-SB tumors. Unsupervised clustering analysis was performed using expression data from a set of 40 genes that distinguished typical T-ALL from ETP-ALL.34 (B) Global gene expression analysis identified 2 gene sets that were up-regulated in either Vav-SB or CD4-SB tumors. Gene-set enrichment analysis showed that genes up-regulated in CD4-SB are enriched in ETP-ALL samples (blue arrows) while the Vav-SB gene set is enriched in typical T-ALL samples.
Comparative oncogenomics of SB-induced T-cell lymphomas. (A) Global gene expression was analyzed in a set of 7 Vav-SB and 7 CD4-SB tumors. Unsupervised clustering analysis was performed using expression data from a set of 40 genes that distinguished typical T-ALL from ETP-ALL.34 (B) Global gene expression analysis identified 2 gene sets that were up-regulated in either Vav-SB or CD4-SB tumors. Gene-set enrichment analysis showed that genes up-regulated in CD4-SB are enriched in ETP-ALL samples (blue arrows) while the Vav-SB gene set is enriched in typical T-ALL samples.
This result was corroborated by other similarities between human ETP-ALL and the CD4-SB lymphoma model. First, human ETP-ALL tumors exhibited increased genomic instability, with respect to changes in both copy number and the size of the affected regions of the genome.34 Tumors from the CD4-SB model were characterized by a large number of mutations per tumor, though these mutations are caused by transposon mutagenesis rather than genomic instability (Figure 5). In addition, roughly half of CD4-SB tumors analyzed were CD4/CD8 negative, and thus represented a more immature immunophenotype, as in the case of ETP-ALL (supplemental Table 1).
Next, we also performed unsupervised clustering based on expression levels of 54 genes showing the greatest differential expression among 14 tumors from the Vav-SB and CD4-SB models (supplemental Figure 5). Tumors from each model clustered together, as did tumors within the CD4-SB model that had a similar genetic profile (eg, Gfi1 or Whsc1). This analysis identified 2 gene sets that were up-regulated in either Vav-SB or CD4-SB tumors (supplemental Figure 5). We then performed gene set enrichment analysis on a set of 55 human T-ALL samples34 to determine whether the mouse gene sets were able to distinguish ETP-ALL and typical T-ALL. Consistent with our prior observations, a subset of the CD4-SB gene set was also up-regulated in human ETP-ALL samples (P = .002) while the Vav-SB gene set was enriched in typical T-ALL samples (P = .006). It is important to note that of the 15 genes showing significant enrichment in either ETP-ALL or typical T-ALL, only one is also found in the ETP gene signature initially used to define the ETP-ALL subtype (Figure 6B). Therefore, this analysis provides additional evidence demonstrating the correlation between the CD4-SB tumor model and human ETP-ALL.
Discussion
Here we describe the results of a unique forward genetic screen in which transposon mutagenesis is tied to distinct developmental time points during T lymphopoiesis. Our results provide direct evidence that the cell of origin strongly influences the genetic progression of the resulting tumor. It currently remains unclear whether these trends will apply to other tumor types, or if these findings reflect some unique property of T cells. Nevertheless, this is the first demonstration of a genetic screen designed to study the role of the tumor cell of origin.
Our results suggest that while inferring the cancer cell of origin based on gene or protein expression may be not be possible, identifying the cell of origin is still an important pursuit. Technological improvements have made it relatively easy to identify somatically acquired mutations in human tumors. However, additional information will be required to understand the biologic context in which these somatic mutations act.
The correlations observed between T-ALL subsets and our SB-induced lymphoma models suggest that subtypes of human cancer could be defined by differences in the cell type from which the tumor arises. The SB-induced CD4-SB mouse model shows several similarities with a subset of human ETP-ALLs. Therefore, we suspect that these tumors share a common cell of origin. This raises the possibility that some human ETP-ALLs are not necessarily derived from early T-cell precursors, but may arise from more differentiated T cells that regain features of earlier T-cell progenitors. It is interesting to note that ∼ 50% of ETP-ALLs described by Coustan-Smith and colleagues showed clonal rearrangements in the TCRB, TCRG, or TCRD genes.34 This is a feature of differentiated T cells, not early T-cell progenitors, and is not consistent with a progenitor cell of origin for this subset of tumors.
