Abstract
Sustained undetectable molecular residual disease (UMRD) is obtained in a minority of patients with chronic myeloid leukemia treated with tyrosine kinase inhibitors. It remains unclear whether these patients are definitively cured of their leukemia or whether leukemic stem cells (LSCs) persist in their BM. We have evaluated the presence of BCR-ABL–expressing marrow LSCs in 6 patients with chronic myeloid leukemia with sustained UMRD induced by IFN-α (n = 3), imatinib mesylate after IFN-α failure (n = 2), and dasatinib after imatinib intolerance (n = 1). Purified CD34+ cells were used for clonogenic and long-term culture-initiating cell assays performed on classic or HOXB4-expressing MS-5 feeders. Using this strategy, we identified BCR-ABL–expressing LSCs in all patients. Interestingly, long-term culture-initiating cell assays with MS-5/HOXB4 stromal feeders increased detected numbers of LSCs in 3 patients. The relation between LSC persistency and a potential risk of disease relapse for patients with durable UMRD (on or off tyrosine kinase inhibitor therapy) warrants further investigation.
Introduction
The introduction of imatinib mesylate (IM) has dramatically improved the treatment of chronic myeloid leukemia (CML).1,2 This tyrosine kinase inhibitor (TKI) has shown superior efficacy to IFN-α plus cytarabine, the former standard therapy.3,4 Second-generation TKIs (dasatinib, nilotinib) were developed to overcome IM resistance or intolerance.5,6 In a minority of patients treated with IM, an undetectable molecular residual disease (UMRD; also named complete molecular response) can be obtained. UMRD was defined as the absence of detectable BCR-ABL mRNA transcript from blood or marrow samples or both with the use of RNA samples of adequate quality.7
In patients achieving a complete cytogenetic response, the persistence of malignant CD34+ cells, CD34+/CD38− cells, CFU-Cs (CFUs in culture) or LTC-ICs (long-term culture-initiating cells) has been shown with the use of RT-PCR or FISH.8-11 For patients who maintain a durable UMRD, there are currently no data about the persistence of leukemic stem cells (LSCs), a fraction known to be intrinsically resistant to TKIs.12-15 The French STIM (STop IMatinib) trial reported the results of IM cessation in patients with sustained UMRD for ≥ 2 years.16 In that work, the molecular relapse-free survival was 41% at 1 year, showing that a molecular relapse occurred in a significant proportion of patients. Detection at the genomic level of BCR-ABL rearrangement in patients with UMRD after IM therapy or stem cell transplantation has shown the presence of residual blood leukemic cells not detected by RT-PCR.17,18 All these data support but do not show the hypothesis of LSC persistency in patients with sustained UMRD induced by TKI or IFN-α or both. In this work, we analyzed the presence of BCR-ABL–expressing hematopoietic stem cells in BM samples from 6 patients with CML with undetectable BCR-ABL transcript in their peripheral blood for > 3 years.
Methods
Patients
Clinical characteristics of the patients are detailed in Table 1. Three patients (patients 1-3) were treated with IFN-α alone for 13, 9, and 6 years, respectively, and were off therapy for > 8 years without relapse. These patients had no detectable BCR-ABL mRNA in their peripheral blood for > 4 years. Patients 4 and 5 were initially treated with IFN-α and then switched to IM, leading to an UMRD. These 2 patients were off IM for ∼ 2 years and had no detectable BCR-ABL mRNA in their peripheral blood for 4 and 5 years, respectively. Patient 6 was first treated with IM and then switched to dasatinib. At the time of the study, she was still on dasatinib therapy with no detectable BCR-ABL transcript in her blood for 3 years. All patients provided informed consent in accordance with the Declaration of Helsinki for participation to the study that was approved by the scientific committee of the Inserm CIC 0802.
