Abstract
Hepcidin controls the levels and distribution of iron, an element whose availability can influence the outcome of infections. We investigated hepcidin regulation by infection-associated cytokines, pathogen-derived molecules, and whole pathogens in vitro and in vivo. We found that IL-22, an effector cytokine implicated in responses to extracellular infections, caused IL-6–independent hepcidin up-regulation in human hepatoma cells, suggesting it might represent an additional inflammatory hepcidin agonist. Like IL-6, IL-22 caused phosphorylation of STAT3 and synergized with BMP6 potentiating hepcidin induction. In human leukocytes, IL-6 caused potent, transient hepcidin up-regulation that was augmented by TGF-β1. Pathogen-derived TLR agonists also stimulated hepcidin, most notably the TLR5 agonist flagellin in an IL-6–dependent manner. In contrast, leukocyte hepcidin induction by heat-killed Candida albicans hyphae was IL-6–independent, but partially TGF-β–dependent. In a murine acute systemic candidiasis model, C albicans strongly stimulated hepcidin, accompanied by a major reduction in transferrin saturation. Similarly, hepcidin was up-regulated with concomitant lowering of serum iron during acute murine Influenza A/PR/8/34 virus (H1N1) infection. This intracellular pathogen also stimulated hepcidin expression in leukocytes and hepatoma cells. Together, these results indicate that hepcidin induction represents a component of the innate immune response to acute infection, with the potential to affect disease pathogenesis.
Introduction
Almost all life forms require iron because of its involvement in basic cellular processes. During infections, pathogens use various means to acquire iron from their hosts, while hosts attempt to withhold it from pathogens.1-3 Iron therefore represents a point of conflict between host and pathogen, and altered iron balance associates with poor outcomes in several infectious diseases, including malaria,4 tuberculosis,5 and HIV-1 infection.6
Systemic iron homeostasis in humans is controlled by the 25 amino acid peptide hormone hepcidin,7 produced by hepatocytes. Hepcidin binds the sole known mammalian iron exporter ferroportin causing its internalization and degradation, thereby inhibiting iron efflux.8 Ferroportin is expressed highly on duodenal enterocytes and macrophages.9 Consequently, when hepcidin levels are high, both dietary iron uptake and recycling of iron from senescent red cells by macrophages are inhibited. Persistently raised hepcidin causes iron redistribution from serum and tissues into the reticuloendothelial system, and anemia often manifests because of reduced iron supply to the bone marrow for erythropoiesis.7 In contrast, inappropriately low hepcidin, as observed during various forms of hereditary haemochromatosis, facilitates excessive dietary iron uptake, eventually leading to toxic accumulation of iron in tissues such as the liver.7 The hepcidin-ferroportin interaction therefore plays a central role in regulating both the quantity and distribution of iron throughout the body. Given the association of iron with infection outcomes, understanding how hepcidin itself is regulated during inflammation and infections is clearly important.
Hepcidin transcription is stimulated by increases in serum iron, in part via the bone morphogenetic protein (BMP) signaling pathway.7 BMPs, members of the TGF-β superfamily, bind BMP receptors in conjunction with coreceptors such as hemojuvelin to enable signaling via SMAD1/5/8 and the common mediator SMAD4.10 Loss of function of hemojuvelin in humans and mice,10,11 and of Bmp612,13 and Smad414 in mice, lead to inappropriately low hepcidin production and consequent iron overloading. Because BMP6 is increased when iron levels in the liver are high,15 the BMP-hepcidin-ferroportin axis represents an important homeostatic mechanism of systemic iron regulation. The fact that patients with hepcidin-resistant ferroportin mutations develop iron overload reveals a lack of redundancy in the control of systemic iron homeostasis.16
Hepcidin is also induced in hepatocytes by inflammatory stimuli, most notably IL-6,17 which signals through IL-6 receptor α (IL-6Rα) and gp130 via the janus kinase (JAK)– STAT3 pathway.18,19 Persistently raised hepcidin during chronic inflammatory conditions such as rheumatoid arthritis and Castleman disease is thought to contribute to the anemia of inflammation.20 Inflammatory mediators are released during many infections, so hepcidin induction during inflammation may represent a host adaptation for withholding iron from serum and tissues after infectious insult, thereby restricting the growth of invading pathogens. Moreover, the lack of redundancy in mechanisms for controlling systemic iron levels and localization16 may reflect the need for a system capable of rapidly redistributing iron during infections.21
Here, we first investigate hepcidin induction in vitro in hepatoma cells and PBMCs—important components of the host's response to invading pathogens—in response to inflammatory/infection-associated stimuli, notably the cytokine IL-22. We then demonstrate hepcidin up-regulation, with associated changes in serum iron parameters, during both fungal and viral infections in vivo. Together, the data suggest that hepcidin induction should be considered part of the innate immune response to various classes of infection.
Methods
Cells and reagents
The hepatoma cell lines Hep3B and HepG2 (ATCC) were maintained in minimal essential medium (Alpha-modification; Sigma-Aldrich) supplemented with 10% FCS (PAA Laboratories), 100 U/mL penicillin, 0.1 mg/mL streptomycin and 2mM L-glutamine (all Sigma-Aldrich) at 37°C, 5% CO2. For experimental treatments, 1-2 × 104 cells seeded in 100 μL media into flat-bottomed 96-well plates, were allowed to adhere for 5-6 hours. After washing with PBS, serum-free media (with penicillin, streptomycin, and L-glutamine, but lacking FCS) was added. Treatments were applied for 18 (cytokine-BMP6 experiments) or 24 (Influenza experiment) hours.
Human PBMCs, isolated by ficoll density gradient centrifugation of heparinized whole blood obtained with informed consent from healthy male volunteers in accordance with the Declaration of Helsinki, were cultured at 37°C, 5% CO2 in 12-well plates at 5 × 106 cells/mL in 1 mL RPMI-1640 (supplemented with penicillin, streptomycin, L-glutamine, and 10% FCS); treatments were added immediately after plating. PBMC cultures with Influenza A/PR/8/34 virus used serum-free media. For all PBMC experiments, at least 3 donors were used; experiments were run in biologic singlicate or duplicate; technical duplicates (qRT-PCR) were run for every sample; data for distinct individuals are represented in figures by different symbols, which are conserved between figure panels.
Anti–human IL-6 and anti–human IL-6 Receptor (anti–IL-6R) neutralizing antibodies, goat IgG, recombinant human Bone Morphogenetic Protein 6 (BMP6), BMP9, IL-6, IL-22, and TGF-β1 (R&D Systems) were reconstituted following the manufacturer's recommendations and used at concentrations indicated in figures/legends. All TLR agonists (Invivogen-Source Biosciences) were reconstituted in sterile endotoxin free water, and used at concentrations indicated in figure legends. SD208 (Alk5/TGF-β pathway inhibitor; Tocris Bioscience) was reconstituted at 20mM in DMSO and used at 1μM.
C albicans preparation
Candida albicans strain SC5314 (kind gift from Neil Gow and Frank Odds, Aberdeen, United Kingdom) was cultured for 18 hours at 30°C in YPD medium. Cells were diluted to 1 × 107 cells/mL after 2 PBS washes. To generate hyphae, 1 × 107 cells/mL were incubated at 37°C in RPMI-1640/10% FCS for 2 hours and washed in PBS. Heat-killed C albicans was generated by heating at 80°C for 40 minutes.
C albicans–PBMC coculture
Candida albicans yeast/hyphae were resuspended in RPMI-1640 media (500 μL at 2 × 106 cells/mL), and added to 5 × 106 PBMCs in 500 μL media (final ratio 5 × 106 PBMC: 1 × 106C albicans in 1 mL media). To remove large aggregates, heat-killed hyphal preparations were passed through 70-μm cell strainers (BD Biosciences) before co-culture; resulting suspensions comprised aggregates containing up to ∼ 50 hyphal units (mean ∼ 20; supplemental Figure 5A-B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Based on this, suspensions containing ∼ 2 × 106 hyphal units/mL were prepared. In one case (filled triangles, Figure 4A-B; supplemental Figure 4C-D), the antifungal agent fungin (Invivogen) was included (20 μg/mL).
Influenza A/PR/8/34 virus preparation
Influenza A/PR/8/34 virus (H1N1) was kindly provided by John Skehel (NIMR, Mill Hill, London, United Kingdom). Virus used in in vitro experiments was tissue culture adapted through 7 passages, plaque-titered in MDCK-SIAT1 cells as described,22,23 and added to cultures in serum-free media at 10 or 100 plaque forming units (PFU) per cell.
Murine C albicans and Influenza A virus infections
C57BL/6 mice obtained from breeding pairs originally purchased from The Jackson Laboratory were maintained on standard diets at the Biomedical Services Unit, John Radcliffe Hospital, Oxford, and used according to established institutional guidelines under the authority of United Kingdom Home Office project licenses. For C albicans infection, male mice were injected intravenously via the tail vein with 5 × 105C albicans SC5314 yeast cells in 100 μL PBS. For Influenza A/PR/8/34 virus infection, 3.5 HAU were administered intranasally under anesthetic to female mice; intranasal PBS administrations were carried out as controls.