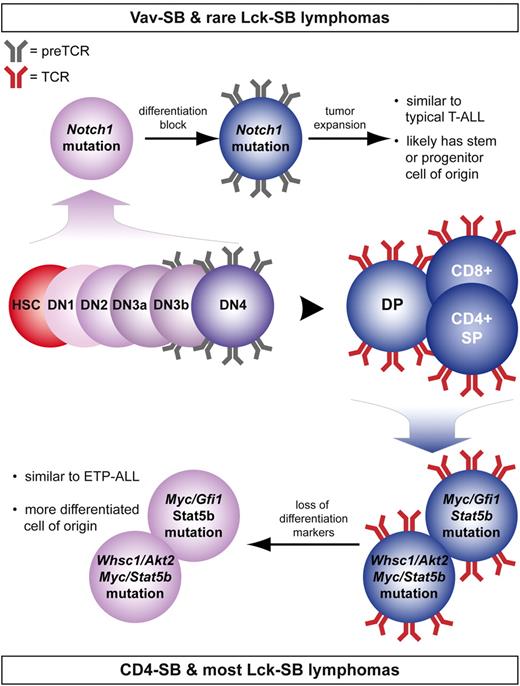
The most dramatic example of the disparity between these models is the frequency of Notch1 mutation (Table 2). One possible explanation for this difference is that lymphomas induced by Notch1 mutation appear to be driven by expression of the pre-TCR, an immature form of the TCR composed of the pTα receptor paired with a rearranged and functional TCRβ chain.37,38 The expression of the pTα receptor is rapidly down-regulated at the double-positive stage of T-cell development (Figure 7).39 By contrast, expression of mutant Notch1 in early lymphopoiesis has been shown to enforce a T-cell fate, although continued Notch activation ultimately causes a block in T-cell development at the double-positive stage.40,41 This would lock the cell in a state that maintains both Notch1 and pTα expression (Figure 7). This specific timing of Notch1 mutation can occur in either the Vav-SB or Lck-SB models, although the timing of Lck-Cre expression provides a narrow window of time in which a cell can acquire a Notch1 mutation before pTα expression is lost. Consistent with this idea, Notch1 mutations in the Lck-SB model are less common than in Vav-SB tumors (Figure 4).
Summary showing the effect of cell of origin on lymphomagenesis. Despite that transposon mutagenesis in Vav-SB mice occurs at all stages of T-cell development, lymphomas that develop in these mice are driven by a largely nonoverlapping set of mutations compared with lymphomas from CD4-SB mice (Table 2). A proposed model is shown to explain the significant differences in these models of T-cell lymphoma.
Summary showing the effect of cell of origin on lymphomagenesis. Despite that transposon mutagenesis in Vav-SB mice occurs at all stages of T-cell development, lymphomas that develop in these mice are driven by a largely nonoverlapping set of mutations compared with lymphomas from CD4-SB mice (Table 2). A proposed model is shown to explain the significant differences in these models of T-cell lymphoma.
The bias we observed for Notch1 mutation in lymphomas with a progenitor cell of origin is also consistent with a previous study in which expression of the Notch1 intracellular domain was shown to specifically transform immature thymocytes after pre-TCR signaling but before TCRα rearrangement.42 In agreement with this report, we found that Vav-SB lymphomas have clonal TCRβ rearrangements (supplemental Table 1), although mutagenesis was initiated much earlier in T-cell development (Figure 1B). As previously mentioned, pre-TCR signaling has been shown to drive proliferation in Notch1-induced lymphoma.37,38 This suggests that the combination of Notch1 activation together with pre-TCR signaling leads to clonal expansion of malignant thymocytes.
However, this model does not sufficiently explain the relative absence of Notch1 mutations in CD4-SB lymphomas. For example, it has been shown that Notch1 activation can drive pTα expression, even in differentiated T cells.43 Nevertheless, Notch1-induced pTα expression in more mature thymocytes is not sufficient to cause transformation.42 It is also important to note the inverse relationship between Notch1 and Myc mutations in the Vav-SB and CD4-SB models. Prior work has shown that Myc is a critical Notch1 target gene required for Notch1-induced transformation.42,44 Interestingly, the degree of Notch1-induced Myc activation is reduced in more mature T cells compared with tumors derived from immature thymocytes.42 This suggests that the ability of Notch1 to induce Myc expression is influenced by the developmental state of the cell. In differentiated T cells, Notch1 activation may not be sufficient to drive high levels of Myc expression and thus transposon-induced overexpression of Myc is selected instead.