Clinical characteristics of the patients
| Patient . | Sex/age, y . | Age at diagnosis, y . | Phase and BCR-ABL transcript at diagnosis . | Treatment 1 . | Treatment 2 . | Treatment discontinuation . | BCR-ABL undetectability (blood samples) . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Treatment . | Duration, y . | Yes/no . | Treatment . | Duration, y . | Yes/no . | Age at discontinuation, y . | Off-therapy duration, y . | Duration, y . | ||||
| 1 | F/66.7 | 43 | CP, b3a2 | IFN-α | 13.3 | No | Yes | 55.4 | 11.3 | 5.1 | ||
| 2 | M/71.8 | 46 | CP, b3a2 | IFN-α | 9.1 | No | Yes | 55.1 | 16.7 | 4.3 | ||
| 3 | F/72.3 | 57.5 | CP, b3a2 | IFN-α | 6.5 | No | Yes | 64 | 8.3 | 5.8 | ||
| 4 | F/69.2 | 58.1 | CP, b3a2 | IFN-α | 4.0 | Yes | IM 400 | 4.6 | Yes | 66.8 | 2.3 | 4.0 |
| 5 | F/70 | 61.6 | CP, b3a2 | IFN-α | 1.3 | Yes | IM 400 | 5.0 | Yes | 68.1 | 1.9 | 5.1 |
| 6 | F/78.5 | 74 | CP, b3a2 | IM 400 | 4 months | Yes | Dasatinib* | 4 | No | 3.2 | ||
| Patient . | Sex/age, y . | Age at diagnosis, y . | Phase and BCR-ABL transcript at diagnosis . | Treatment 1 . | Treatment 2 . | Treatment discontinuation . | BCR-ABL undetectability (blood samples) . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Treatment . | Duration, y . | Yes/no . | Treatment . | Duration, y . | Yes/no . | Age at discontinuation, y . | Off-therapy duration, y . | Duration, y . | ||||
| 1 | F/66.7 | 43 | CP, b3a2 | IFN-α | 13.3 | No | Yes | 55.4 | 11.3 | 5.1 | ||
| 2 | M/71.8 | 46 | CP, b3a2 | IFN-α | 9.1 | No | Yes | 55.1 | 16.7 | 4.3 | ||
| 3 | F/72.3 | 57.5 | CP, b3a2 | IFN-α | 6.5 | No | Yes | 64 | 8.3 | 5.8 | ||
| 4 | F/69.2 | 58.1 | CP, b3a2 | IFN-α | 4.0 | Yes | IM 400 | 4.6 | Yes | 66.8 | 2.3 | 4.0 |
| 5 | F/70 | 61.6 | CP, b3a2 | IFN-α | 1.3 | Yes | IM 400 | 5.0 | Yes | 68.1 | 1.9 | 5.1 |
| 6 | F/78.5 | 74 | CP, b3a2 | IM 400 | 4 months | Yes | Dasatinib* | 4 | No | 3.2 | ||
CP indicates chronic phase; and IM 400, imatinib 400 mg daily.
Dasatinib 70 mg twice daily and then 100 mg twice daily.
Design of the study
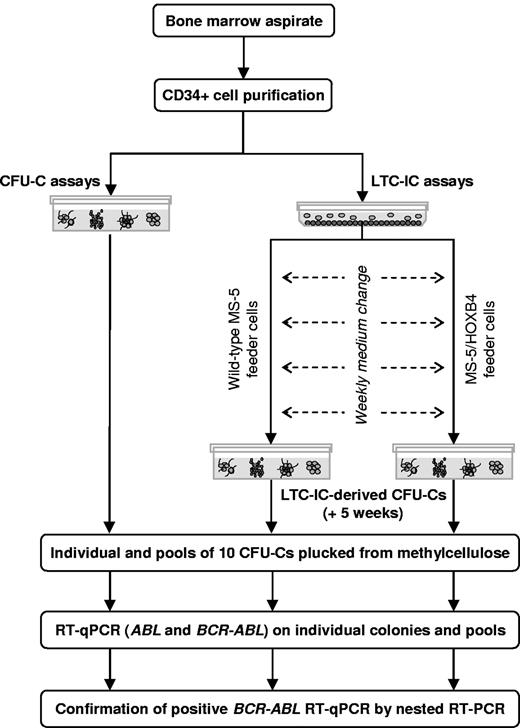
BM samples were collected, and CD34+ cells were purified. After performing a clonogenic assay, CD34+ cells were used in LTC-IC assays (Figure 1). To screen the largest number of CFU-Cs and LTC-IC–derived CFU-Cs from each patient, ∼ 20 individual colonies and ≤ 20 pools of 10 colonies were plucked from clonogenic assays at day 0 and from LTC-IC–derived progenitors at week 5. Individual and pooled CFU-Cs were tested for the presence of BCR-ABL mRNA by reverse transcription quantitative real-time PCR (RT-qPCR) and nested RT-PCR.
Analysis of BCR-ABL–expressing cells in progenitor and stem cell compartments, design of the study.
Analysis of BCR-ABL–expressing cells in progenitor and stem cell compartments, design of the study.