Mice were terminated by asphyxiation after 24 hours (C albicans) or 3 days (Influenza virus). Blood was taken by cardiac puncture and serum separated using BD microtainers (Bunzl Healthcare). For subsequent RNA extraction, liver explants (∼ 2 mm3) were preserved in RNAlater (QIAGEN); whole lung (Influenza only) was immediately lysed in 4 mL RLT lysis buffer containing 10 μL/mL β-mercaptoethanol (see “RNA extraction and cDNA synthesis”) using a TissueRuptor (both QIAGEN).
RNA extraction and cDNA synthesis
Hepatoma cells.
Cells were lysed within 96-well plates after washing with cold PBS. Lysate RNA was reverse transcribed using the TaqMan Gene Expression Cells-to-CT kit (Applied Biosystems), following the manufacturer's protocol.
PBMCs and murine tissues.
RNA was extracted using the RNeasy Mini Kit (QIAGEN) following the manufacturer's protocols. Pooled adherent and nonadherent PBMC lysates were homogenized with QIAshredders (QIAGEN). Murine liver explants were TissueRuptor homogenized (10 seconds in 600 μL Buffer RLT). RNA was extracted from 350 μL RLT lysate for both lung (see “Murine C albicans and influenza A virus infections”) and liver samples. RNA concentrations were measured by NanoDrop. RNA was reverse transcribed using the High Capacity RNA-to-cDNA kit (Applied Biosystems) following the manufacturer's recommendations.
Quantitative real-time PCR (qRT-PCR)
Gene expression levels were quantified by qRT-PCR using inventoried TaqMan Gene Expression assays (supplemental Table 1) and TaqMan Gene Expression Master Mix (Applied Biosystems), using the manufacturer's protocol. Technical duplicates were performed. Changes in gene expression relative to endogenous controls (GAPDH [human]/Hprt1 [mouse]) were calculated using the formula 2−ΔCT. Fold-changes in expression relative to control untreated samples were calculated according to the 2−ΔΔCT method. Variations in control untreated hepcidin mRNA levels between experi-ments were observed in hepatoma cell lines and between PBMC donors; however, patterns of induction after stimulation were highly consistent for both cell types.
Immunoblotting
Hepatoma cells, treated in serum-free media for 40 minutes with BMP6 and/or IL-6/IL-22, were lysed on ice for 30 minutes in NP40 1% buffer with protease (1:500) and phosphatase inhibitors (1:100; both Sigma-Aldrich); lysate supernatants were stored at −80°C until required. Lysates, with protein concentrations normalized using the BCA assay (Thermo-Fisher), were run on 12% SDS-PAGE gels, blotted onto nitrocellulose membranes (GE Healthcare) and blocked in PBS/milk (5% wt/vol). Membranes were incubated overnight at 4°C in TBS-TWEEN/BSA (5% wt/vol) with: mouse anti–β-actin (1:10 000, Sigma-Aldrich); rabbit anti-pSMAD1/5/8 (1:1000); rabbit anti-SMAD1 (1:500); rabbit anti-pSTAT3(Tyr705; 1:2000); rabbit anti-STAT3 (1:2000; all from Cell Signaling). After washing, they were incubated with appropriate secondary antibodies for 1 hour at room temperature: goat anti–mouse/HRP (1:2000; Dako); donkey anti–rabbit/HRP (1:3000; Santa Cruz Biotechnology). Immunoblots were developed using ECL reagent (GE Healthcare).
Transferrin saturations
Serum iron and unsaturated iron binding capacity were quantified by the Veterinary Laboratories Agency; transferrin saturations were calculated conventionally.
Statistics
Data were processed and analyzed using Microsoft Excel and GraphPad Prism Version 5.0.1 (GraphPad Software Inc). Statistical tests used are indicated in figure legends; nonparametric tests were used, except where datasets passed the Kolmogorov-Smirnov test for normality. P values < .05 were considered statistically significant.
Results
IL-22 induces hepcidin and synergises with BMP6 in hepatoma cells
Recent evidence indicates that costimulation of the hepatoma cell lines with agonists of the BMP-SMAD pathway and JAK-STAT3 pathway (IL-6) causes synergistic up-regulation of hepcidin mRNA.24-26 Similarly, we found that IL-6 and BMP6, the BMP critical for maintaining iron homeostasis in mice,12,13 synergized in inducing hepcidin on cotreatment of Hep3B and HepG2 hepatoma cells (Figure 1A-B). In contrast TGF-β1, which also signals via SMAD4, failed to notably up-regulate hepcidin in hepatoma cells consistent with previous observations10,24 ; similarly, IL-6–mediated hepcidin induction was not enhanced by TGF-β1 (supplemental Figure 1A-B).
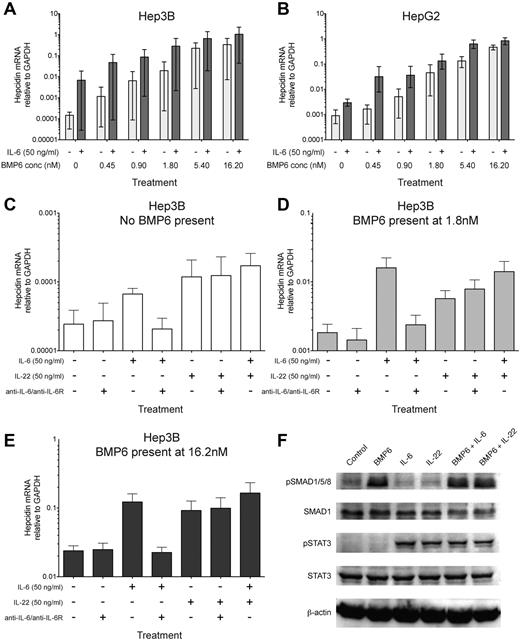
IL-22, like IL-6, synergizes with BMP6 in inducing hepcidin and causes STAT3 phosphorylation in hepatoma cells. (A-B) BMP6 synergises with IL-6 in inducing hepcidin in Hep3B and HepG2 cells. qRT-PCR measurement of hepcidin mRNA expression relative to GAPDH in (A) Hep3B and (B) HepG2 hepatoma cells cultured for 18 hours in serum-free media with a titration of recombinant human BMP6 in the presence (dark gray bars) or absence (light gray bars) of recombinant human IL-6 (50 ng/mL, ∼ 2.4nM). The mean and range of 3 independent experiments are plotted; a significant synergistic effect of IL-6 was observed for both cell types (P < .001, Wilcoxon matched-pairs signed rank test, accounting for each condition from each of the 3 experiments), despite the variation in baseline hepcidin levels between the 3 experiments. (C-E) IL-22 stimulates hepcidin up-regulation in Hep3B cells independently of IL-6. Hepcidin expression relative to GAPDH in Hep3B cells cultured with combinations of recombinant human BMP6 (low and high concentrations, 1.8nM or 16.2nM, respectively), IL-6 (50 ng/mL), IL-22 (50 ng/mL, ∼ 3nM), and a mixture of neutralizing anti–IL-6 and anti–IL-6R antibodies (termed anti-IL6/R, 10 μg/mL each). The effects of IL-6, IL-22, and anti-IL6/R on hepcidin induction in the absence of BMP6, or presence of 1.8nM or 16.2nM BMP6 are shown in panels C through E, respectively. Note that for clarity the ranges of y-axis scales in panels C through E are not identical. (F) Immunoblots of Hep3B cells treated with BMP6 and/or IL-6/IL-22. Immunoblots for phospho-SMAD1/5/8 (pSMAD1/5/8) and phospho-STAT3 (pSTAT3) in Hep3B cell lysates after 40 minutes stimulation with BMP6 (1.8nM) and/or IL-6 (50 ng/mL) or IL-22 (50 ng/mL); equal loading is demonstrated by immunoblotting for SMAD1, STAT3, and β-actin. Data are representative of 3 independent experiments.
IL-22, like IL-6, synergizes with BMP6 in inducing hepcidin and causes STAT3 phosphorylation in hepatoma cells. (A-B) BMP6 synergises with IL-6 in inducing hepcidin in Hep3B and HepG2 cells. qRT-PCR measurement of hepcidin mRNA expression relative to GAPDH in (A) Hep3B and (B) HepG2 hepatoma cells cultured for 18 hours in serum-free media with a titration of recombinant human BMP6 in the presence (dark gray bars) or absence (light gray bars) of recombinant human IL-6 (50 ng/mL, ∼ 2.4nM). The mean and range of 3 independent experiments are plotted; a significant synergistic effect of IL-6 was observed for both cell types (P < .001, Wilcoxon matched-pairs signed rank test, accounting for each condition from each of the 3 experiments), despite the variation in baseline hepcidin levels between the 3 experiments. (C-E) IL-22 stimulates hepcidin up-regulation in Hep3B cells independently of IL-6. Hepcidin expression relative to GAPDH in Hep3B cells cultured with combinations of recombinant human BMP6 (low and high concentrations, 1.8nM or 16.2nM, respectively), IL-6 (50 ng/mL), IL-22 (50 ng/mL, ∼ 3nM), and a mixture of neutralizing anti–IL-6 and anti–IL-6R antibodies (termed anti-IL6/R, 10 μg/mL each). The effects of IL-6, IL-22, and anti-IL6/R on hepcidin induction in the absence of BMP6, or presence of 1.8nM or 16.2nM BMP6 are shown in panels C through E, respectively. Note that for clarity the ranges of y-axis scales in panels C through E are not identical. (F) Immunoblots of Hep3B cells treated with BMP6 and/or IL-6/IL-22. Immunoblots for phospho-SMAD1/5/8 (pSMAD1/5/8) and phospho-STAT3 (pSTAT3) in Hep3B cell lysates after 40 minutes stimulation with BMP6 (1.8nM) and/or IL-6 (50 ng/mL) or IL-22 (50 ng/mL); equal loading is demonstrated by immunoblotting for SMAD1, STAT3, and β-actin. Data are representative of 3 independent experiments.