The genetic profiles of CD4-SB tumors also implicate WHSC1 and GFI1 in the development of ETP-ALL, if not by direct mutation then as downstream targets of pathways activated in ETP-ALL. Interestingly, ∼ 25% of multiple myeloma cases harbor a t(4;14) translocation that leads to the ectopic expression of WHSC1.45 Aberrant expression of WHSC1 has also been recently associated with tumor aggressiveness in 15 different forms of cancer.46 The Whsc1 protein is a histone methyltransferase that functions in combination with a variety of transcription factors and plays a critical role during development of different tissues.29 In addition, Gfi1 has been shown to play a role in maintenance of hematopoietic stem cells.47 Interestingly, Gfi1 regulates transcription by repressing gene expression by recruiting histone modifying enzymes to target genes.48,49
Many of the unique observations in these experiments were made possible by the ability to control the timing of mutagenesis in our SB-induced models of lymphoma. One of the more interesting observations was that tumors initiated in more differentiated cells acquire more driver mutations and show greater genetic complexity than those initiated in stem cells. This concept is not novel, as mathematical models have made a similar prediction.50 However, these results provide the first direct experimental evidence in support of this hypothesis.
Finally, our data clearly show that different genetic mechanisms can drive transformation in disparate cell types. Although our experiments focused on T-cell lymphoma, it is reasonable to assume that cell of origin will also play an important role in genetic selection in other tumor types as well. Therefore, construction of a mouse model that faithfully recapitulates the genetic events seen in a specific form of human cancer will require introducing specific gene mutations into the appropriate cell of origin. In some cases, this may require development of novel tools (ie, Cre-transgenic strains) to facilitate the targeting of rare somatic cell populations. Although this may prove to be a technical challenge, our findings suggest that this may be required to generate mouse models of cancer that better mimic the biology of human cancer.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Christine Blaumueller for her assistance in editing the manuscript.
This work was funded by the National Cancer Institute (5R01CA130867).
National Institutes of Health
Authorship
Contribution: K.E.B.-V. designed and performed research, contributed vital new reagents or analytical tools, collected, analyzed, and interpreted data, and performed statistical analysis; K.N. analyzed and interpreted data, and performed statistical analysis; B.T.B. analyzed and interpreted data, and performed statistical analysis; L.H. designed and performed research, and collected, analyzed, and interpreted data; J.M. analyzed and interpreted data, and performed statistical analysis; O.Z. designed and performed research, and collected data; N.A.J. contributed reagents; N.G.C. contributed reagents; D.K.M. and C.M.K. designed and performed research, and collected data; C.G.M. designed and performed research; T.E.S. analyzed and interpreted data, and performed statistical analysis; and A.J.D. designed research, analyzed and interpreted data, performed statistical analysis, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Adam J. Dupuy, University of Iowa, 1-400A BSB, 51 Newton Rd, Iowa City, IA 52242; e-mail: adam-dupuy@uiowa.edu.

![Figure 1. Sleeping Beauty (SB)–induced models of T-cell lymphoma that initiate at distinct developmental time points. (A) An overview of the SB mutagenesis screen to identify driver mutations is shown. (B) Three different Cre-transgenic strains were used to induce SB transposase expression, and thus transposon mutagenesis, at distinct stages of differentiation. (C) A Mantel-Cox log-rank test indicated showed that tumor latency varied significantly between all 3 models (Vav-SB vs Lck-SB [P < .0001], Vav-SB vs CD4-SV [P < .001], Lck-SB vs CD4-SB [P = .026]). Tumors developed rapidly in the Vav-SB model (avg = 11 weeks), but much more slowly in the Lck-SB (avg = 45 weeks) and CD4-SB (avg = 49 weeks) models. DN indicates CD4/CD8 double-negative; ISP, immature single positive; and DP = CD4/CD8 double positive.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/17/10.1182_blood-2011-03-343947/4/m_zh89991179100001.jpeg?Expires=1765993515&Signature=Bj3xberGL0AmIbr4CIvQrBEHbIv5iIO8C3DtZcjM7OgUSnG8ZfKcKSUcrtzVJRUt~hlDKi4sDA3pmWvnrEfzYQX-e3kU~c33~0XrYt7AfVgPA0u7GdpM3HQMHvvxIFV~LVhNtrk6-gDlcBh7-4yOivCcZJVRMUIoS0kLVPuwDO4tIi9iQEtUb-9seZkWgSCv240WsvRGaT-KB3mePGbcT~B4PTiL25K5-gFVGPidqO5ipQkzV02DZgiq0DWqsEJW8HaI3Q-fpYmkL7Gt99aXvaWog2o0avAsazcq2UT1l6D5Hclv3D2BVYCuiVq6WI47975pv8KgtFPIIwYgfb8mIQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)