In vitro hematopoietic progenitor and stem cell assays
CD34+ cells were purified with an immunomagnetic cell sorting system (Miltenyi Biotech). CFU-C assays (at day 0) were performed by plating ∼ 5000 CD34+ cells on semisolid methylcellulose Methocult H4435 medium (Stem Cell Technologies). After 14 days of culture, hematopoietic colonies were enumerated, and a large fraction or the almost totality of CFU-Cs were plucked from methylcellulose and put (individually or by pools of 10 colonies) into RNA extraction buffer (Arcturus Bioscience). Long-term cultures were initiated with ∼ 60 000 CD34+ cells on murine MS-5 feeder cells as previously described.19 In addition, in 4 patients, LTC-IC experiments were performed on HOXB4-expressing MS-5 stromal cells (MS-5/HOXB4).20
Detection of BCR-ABL mRNA on individual and pooled colonies
Total RNA was extracted from individual and pooled colonies with the use of the PicoPure RNA isolation kit (Arcturus Bioscience) and totally reverse transcribed with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The presence of BCR-ABL transcripts was analyzed by RT-qPCR according to the Europe Against Cancer protocol.21 Each plate consisted of cDNA samples from hematopoietic colonies, no template controls, BCR-ABL–positive and –negative controls. Real-time PCR reaction consists of 45 cycles, each assay was done in duplicate, and Ct (cycle threshold) data from both were collected. ABL amplification was used to assess the presence of amplifiable cDNA. The occurrence of BCR-ABL mRNA was controlled by a 2-step RT-PCR protocol that used nested primers.22 For all patients, BCR-ABL rearrangements identified by nested RT-PCR were characterized by direct sequencing with the use of the Big Dye Terminator Version 3.0 kit (Applied Biosystems).
Results and discussion
All patients included in this work were BCR-ABL negative in their peripheral blood for > 3 years (with ≥ 20 000 copies of ABL mRNA). At the time of study, some bulk CD34+ cells (patients 3 and 5) and CD34− cell fractions (patients 2, 3, and 4) isolated from the BM were also tested by RT-qPCR. In these samples, there was no detectable BCR-ABL transcript. We then evaluated BCR-ABL expression in individual and pooled CFU-Cs derived from clonogenic and LTC-IC assays (supplemental Tables 1-3, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). A total of 2000 hematopoietic colonies were screened by RT-qPCR. All BCR-ABL–positive colonies and some –negative ones were then analyzed by nested RT-PCR (supplemental Figures 1-2). To be stringent, only the colonies in which BCR-ABL mRNA transcripts were detected by RT-qPCR and confirmed by nested RT-PCR were definitively considered as positive (supplemental Table 4). A b3a2 rearrangement, similar to the original chimeric mRNA identified at diagnosis, was observed in all BCR-ABL–positive colonies.
For RT-qPCR experiments, necessarily performed on a limited number of cells (500-4000, according to the size of the colonies), median Cts for ABL control were 29.9 and 30.5 (BCR-ABL–negative and –positive CFU-Cs, respectively), and 36.7 for BCR-ABL. These parameters might reflect both the small number of hematopoietic cells and the variable transcription of BCR-ABL in Ph1 hematopoietic progenitors, because it has previously been documented.23 The 66 RT-qPCR assays performed in this study were validated by the constant absence of fluorescent signal for the no template controls and BCR-ABL–negative controls after 45 amplification cycles and the repeatability of Ct values for duplicates (even for high Cts). Therefore, the procedure used ensures the reliability of our analysis, with, however, a potential risk of underestimating the number of BCR-ABL–expressing CFU-Cs.
The analysis of BCR-ABL mRNA expression in individual and pooled clonogenic progenitors and LSCs is summarized in Table 2. It should be noted that a BCR-ABL–expressing pool could represent ≥ 1 (≤ 10) positive colonies. Hence, the number of BCR-ABL–positive CFU-Cs and LTC-IC–derived stem cells may commonly be underestimated. As can be seen in the Table 2, patients treated with IFN-α alone (patients 1-3) and in sustained molecular remission clearly showed evidence of BCR-ABL–expressing CFU-Cs as well as LTC-ICs. In patients 1 and 2, only one pool of progenitors expressed BCR-ABL, whereas several positive LSCs were detectable. Remarkably, in patient 2 who was off therapy for 16 years, 6 individual LTC-IC–derived CFU-Cs of 79 expressed BCR-ABL. It should be noted that patient 3, who presented the shortest off-therapy period, harbored the largest number of BCR-ABL–positive CFU-Cs at day 0. For patients treated successively with IFN-α then IM (patients 4 and 5), BCR-ABL–expressing cells were only observed in clonogenic assays but not in LTC-IC–derived progenitors with the use of wild-type MS-5 feeder cells. However, the presence of clonogenic cells harboring BCR-ABL rearrangement in these patients clearly indicated the involvement of a more primitive stem cell compartment. Interestingly, in patient 5, who had an UMRD for > 4 years, the use of MS-5/HOXB4 feeder cells allowed the detection of 2 BCR-ABL–expressing LTC-IC–derived progenitors, suggesting the amplification or self-renewal or both of an otherwise quiescent stem cell. It is noticeable that, in the UMRD context, the use of LTC-IC assays with MS-5/HOXB4 stromal feeders increased detected numbers of LSCs in 3 patients (2,3 and 5). In patient 6, treated only by TKIs and with a sustained UMRD of > 3 years, a massive involvement of LTC-IC–derived progenitors expressing BCR-ABL was observed.