IL-22 is an IL-10 cytokine family member implicated in mucosal defense against extracellular pathogens.27,28 After binding the heterodimeric IL-10R2/IL-22R1 receptor on tissue-resident cells such as epithelial cells and hepatocytes, IL-22 initiates transcription of effector molecules including anti-microbial peptides.28-30 Hepcidin, initially characterized as an anti-microbial peptide,31 rapidly reduces serum iron,21 which is required for the growth of extracellular pathogens. We therefore hypothesized that IL-22 would up-regulate hepcidin in hepatoma cells. IL-22 stimulated hepcidin mRNA production in Hep3B (Figure 1C-E) and HepG2 cells (supplemental Figure 1C) and like IL-6, synergized with BMP6 at various doses, potentiating hepcidin induction (Figure 1C-E, supplemental Figure 1C). The up-regulation was IL-6–independent, since antibody-mediated IL-6 neutralization failed to abrogate the effect (Figure 1C-E). Like IL-6, IL-22 treatment led to STAT3, but not SMAD1/5/8 phosphorylation in Hep3B (Figure 1F) and HepG2 (supplemental Figure 1D) cells within 40 minutes; stimulation with BMP6 enhanced SMAD1/5/8 phosphorylation but did not alter phosphorylation of STAT3. Together, these data suggest IL-22–mediated hepcidin induction involves a signaling pathway distinct from, but convergent with that of IL-6.
Synergy between SMAD and STAT3 pathway agonists in PBMCs
Circulating PBMCs contribute to the host's defense against invading microorganisms. Cells within this compartment recognize pathogens, respond by secreting inflammatory cytokines and chemokines, and ultimately drive the development of adaptive immune responses. Given the influence of iron on infection outcomes, we investigated whether PBMCs up-regulated hepcidin in response to cytokines and pathogenic stimuli.
We first stimulated fresh PBMCs with IL-6 and/or TGF-β1. In this and subsequent experiments, basal hepcidin mRNA expression levels increased with time in ex vivo culture (Figure 2A) and also varied between individuals, typically being in the range 0.01-0.1% of the level of GAPDH mRNA. Despite this variation, the observed patterns of hepcidin induction by various stimuli were typically conserved between individuals.
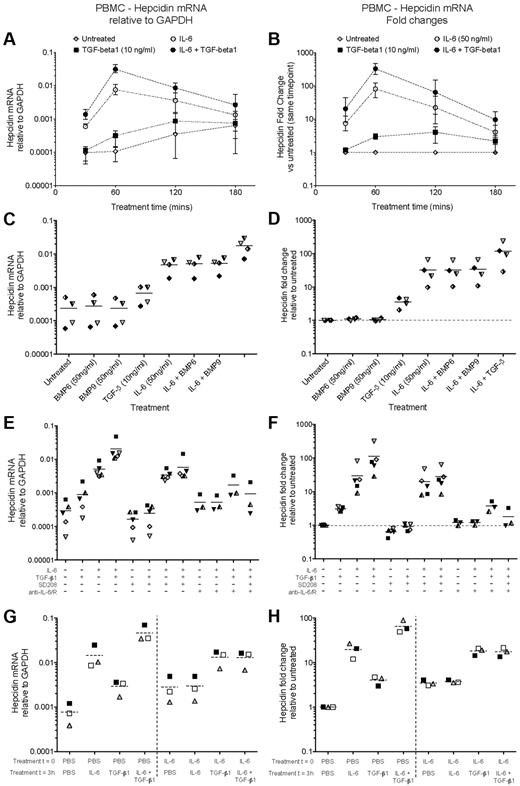
IL-6 and TGF-β1, but not BMPs, induce hepcidin mRNA in human PBMCs. (A-B) Time course of hepcidin induction by IL-6 and TGF-β1. qRT-PCR measurement of hepcidin mRNA induction in 5 × 106 PBMCs cultured for various times in the presence or absence of IL-6 (50 ng/mL) and TGF-β1 (10 ng/mL). Data are presented as (A) expression relative to GAPDH, and (B) fold-change in hepcidin expression relative to the untreated sample from the same timepoint. Datapoints represent means and ranges for PBMCs obtained from 3 donors, run in singlicate. (C-D) BMP6 and BMP9 do not induce hepcidin. Hepcidin mRNA expression in PBMCs cultured for 1 hour with BMP6, BMP9 (both 50 ng/mL), or TGF-β1 (10 ng/mL) in the presence or absence of IL-6 (50 ng/mL). Data are presented as (C) expression relative to GAPDH, and (D) fold-change in hepcidin expression relative to the untreated sample from the same individual. Dashed lines represent the mean values for PBMCs obtained from 4 donors, indicated by different symbols. (E, F) Small molecule and antibody-mediated inhibition of TGF-β1– and IL-6–mediated hepcidin induction. Hepcidin mRNA expression in PBMCs (N = 5 donors) cultured for 1 hour with combinations of IL-6 (50 ng/mL), TGF-β1 (10 ng/mL), the small molecule Alk5 (TGF-β receptor) inhibitor SD208 (1μM), and a mixture of neutralizing anti–IL-6 and anti–IL-6R antibodies (10 μg/mL each). Data are presented as (E) relative to GAPDH and (F) as fold-changes as in panels C, D. Data for anti–IL-6 treatment were not obtained for 2 PBMC preparations. (G-H) IL-6 treatment of human PBMCs renders them refractory to hepcidin induction by subsequent retreatment with IL-6, but not TGF-β1. Hepcidin mRNA expression in PBMCs (N = 3 donors) cultured for a total of 4 hours: at time zero, cells were treated with or without IL-6 (50 ng/mL); after 3 hours, they were treated with either IL-6 (50 ng/mL), TGF-β1 (10 ng/mL) or both. RNA was extracted after 4 hours. Data are presented as (G) relative to GAPDH and (H) fold-changes relative to the untreated sample from the same individual.
IL-6 and TGF-β1, but not BMPs, induce hepcidin mRNA in human PBMCs. (A-B) Time course of hepcidin induction by IL-6 and TGF-β1. qRT-PCR measurement of hepcidin mRNA induction in 5 × 106 PBMCs cultured for various times in the presence or absence of IL-6 (50 ng/mL) and TGF-β1 (10 ng/mL). Data are presented as (A) expression relative to GAPDH, and (B) fold-change in hepcidin expression relative to the untreated sample from the same timepoint. Datapoints represent means and ranges for PBMCs obtained from 3 donors, run in singlicate. (C-D) BMP6 and BMP9 do not induce hepcidin. Hepcidin mRNA expression in PBMCs cultured for 1 hour with BMP6, BMP9 (both 50 ng/mL), or TGF-β1 (10 ng/mL) in the presence or absence of IL-6 (50 ng/mL). Data are presented as (C) expression relative to GAPDH, and (D) fold-change in hepcidin expression relative to the untreated sample from the same individual. Dashed lines represent the mean values for PBMCs obtained from 4 donors, indicated by different symbols. (E, F) Small molecule and antibody-mediated inhibition of TGF-β1– and IL-6–mediated hepcidin induction. Hepcidin mRNA expression in PBMCs (N = 5 donors) cultured for 1 hour with combinations of IL-6 (50 ng/mL), TGF-β1 (10 ng/mL), the small molecule Alk5 (TGF-β receptor) inhibitor SD208 (1μM), and a mixture of neutralizing anti–IL-6 and anti–IL-6R antibodies (10 μg/mL each). Data are presented as (E) relative to GAPDH and (F) as fold-changes as in panels C, D. Data for anti–IL-6 treatment were not obtained for 2 PBMC preparations. (G-H) IL-6 treatment of human PBMCs renders them refractory to hepcidin induction by subsequent retreatment with IL-6, but not TGF-β1. Hepcidin mRNA expression in PBMCs (N = 3 donors) cultured for a total of 4 hours: at time zero, cells were treated with or without IL-6 (50 ng/mL); after 3 hours, they were treated with either IL-6 (50 ng/mL), TGF-β1 (10 ng/mL) or both. RNA was extracted after 4 hours. Data are presented as (G) relative to GAPDH and (H) fold-changes relative to the untreated sample from the same individual.
In PBMCs, IL-6 induced potent, yet transient, hepcidin mRNA up-regulation, peaking after 1 hour at a mean ∼ 80-fold higher than untreated samples, returning to near baseline levels by 3 hours after treatment (Figure 2A-B). Other genes, notably SOCS3 and SOCS1, feedback inhibitors of the IL-6Rα/gp130–JAK-STAT3 pathway show similar kinetics of induction by IL-6.32 Hepcidin and SOCS3 mRNA were induced strongly in PBMCs 1 hour after stimulation by IL-6 concentrations as low as 2 ng/mL (supplemental Figure 2A-B). Hematopoietic cells typically do not express IL-22R1,29 and consistent with this, IL-22 failed to induce hepcidin in PBMCs (supplemental Figure 2C).
TGF-β1 induced hepcidin modestly (∼ 3-fold) in PBMCs, and also synergized with IL-6, augmenting hepcidin transcription to ∼ 3% of the level of GAPDH 1 hour after stimulation (Figure 2A), corresponding to a mean hepcidin up-regulation of > 300-fold (Figure 2B). In contrast, BMP6 and BMP9 had no effect on hepcidin transcription in PBMCs, nor did they influence the response to IL-6 (Figure 2C-D). Together, these data are suggestive of synergy between STAT3 and SMAD signaling as in hepatoma cells; however in PBMCs, TGF-β1 rather than BMPs induces hepcidin.
The stimulatory effects of IL-6 and TGF-β1 could be specifically inhibited with neutralizing anti–IL-6/anti–IL-6R antibodies, and with the Alk5 TGF-β receptor 1 kinase inhibitor SD208, respectively (Figure 2E-F). SD208 alone effected a modest reduction in hepcidin, reminiscent of the effect of the BMP-SMAD pathway inhibitor dorsomorphin on baseline hepcidin expression in hepatoma cells.33
IL-6 treatment renders PBMCs refractory to subsequent IL-6, but not TGF-β1 stimulation
The transience of the IL-6 hepcidin-inducing signal is consistent with feedback inhibition of signaling after stimulation. Feedback inhibition of BMP-SMAD–mediated hepcidin activation in hepatocytes by the inhibitory SMAD, SMAD7, has recently been demonstrated.34 SOCS3, induced rapidly after IL-6 stimulation (supplemental Figure 2B), is a known feedback inhibitor of the STAT3 pathway and therefore analogously could be implicated.32,35 Consequently, we considered whether initial treatment of PBMCs with IL-6 would cause desensitization to subsequent stimulation.35 PBMCs were treated at time zero with IL-6 or PBS, and 3 hours later IL-6 and/or TGF-β1 were applied. RNA was extracted after a further hour. PBMCs stimulated at time zero with IL-6 were indeed refractory to further IL-6 treatment 3 hours later (Figure 2G-H). Likewise, SOCS3 induction by IL-6 was abrogated in IL-6–pretreated cells (supplemental Figure 2E-F). However, TGF-β1 effected a larger increase in hepcidin mRNA in IL-6–pretreated PBMCs than in PBS-pretreated cells (Figure 2G-H); TGF-β1 as expected had no effect on SOCS3 expression (supplemental Figure 2E-F). These data suggest that STAT3 pathway responsiveness is not required for sensitivity to TGF-β.
Induction of hepcidin in PBMC by agonists of cell-surface expressed TLRs and NOD2
Pattern recognition receptors (PRRs) including Toll-like receptors (TLRs) and Nod-like receptors (NLRs) expressed by innate immune cells including PBMCs detect conserved pathogenic motifs produced by invading microorganisms. PRR stimulation induces inflammatory cytokines and chemokines, and initiates immune responses.36 Different PRRs surveil distinct cellular and extracellular environs, and encounter pathogenic molecules derived from different classes of infections: cell-surface expressed TLR1/2/4/5/6 principally detect motifs derived from extracellular bacteria and fungi; endosomally-expressed TLR3/7/8/9 detect nucleic acids, often virally-derived; and NLRs detect pathogenic material in the cytoplasm. We investigated whether TLR/NLR agonists induced hepcidin in PBMCs, providing a link between the response to infection and modulation of iron.
PBMCs were first cultured with an array of bacterial-type TLR agonists, known to bind cell surface TLRs (Figure 3A; supplemental Figure 3A). The TLR5 agonist flagellin (from Salmonella typhimurium) consistently induced the largest fold-increase (mean ∼ 8-fold) in hepcidin mRNA in PBMCs; more modest increases were stimulated by Pam3CSK4 (TLR1/2 agonist), FSL1 (TLR6/2) and in most cases by LPS (TLR4). Heat-killled Listeria monocytogenes (HKLM, TLR2) and Muramyl Dipeptide (MDP, NOD2 agonist) did not significantly up-regulate hepcidin. This hierarchy did not correlate with IL-6 induction in the same cells (Figure 3B; supplemental Figure 3B): LPS and Pam3CSK4 induced IL-6 mRNA more strongly than flagellin; the level induced by FSL1 was lower. Unlike IL-6–mediated hepcidin expression that peaked 1 hour after treatment, hepcidin and IL-6 induction by flagellin and LPS were highest after ∼ 6 hours (Figure 3C-D; supplemental Figure 3C-D). Bacillus subtilis derived flagellin, like S typhimurium flagellin, also induced hepcidin and IL-6 mRNA expression at concentrations > 100 ng/mL (Figure 3E-F).
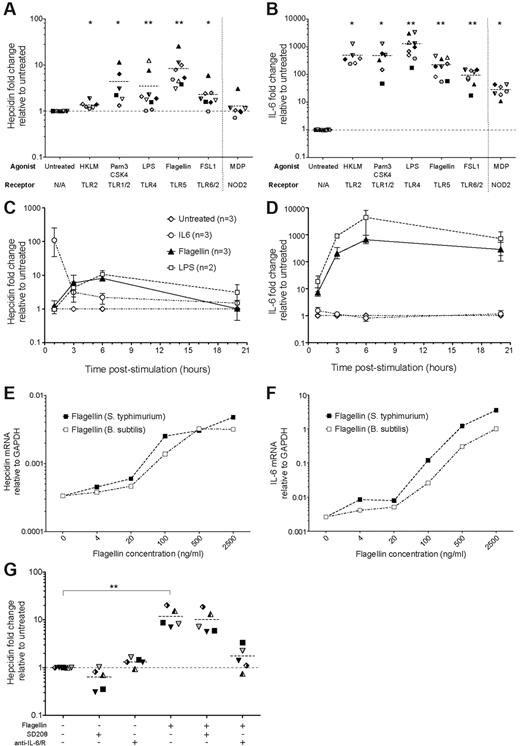
Hepcidin induction in human PBMCs by bacterial and fungal-related TLR agonists. (A-B) Hepcidin and IL-6 induction. qRT-PCR measurement of (A) hepcidin and (B) IL-6 mRNA in PBMCs cultured for 3 hours in the presence of various TLR agonists derived from bacteria or fungi, or their mimics: the TLR2 agonist Heat-killed L monocytogenes (HKLM, 7-8 × 107 cells/mL); the TLR1/2 agonist Pam3CSK4, a synthetic bacterial lipopeptide mimic (0.5 μg/mL); the TLR4 agonist lipopolysaccharide (LPS) derived from E coli K12 (1 μg/mL); the TLR5 agonist S typhimurium flagellin, a flagellar motor protein (0.5 μg/mL); and the TLR6/2 agonist FSL1, a synthetic Mycoplasma lipoprotein mimic (0.5 μg/mL). Muramyl dipeptide (MDP), a bioactive motif found within bacterial peptidoglycan and agonist for the cytoplasmically-expressed NLR NOD2 was also tested, separated from TLR agonists by a vertical dotted line. Data are presented as fold-changes in expression relative to untreated samples from the same individual (N = 9 PBMC donors, each symbol represents one PBMC preparation; not every individual was tested with every TLR agonist; datapoints represent, in the majority of cases, mean values of biologic duplicates; dashed line represents the mean for each dataset). *P < .05, **P < .01, Wilcoxon matched pairs signed-rank test, comparing with untreated samples: note that to account for variability in baseline hepcidin levels between individuals, statistics were generated based on the data presented in supplemental Figure 3A-B (see supplemental Methods). (C-D) Time course of hepcidin and IL-6 induction by Flagellin and LPS. Fold-changes in (C) hepcidin and (D) IL-6 expression (relative to untreated samples from the same timepoint and individual) in PBMCs (N = 3 donors) treated with LPS (1μg/ml) Flagellin (0.5 μg/ml) or IL-6 (50 ng/mL) for 1, 3, 6, or 20 hours. Mean and range are shown. (E-F) Hepcidin and IL-6 induction in PBMCs by titrations of S. typhimurium and B subtilis flagellins. (E) Hepcidin and (F) IL-6 mRNA expression relative to GAPDH in PBMCs (N = 1 donor) cultured for 3 hours with increasing concentrations of flagellins derived from S typhimurium (purple line) and B subtilis (red line). (G) Inhibition of flagellin-mediated hepcidin induction in PBMCs. Fold-changes in hepcidin mRNA expression (relative to untreated samples from the same individual; N = 5 donors) in PBMCs cultured for 3 hours in the presence or absence of: flagellin (0.5 μg/mL); the Alk5/TGF-β pathway inhibitor, SD208 (1μM); and a mix of neutralizing anti–IL-6 and anti–IL-6R antibodies (10 μg/mL each, added 15 minutes before addition of flagellin/SD208 to the cultures). P < .001, Friedman test; **P < .01 indicates Dunn multiple comparison posttest.
Hepcidin induction in human PBMCs by bacterial and fungal-related TLR agonists. (A-B) Hepcidin and IL-6 induction. qRT-PCR measurement of (A) hepcidin and (B) IL-6 mRNA in PBMCs cultured for 3 hours in the presence of various TLR agonists derived from bacteria or fungi, or their mimics: the TLR2 agonist Heat-killed L monocytogenes (HKLM, 7-8 × 107 cells/mL); the TLR1/2 agonist Pam3CSK4, a synthetic bacterial lipopeptide mimic (0.5 μg/mL); the TLR4 agonist lipopolysaccharide (LPS) derived from E coli K12 (1 μg/mL); the TLR5 agonist S typhimurium flagellin, a flagellar motor protein (0.5 μg/mL); and the TLR6/2 agonist FSL1, a synthetic Mycoplasma lipoprotein mimic (0.5 μg/mL). Muramyl dipeptide (MDP), a bioactive motif found within bacterial peptidoglycan and agonist for the cytoplasmically-expressed NLR NOD2 was also tested, separated from TLR agonists by a vertical dotted line. Data are presented as fold-changes in expression relative to untreated samples from the same individual (N = 9 PBMC donors, each symbol represents one PBMC preparation; not every individual was tested with every TLR agonist; datapoints represent, in the majority of cases, mean values of biologic duplicates; dashed line represents the mean for each dataset). *P < .05, **P < .01, Wilcoxon matched pairs signed-rank test, comparing with untreated samples: note that to account for variability in baseline hepcidin levels between individuals, statistics were generated based on the data presented in supplemental Figure 3A-B (see supplemental Methods). (C-D) Time course of hepcidin and IL-6 induction by Flagellin and LPS. Fold-changes in (C) hepcidin and (D) IL-6 expression (relative to untreated samples from the same timepoint and individual) in PBMCs (N = 3 donors) treated with LPS (1μg/ml) Flagellin (0.5 μg/ml) or IL-6 (50 ng/mL) for 1, 3, 6, or 20 hours. Mean and range are shown. (E-F) Hepcidin and IL-6 induction in PBMCs by titrations of S. typhimurium and B subtilis flagellins. (E) Hepcidin and (F) IL-6 mRNA expression relative to GAPDH in PBMCs (N = 1 donor) cultured for 3 hours with increasing concentrations of flagellins derived from S typhimurium (purple line) and B subtilis (red line). (G) Inhibition of flagellin-mediated hepcidin induction in PBMCs. Fold-changes in hepcidin mRNA expression (relative to untreated samples from the same individual; N = 5 donors) in PBMCs cultured for 3 hours in the presence or absence of: flagellin (0.5 μg/mL); the Alk5/TGF-β pathway inhibitor, SD208 (1μM); and a mix of neutralizing anti–IL-6 and anti–IL-6R antibodies (10 μg/mL each, added 15 minutes before addition of flagellin/SD208 to the cultures). P < .001, Friedman test; **P < .01 indicates Dunn multiple comparison posttest.
Antibody-mediated neutralization of IL-6 significantly reduced flagellin-mediated hepcidin induction, indicating IL-6–dependence (Figure 3G; supplemental Figure 3E). In contrast, SD208 had little effect (Figure 3G), suggesting this pathway does not contribute to the effect of flagellin. Neither anti–IL-6/anti–IL-6R nor SD208 treatment significantly affected IL-6 production by PBMCs in response to flagellin (supplemental Figure 3F).
Induction of hepcidin by C albicans
In contrast to purified TLR agonists, whole pathogens are likely to express combinations of host receptor agonists, allowing interactions between different signaling pathways.37 C albicans causes a spectrum of clinical disease in immunocompromised hosts ranging from chronic mucocutaneous candidiasis to candidaemia and invasive candidiasis, and its control is associated with effective Th17 T cell responses.38 Th17 responses are themselves directed by a cytokine milieu that includes both IL-6 and TGF-β.39,40 Because these cytokines potently up-regulate hepcidin in PBMCs, we hypothesized that hepcidin induction might represent a component of the host response to C albicans infection.
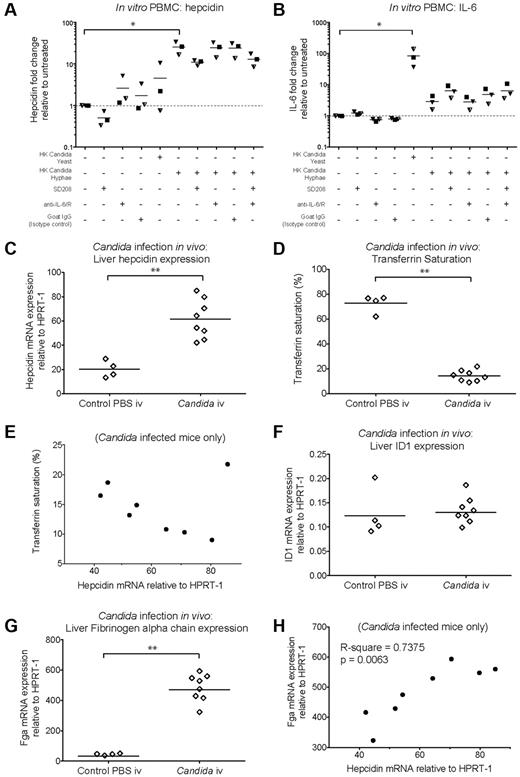
Heat-killed C albicans yeast cells and hyphae (supplemental Figure 4A-B) were cocultured for 22 hours with PBMCs. Heat-killed C. albicans hyphae caused significant hepcidin up-regulation in PBMCs, while heat-killed yeast induced hepcidin less notably (Figure 4A; supplemental Figure 4C). However, yeast caused greater up-regulation of PBMC IL-6 mRNA than hyphae, which only yielded an approximate 4-fold increase relative to untreated PBMCs (Figure 4B; supplemental Figure 4D). In contrast, the levels of IL-6 mRNA induced by the TLR agonists LPS and Flagellin were orders of magnitude higher (Figure 3D). Anti–IL-6/anti–IL-6R neutralizing antibodies did not prevent hepcidin up-regulation by hyphae suggesting a lack of IL-6 involvement whereas SD208 partially abrogated this response (Figure 4A; supplemental Figure 4C). These data suggest that the mechanism of hepcidin induction in PBMCs in response to heat-killed C albicans hyphae differs from that of the response to bacterial flagellin (Figure 3G).
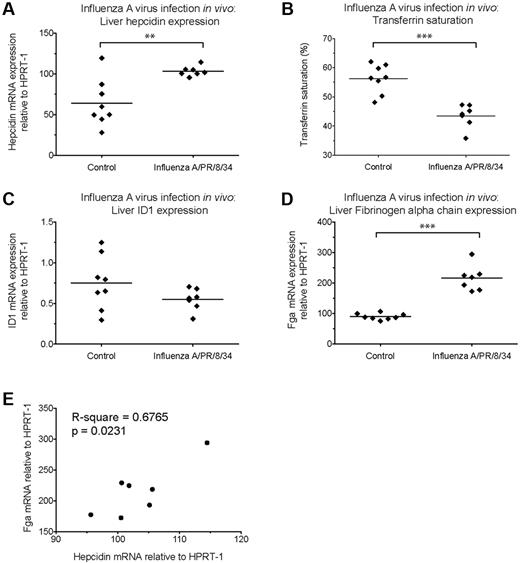
Hepcidin induction by C albicans in vitro and in vivo. (A-B) Hepcidin and IL-6 induction in PBMCs after coculture with heat-killed C albicans. qRT-PCR measurement of fold-changes in (A) hepcidin and (B) IL-6 mRNA (relative to untreated controls from the same individual) in PBMCs (N = 3 donors, represented by different symbols) cultured for 22 hours in the presence of heat-killed C albicans yeast or hyphae (PBMC:Candida ratio = 5:1), with or without the Alk5/TGF-β1 pathway inhibitor, SD208 (1μM), and a mix of neutralizing anti–IL-6 and anti–IL6-R antibodies (10 μg/mL each, added 15 minutes before addition of C albicans or SD208 to cultures); goat IgG was used as an isotype control (20 μg/mL). Means are shown; P < .01, Friedman test; *P < .05 indicate Dunn multiple comparison test. (C-H) Hepcidin induction and effects on serum iron during acute systemic C albicans infection of mice. qRT-PCR measurement of (C) Hepcidin (Hamp1), (F) Id1, and (G) Fibrinogen α chain (Fga) mRNA expression relative to the endogenous control gene Hprt1 in livers of 8-week-old male C57BL/6 mice terminated 24 hours after intravenous infection with 5 × 105C albicans strain SC5314 yeast cells (N = 8) or PBS (N = 4). Serum was taken at termination for calculation of (D) transferrin saturation. **P < .01, Mann-Whitney test. (E,H) Pearson correlation between (E) liver hepcidin mRNA and transferrin saturation, and between (H) liver hepcidin and Fga mRNA levels considering infected mice only.
Hepcidin induction by C albicans in vitro and in vivo. (A-B) Hepcidin and IL-6 induction in PBMCs after coculture with heat-killed C albicans. qRT-PCR measurement of fold-changes in (A) hepcidin and (B) IL-6 mRNA (relative to untreated controls from the same individual) in PBMCs (N = 3 donors, represented by different symbols) cultured for 22 hours in the presence of heat-killed C albicans yeast or hyphae (PBMC:Candida ratio = 5:1), with or without the Alk5/TGF-β1 pathway inhibitor, SD208 (1μM), and a mix of neutralizing anti–IL-6 and anti–IL6-R antibodies (10 μg/mL each, added 15 minutes before addition of C albicans or SD208 to cultures); goat IgG was used as an isotype control (20 μg/mL). Means are shown; P < .01, Friedman test; *P < .05 indicate Dunn multiple comparison test. (C-H) Hepcidin induction and effects on serum iron during acute systemic C albicans infection of mice. qRT-PCR measurement of (C) Hepcidin (Hamp1), (F) Id1, and (G) Fibrinogen α chain (Fga) mRNA expression relative to the endogenous control gene Hprt1 in livers of 8-week-old male C57BL/6 mice terminated 24 hours after intravenous infection with 5 × 105C albicans strain SC5314 yeast cells (N = 8) or PBS (N = 4). Serum was taken at termination for calculation of (D) transferrin saturation. **P < .01, Mann-Whitney test. (E,H) Pearson correlation between (E) liver hepcidin mRNA and transferrin saturation, and between (H) liver hepcidin and Fga mRNA levels considering infected mice only.
We next investigated whether C albicans could induce hepcidin in vivo in a mouse model of systemic candidiasis. Hepcidin was significantly up-regulated in the livers of male C57BL/6 mice 24 hours after intravenous infection with C albicans SC5314 yeast (Figure 4C). Infection was associated with a dramatic reduction in transferrin saturation, from ∼ 70% in PBS-injected controls to 15% in infected animals (Figure 4D), consistent with a rapid and potent effect of elicited hepcidin on iron localization.21 We then considered whether the extent of hepcidin induction correlated with transferrin saturation in the infected mice. Although there was no significant correlation when all infected mice were considered (Figure 4E), a highly significant correlation was observed if the probable outlier was removed from the analysis (P = .0022, R-square = 0.87: Pearson correlation), in agreement with the level of induced hepcidin influencing the degree of iron redistribution.
To investigate whether Smad and Stat3 pathways might be involved in the observed hepcidin up-regulation, we assessed liver induction of other genes dependent on those pathways for their transcription. The Bmp-Smad–dependent gene Id1 was not up-regulated in concert with hepcidin (Figure 4F) suggesting a lack of involvement of this pathway in the hepcidin response to acute systemic C albicans infection. Conversely, the Stat3-dependent acute phase gene, Fga (encoding fibrinogen α chain) was strongly induced in infected mice (Figure 4G) and the degree of hepcidin up-regulation in infected animals correlated with the level of Fga induction (Figure 4H). Similarly, liver Il6 mRNA was induced in infected mice (supplemental Figure 4E). These data are consistent with Stat3 pathway activation being the major contributor to liver hepcidin induction in this context.
Effect of viral-like TLR agonists and live Influenza A virus on hepcidin induction
We then considered whether intracellular pathogens, especially viruses, might induce hepcidin. Pathogen-derived nucleic acid ligands are sensed by a different set of TLRs within endosomes rather than at the plasma membrane. Unlike the bacterial-type TLR agonists, a set of viral-type TLR agonists at best only weakly induced hepcidin in PBMCs (Figure 5A; supplemental Figure 5A). These agonists only modestly induced IL-6 relative to the levels induced by LPS, Pam3CSK4 or Flagellin (Figure 5B; supplemental Figure 5B). The comparison of this data with that in Figure 3 suggests that hepcidin induction by TLRs is more potently achieved after detection of pathogen-derived components at the cell surface rather than in endosomes.
Hepcidin and IL-6 induction by viral-like TLR agonists and whole live Influenza A virus. (A-B) Hepcidin and IL-6 induction in PBMCs by viral-like TLR agonists. qRT-PCR measurement or fold-changes in (A) hepcidin and (B) IL-6 mRNA expression relative to untreated samples from the same individual in PBMCs (N = 5 donors, represented by different symbols) cultured for 3 hours in the presence of various viral-like TLR agonists: the TLR3 agonist poly(I:C), a synthetic mimic of viral double-stranded RNA (10 μg/mL); the TLR7 agonist R837 (Imiquimod, 1 μg/mL); the TLR8 agonist ssRNA40, representing viral single-stranded RNA (4 μg/mL); and the TLR9 agonist ODN2006, a synthetic unmethylated CpG-containing oligonucleotide (2μM [except individual denoted by black diamond, 1.25μM]). The dashed line represents the mean for each dataset; not every individual was tested with every TLR agonist. (C-D) Hepcidin and IL-6 induction by live Influenza A virus in PBMCs. Fold-changes in (C) hepcidin and (D) IL-6 mRNA expression in PBMCs (N = 10 donors) cultured for 3 hours in the presence of Influenza A virus (A/PR/8/34) in serum-free media (10 PFU per PBMC). ***P < .001, paired t test (C); **P < .01, Wilcoxon matched pairs test (D). (E) Effect of anti–IL-6/anti–IL-6R antibodies on Influenza A virus mediated hepcidin induction. Fold-changes in hepcidin mRNA expression in a subset of the PBMC samples from panel C also cultured with and without Influenza A virus in the presence of a mix of neutralizing anti–IL-6 and anti–IL6-R antibodies (10 μg/mL each, added 15 minutes before addition of virus to cultures). Values for Influenza A virus alone are those from panel C. P < .001, Friedman test; **P < .01, ***P < .001 indicate Dunn multiple comparison test. (F) Effect of the TGF-β pathway inhibitor SD208 on Influenza A virus mediated hepcidin induction. Fold-changes in hepcidin mRNA expression in a subset of the PBMC samples from panel C also cultured with and without Influenza A virus in the presence of SD208 (1 μM). Values for Influenza A virus alone are those from panel C. P < .001, 1-way repeated measures ANOVA; *P < .05 indicates significant Bonferroni posttest. (G-H) Hepcidin induction by live Influenza A virus in Hep3B and HepG2 cells. Fold-changes in hepcidin mRNA expression (relative to untreated cells) in (G) Hep3B cells and (H) HepG2 cells treated with Influenza A virus (A/PR/8/34; 100:1 PFU:Hep3B, and 10:1 PFU:HepG2) or recombinant human BMP6 (18nM) for 24 hours. Note that despite equivalent cell numbers, the quantities of GAPDH mRNA detected in infected cultures were ∼ 3-fold lower in infected cells than in untreated controls (data not shown), yet amounts of hepcidin mRNA were ∼ 30-fold greater, corresponding to the displayed ∼ 100-fold increases in infected cells. Data represent the mean + SD of single experiments carried out in biologic triplicate. **P < .01, t test (Gaussian distribution assumed).
Hepcidin and IL-6 induction by viral-like TLR agonists and whole live Influenza A virus. (A-B) Hepcidin and IL-6 induction in PBMCs by viral-like TLR agonists. qRT-PCR measurement or fold-changes in (A) hepcidin and (B) IL-6 mRNA expression relative to untreated samples from the same individual in PBMCs (N = 5 donors, represented by different symbols) cultured for 3 hours in the presence of various viral-like TLR agonists: the TLR3 agonist poly(I:C), a synthetic mimic of viral double-stranded RNA (10 μg/mL); the TLR7 agonist R837 (Imiquimod, 1 μg/mL); the TLR8 agonist ssRNA40, representing viral single-stranded RNA (4 μg/mL); and the TLR9 agonist ODN2006, a synthetic unmethylated CpG-containing oligonucleotide (2μM [except individual denoted by black diamond, 1.25μM]). The dashed line represents the mean for each dataset; not every individual was tested with every TLR agonist. (C-D) Hepcidin and IL-6 induction by live Influenza A virus in PBMCs. Fold-changes in (C) hepcidin and (D) IL-6 mRNA expression in PBMCs (N = 10 donors) cultured for 3 hours in the presence of Influenza A virus (A/PR/8/34) in serum-free media (10 PFU per PBMC). ***P < .001, paired t test (C); **P < .01, Wilcoxon matched pairs test (D). (E) Effect of anti–IL-6/anti–IL-6R antibodies on Influenza A virus mediated hepcidin induction. Fold-changes in hepcidin mRNA expression in a subset of the PBMC samples from panel C also cultured with and without Influenza A virus in the presence of a mix of neutralizing anti–IL-6 and anti–IL6-R antibodies (10 μg/mL each, added 15 minutes before addition of virus to cultures). Values for Influenza A virus alone are those from panel C. P < .001, Friedman test; **P < .01, ***P < .001 indicate Dunn multiple comparison test. (F) Effect of the TGF-β pathway inhibitor SD208 on Influenza A virus mediated hepcidin induction. Fold-changes in hepcidin mRNA expression in a subset of the PBMC samples from panel C also cultured with and without Influenza A virus in the presence of SD208 (1 μM). Values for Influenza A virus alone are those from panel C. P < .001, 1-way repeated measures ANOVA; *P < .05 indicates significant Bonferroni posttest. (G-H) Hepcidin induction by live Influenza A virus in Hep3B and HepG2 cells. Fold-changes in hepcidin mRNA expression (relative to untreated cells) in (G) Hep3B cells and (H) HepG2 cells treated with Influenza A virus (A/PR/8/34; 100:1 PFU:Hep3B, and 10:1 PFU:HepG2) or recombinant human BMP6 (18nM) for 24 hours. Note that despite equivalent cell numbers, the quantities of GAPDH mRNA detected in infected cultures were ∼ 3-fold lower in infected cells than in untreated controls (data not shown), yet amounts of hepcidin mRNA were ∼ 30-fold greater, corresponding to the displayed ∼ 100-fold increases in infected cells. Data represent the mean + SD of single experiments carried out in biologic triplicate. **P < .01, t test (Gaussian distribution assumed).
We next considered whether a live virus could stimulate hepcidin up-regulation in PBMCs. In the majority of PBMC isolates, cocultured Influenza A virus (A/PR/8/34) induced hepcidin mRNA (mean ∼ 5-fold increase), and in every case, IL-6 mRNA was up-regulated although not to the same extent as by LPS or flagellin (Figure 5C-D; supplemental Figure 5C-D). Although antibody-mediated IL-6 neutralization significantly reduced the level of hepcidin induced, inhibition was only partial despite the clear stimulation of IL-6 mRNA, suggesting other factors to be involved in Influenza-mediated hepcidin up-regulation in PBMCs (Figure 5E; supplemental Figure 5E). SD208 did not significantly inhibit hepcidin production (Figure 5F; supplemental Figure 5F).
Of the TLR agonists tested, neither those targeting extracellular nor endosomal TLRs up-regulated hepcidin in Hep3B cells (data not shown). However, because Influenza A/PR/8/34 virus infects multiple cell types, we investigated whether infection of hepatoma cells would stimulate hepcidin. We found that infection of Hep3B and HepG2 cells with 100 or 10 PFU/cell, respectively, for 24 hours caused significant hepcidin up-regulation, corresponding overall to approximate 100-fold increases (Figure 5G-H). Together, these data indicate that an intracellular infection, besides extracellular pathogen detection, can also stimulate hepcidin induction in both leukocytes and liver-derived cells.
We then investigated whether Influenza virus infection might induce hepcidin in vivo. The peak cytokine response in the lung after intranasal infection of mice with a highly pathogenic dose of Influenza A/PR/8/34 virus occurs around 3 days after infection. Mice infected with such a dose incurred significant weight loss by this time (supplemental Figure 6A). We observed marked Il6 mRNA induction in the lungs of infected mice (supplemental Figure 6B), accompanied by significantly raised liver hepcidin mRNA (Figure 6A) and decreased transferrin saturations (Figure 6B). Id1 mRNA remained unchanged (Figure 6C), but Fga induction was evident (Figure 6D), again correlating with hepcidin mRNA levels in infected animals (Figure 6E). As for C albicans infection, this is consistent with infection-associated hepcidin induction occurring through stimulation of the Stat3 pathway.
Hepcidin induction and effects on serum iron during acute Influenza virus A/PR/8/34 infection of mice. qRT-PCR measurement of (A) Hepcidin (Hamp1), (C) Id1 and (D) Fibrinogen α chain (Fga) mRNA expression relative to the endogenous control gene Hprt1 in the livers of 6-week-old female C57BL/6 mice terminated 3 days after intranasal infection with 3.5 HAU Influenza A virus (A/PR/8/34) (n = 7 mice) or mock-infected (PBS; n = 8 mice). Serum was taken at termination for calculation of (B) transferrin saturation. **P < .01, ***P < .001, t test. (E) Pearson correlation between hepcidin and Fga mRNA levels in livers of infected mice.
Hepcidin induction and effects on serum iron during acute Influenza virus A/PR/8/34 infection of mice. qRT-PCR measurement of (A) Hepcidin (Hamp1), (C) Id1 and (D) Fibrinogen α chain (Fga) mRNA expression relative to the endogenous control gene Hprt1 in the livers of 6-week-old female C57BL/6 mice terminated 3 days after intranasal infection with 3.5 HAU Influenza A virus (A/PR/8/34) (n = 7 mice) or mock-infected (PBS; n = 8 mice). Serum was taken at termination for calculation of (B) transferrin saturation. **P < .01, ***P < .001, t test. (E) Pearson correlation between hepcidin and Fga mRNA levels in livers of infected mice.
Discussion
Hepcidin production during innate immune responses to infections may enable a host to withhold iron from extracellular pathogens. By inhibiting ferroportin function in macrophages and enterocytes, hepcidin reduces iron availability in serum,21 potentially limiting pathogen growth therein. Here, we demonstrate that varied inflammatory and infectious stimuli induce hepcidin in hepatoma cells and PBMCs in vitro. We then show that hepcidin up-regulation, with concomitantly reduced transferrin saturation, represents a component of the acute response to both C albicans and Influenza A virus infections in vivo.
Hepatoma cell lines, although nonresponsive to iron-loaded transferrin, are commonly used as hepatocyte models for investigating hepcidin regulation as they display functional BMP-SMAD and IL-6–STAT3 pathways.24-26 IL-6–mediated hepcidin induction is well-characterized,17,18 yet the hepatocyte response during inflammation may be modulated by other factors including BMP2/9 and TNF-α.17,24,25 We found that IL-22 is an IL-6–independent hepcidin inducer in hepatoma cells, which like IL-6 causes STAT3 (but not SMAD1/5/8) phosphorylation and synergizes with BMP6 potentiating hepcidin expression,30 consistent with signaling through the well-described STAT3 element in the hepcidin promoter.18,19 IL-22 is an effector cytokine, produced during responses against extracellular pathogens, that stimulates anti-microbial responses in tissues, and acts on hepatocytes.27-30 Serum IL-22 is also increased in multiple myeloma41 and Crohn's disease,30 and so may, like IL-6, contribute to hepcidin induction and the etiology of anemia in such contexts.24
TGF-β1 fails to stimulate hepcidin significantly in human hepatoma cells, yet unlike BMP6 or BMP9, modestly induces hepcidin in PBMCs. Moreover, TGF-β1 potentiates the strong but short-lived PBMC hepcidin response to IL-6, suggestive of synergy between SMAD and STAT3 pathways in these cells. IL-6 treatment rendered PBMCs refractory to subsequent IL-6, but not TGF-β1, stimulation. Induction of STAT3 pathway feedback inhibitors such as SOCS3, up-regulated both by IL-6 and other cytokines32,35 could explain this observation. These data and those previously published suggest that the precise make-up of the inflammatory cytokine milieu controls hepcidin expression in both liver-derived cells and leukocytes.42,43 Although liver-synthesized hepcidin maintains iron homeostasis, the relative roles of hepatocyte- and nonhepatocyte-derived hepcidin in responses to different infections remain to be clarified. Baseline hepcidin mRNA levels are orders of magnitude higher in human liver tissue (typically exceeding the level of GAPDH mRNA) than in PBMCs. However, acute increases in hepcidin production by leukocytes at infection sites may still affect local iron availability to proliferating pathogens early in infection and therefore be physiologically important. The observation of autocrine down-regulation of transfected ferroportin by LPS-induced hepcidin in THP-1 monocytes is consistent with this hypothesis.42
To more closely mimic the PBMC response to pathogenic stimuli, we investigated hepcidin up-regulation by an array of bacterial-like agonists of cell-surface expressed TLRs and NOD2. LPS induces hepcidin in bone marrow derived murine macrophages and primary human monocytes.42,44 In addition, IL-6–independent, TLR2-mediated hepcidin up-regulation has been described in Borrelia burgdorferi–infected mice.45 Flagellin and less notably LPS, FSL1 and Pam3CSK4, induced hepcidin in PBMCs. Flagellin, derived from bacterial flagella, induces innate immune responses through TLR5.36 Flagellin is necessary for bacterial motility and so contributes to nutrient acquisition; hepcidin induction by flagellin may represent a host response that would reduce nutrient availability by lowering serum iron. Neutralizing antibodies targeting IL-6/IL-6R abrogated hepcidin induction by flagellin indicating IL-6-dependence, while the TGF-β pathway inhibitor SD208 had no effect on flagellin-mediated hepcidin induction.
We next investigated hepcidin induction by whole pathogens in vitro and in vivo, first considering the fungal pathogen C albicans. Iron supplementation associates with increased fungal burden and poor outcomes during systemic C albicans infection in mice.46 We found that heat-killed C albicans hyphae stimulated hepcidin in PBMCs after overnight coculture, in a partially TGF-β–dependent, but IL-6–independent manner, suggesting pathways of hepcidin induction may vary depending on the encountered pathogen. Moreover, liver hepcidin was significantly up-regulated in vivo during acute systemic C albicans infection, concomitant with dramatic reductions in transferrin saturation, equivalent in magnitude to that observed after injection of high concentrations of synthetic hepcidin into mice.21 Hepcidin induction during C albicans infection correlated with up-regulation of the Stat3-dependent gene Fga, but not the Bmp-Smad–dependent Id1 gene, highly suggestive of Stat3 pathway involvement.
Effective control of C albicans infection in vivo is associated with Th17 responses38 – mucosal T-cell responses conferring immunity against extracellular pathogens – specified in part by the combination of IL-6 and TGF-β1.39,40 Various cytokines including IL-22 are associated with these responses,28,38 and IL-22 itself mediates significant protective effects during the early phases of C albicans infections in mice.27 IL-22, and the combination of IL-6 and TGF-β1, as shown here, also induce hepcidin. Given this commonality of cytokines, and the observed hepcidin induction during systemic C albicans infections, the initiation and effector phases of Th17-type immune responses to such pathogens may be accompanied by hepcidin-mediated iron redistribution. In addition, elicited hepcidin has been reported to modulate immune responses directly.47 A rapid and potent hepcidin response during acute extracellular infections could cause iron withdrawal from serum, thereby slowing pathogen growth and buying time for the development of efficacious adaptive immune responses.
Intracellular infections are detected by a distinct set of pathogen recognition receptors. We found that ligands of endosomally-expressed TLRs, typically expressed by viruses or intracellular bacteria, at best weakly induced hepcidin. Hepcidin up-regulation during viral infections in human cells has to our knowledge never been formally demonstrated, although hepatitis C virus infection is associated with reduced serum hepcidin.48 We showed that live Influenza A virus induced hepcidin in both human PBMCs and hepatoma cells. PBMC induction was not completely inhibited by either SD208 or anti–IL-6/anti–IL-6R antibodies (despite robust induction of IL-6 mRNA). The discrepancy between hepcidin regulation by live virus compared with viral-type TLR agonists could reflect activation of nonTLR pathways. In hepatoma cells, infection resulted in increases in hepcidin mRNA greater than those induced by high IL-6 concentrations. Although PBMCs and hepatocytes are not the most notable target cells for Influenza A virus in vivo, these data demonstrate that hepcidin can be up-regulated by intracellular as well as extracellular infections. Furthermore, a highly pathogenic dose of the same virus caused significant liver hepcidin induction in mice, likely mediated by elicited cytokines stimulating the Stat3-pathway (as the Stat3-regulated gene Fga was also strongly up-regulated), and accompanied by reduced transferrin saturations. One report monitoring patients with acute influenza observed a rapid reduction of serum iron,49 suggestive of a similar hepcidin response in humans in the context of natural infection. Hepcidin induction and consequent iron redistribution during viral infection could influence viral replication (since viral life-cycles typically involve iron-dependent steps50 ) or the development of secondary infections.
In summary, our in vitro data demonstrate that pathways associated with the innate immune response, especially to extracellular pathogens, lead to hepcidin induction in both liver- and nonliver-derived cell types. The importance of IL-6 in inflammation-associated hepcidin up-regulation is well described, yet our data indicate that other cytokines such as TGF-β1 and IL-22 may also play key roles in integrating early immune responses with hepcidin synthesis. The association between hepcidin induction and reductions in serum iron during the acute phase of both systemic C albicans and Influenza A virus infections in vivo further supports the hypothesis that alterations to host iron represent a feature of the innate response to infection, which may contribute to disease pathogenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Vincenzo Cerundolo, Paolo Polzella, Giorgio Napolitani, Kevin Shenderov, John Baker, and Oliver Brain (MRC Human Immunology Unit, Oxford, United Kingdom) for reagents and helpful discussions. They also thank Prof N. Gow and Prof F. Odds (Aberdeen, United Kingdom) for the C albicans strain SC5314, Prof J. Skehel for supplying Influenza A virus (A/PR/8/34). This work was funded by the United Kingdom Medical Research Council and the Wellcome Trust. H.D. is a Beit Memorial Fellow for Medical Research and an MRC new investigator.
Authorship
Contribution: A.E.A., U.G., L.-P.H., A.R.M.T., and H.D. designed the experiments; A.E.A., L.A.E., U.G., S.C., N.S., and T.A.S. performed the experiments; A.E.A. and H.D. wrote and edited the manuscript; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Hal Drakesmith, Molecular Immunology Group, Weatherall Institute of Molecular Medicine, University of Oxford, John Radcliffe Hospital, Oxford, OX3 9DS, United Kingdom; e-mail: hdrakes@hammer.imm.ox.ac.uk.

![Figure 5. Hepcidin and IL-6 induction by viral-like TLR agonists and whole live Influenza A virus. (A-B) Hepcidin and IL-6 induction in PBMCs by viral-like TLR agonists. qRT-PCR measurement or fold-changes in (A) hepcidin and (B) IL-6 mRNA expression relative to untreated samples from the same individual in PBMCs (N = 5 donors, represented by different symbols) cultured for 3 hours in the presence of various viral-like TLR agonists: the TLR3 agonist poly(I:C), a synthetic mimic of viral double-stranded RNA (10 μg/mL); the TLR7 agonist R837 (Imiquimod, 1 μg/mL); the TLR8 agonist ssRNA40, representing viral single-stranded RNA (4 μg/mL); and the TLR9 agonist ODN2006, a synthetic unmethylated CpG-containing oligonucleotide (2μM [except individual denoted by black diamond, 1.25μM]). The dashed line represents the mean for each dataset; not every individual was tested with every TLR agonist. (C-D) Hepcidin and IL-6 induction by live Influenza A virus in PBMCs. Fold-changes in (C) hepcidin and (D) IL-6 mRNA expression in PBMCs (N = 10 donors) cultured for 3 hours in the presence of Influenza A virus (A/PR/8/34) in serum-free media (10 PFU per PBMC). ***P < .001, paired t test (C); **P < .01, Wilcoxon matched pairs test (D). (E) Effect of anti–IL-6/anti–IL-6R antibodies on Influenza A virus mediated hepcidin induction. Fold-changes in hepcidin mRNA expression in a subset of the PBMC samples from panel C also cultured with and without Influenza A virus in the presence of a mix of neutralizing anti–IL-6 and anti–IL6-R antibodies (10 μg/mL each, added 15 minutes before addition of virus to cultures). Values for Influenza A virus alone are those from panel C. P < .001, Friedman test; **P < .01, ***P < .001 indicate Dunn multiple comparison test. (F) Effect of the TGF-β pathway inhibitor SD208 on Influenza A virus mediated hepcidin induction. Fold-changes in hepcidin mRNA expression in a subset of the PBMC samples from panel C also cultured with and without Influenza A virus in the presence of SD208 (1 μM). Values for Influenza A virus alone are those from panel C. P < .001, 1-way repeated measures ANOVA; *P < .05 indicates significant Bonferroni posttest. (G-H) Hepcidin induction by live Influenza A virus in Hep3B and HepG2 cells. Fold-changes in hepcidin mRNA expression (relative to untreated cells) in (G) Hep3B cells and (H) HepG2 cells treated with Influenza A virus (A/PR/8/34; 100:1 PFU:Hep3B, and 10:1 PFU:HepG2) or recombinant human BMP6 (18nM) for 24 hours. Note that despite equivalent cell numbers, the quantities of GAPDH mRNA detected in infected cultures were ∼ 3-fold lower in infected cells than in untreated controls (data not shown), yet amounts of hepcidin mRNA were ∼ 30-fold greater, corresponding to the displayed ∼ 100-fold increases in infected cells. Data represent the mean + SD of single experiments carried out in biologic triplicate. **P < .01, t test (Gaussian distribution assumed).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/15/10.1182_blood-2011-04-351957/4/m_zh89991180010005.jpeg?Expires=1765971661&Signature=H-QlEuCMZxrqGSA2KilnnK6BRV97pLW0ltzSxiC3xkbcQJIa5JC83k5LDXyKRueN-6UEYpNLPfCNbz~S~H80W3db1qfXcSwWDbt1Nh8QlS9qnU7bggySPZjqdtxvZaxcjcZTyakeZXJgfF5STvp8QMuhshlBY9RpQtKwmz1W3sOlH2N2RaUB-06lk~5Aow3hefhJroF-ZQW3qhZJ0faTbCYBXXlOn6xYtPyYJLH-J92MkqbEGxCkjtsClArXzTWHoUcD4Js9XLywi4IZT0hwuroyUlfGLKf0Bb5EFYSnUTZka~soTkt88ND06jf9sw0NA3O0sRatrpNlbXnjXe2zGw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)