Analysis of BCR-ABL-expressing cells in CFU-C and LTC-IC assays
| Patient . | Clonogenic assays at day 0* . | Clonogenic assays on LTC-IC (week 5)† . | Tested colonies . | ||||
|---|---|---|---|---|---|---|---|
| Individual CFU-Cs . | Pools of 10 CFU-Cs . | Individual LTC-ICs . | Pools of 10 LTC-ICs . | ||||
| MS-5 . | MS-5/HOXB4 . | MS-5 . | MS-5/HOXB4 . | ||||
| 1 | 0/20 | 1/18 | 4/31 | 1/8 | 311 | ||
| 2 | 0/20 | 1/19 | 2/39 | 4/40 | ND | ND | 289 |
| 3 | 9/19 | 11/16 | 1/30 | 9/30 | ND | ND | 239 |
| 4 | 2/20 | 0/19 | 0/20 | 0/19 | 420 | ||
| 5 | 1/20 | 0/20 | 0/20 | 2/17 | 0/20 | 0/20 | 657 |
| 6 | 1/17 | ND | 23/24 | 12/43 | ND | ND | 84 |
| Tested colonies | 116 | 920 | 164 | 130 | 470 | 200 | 2000 |
| Patient . | Clonogenic assays at day 0* . | Clonogenic assays on LTC-IC (week 5)† . | Tested colonies . | ||||
|---|---|---|---|---|---|---|---|
| Individual CFU-Cs . | Pools of 10 CFU-Cs . | Individual LTC-ICs . | Pools of 10 LTC-ICs . | ||||
| MS-5 . | MS-5/HOXB4 . | MS-5 . | MS-5/HOXB4 . | ||||
| 1 | 0/20 | 1/18 | 4/31 | 1/8 | 311 | ||
| 2 | 0/20 | 1/19 | 2/39 | 4/40 | ND | ND | 289 |
| 3 | 9/19 | 11/16 | 1/30 | 9/30 | ND | ND | 239 |
| 4 | 2/20 | 0/19 | 0/20 | 0/19 | 420 | ||
| 5 | 1/20 | 0/20 | 0/20 | 2/17 | 0/20 | 0/20 | 657 |
| 6 | 1/17 | ND | 23/24 | 12/43 | ND | ND | 84 |
| Tested colonies | 116 | 920 | 164 | 130 | 470 | 200 | 2000 |
ND indicates not done (insufficient clonogenic or LTC-IC–derived CFU-C growth).
Number of BCR-ABL–positive CFU-Cs per tested CFU-Cs.
Number of BCR-ABL–positive LTC-ICs per tested LTC-ICs.
The present study, necessarily performed on a small fraction of CD34+ cells, shows the long-term persistence of a considerable amount of BCR-ABL–expressing stem cells in several patients in durable UMRD. The French STIM trial showed that ∼ 40% of such patients do not relapse at 12 months of follow-up.16 Two patients reported in the present study (patients 4 and 5) were part of this trial, and some BCR-ABL–expressing hematopoietic progenitors were detected in their BM. It remains to be established whether the persistence of BCR-ABL–expressing LSCs in these patients will correlate with a potential disease relapse in the future. Moreover, it has to be determined why and how a significant proportion of patients who achieved long-term remissions with IFN-α alone, IFN-α then IM, or IM alone do not relapse after discontinuation of therapy.
The persistence of residual BCR-ABL–expressing stem cells (cycling or quiescent) in BM represents a theoretical risk of disease recurrence. This would suggest that the characterization of LSCs by functional assays could be of interest in clinical practice. Indeed, even if it is not necessary to kill the last leukemic cell, as recently discussed,24 it might be crucial to eradicate the last LSC.
Detection of BCR-ABL–expressing clonogenic and LTC-IC–derived progenitors in patients with sustained UMRD is suggestive of highly active mechanisms of quiescence that could be operational in vivo. It has been shown that the most primitive quiescent LSC compartment express high levels of BCR-ABL and is intrinsically refractory to all TKIs used in clinical practice.12,15 Moreover, a recent work showed that the survival of Philadelphia-positive LSCs was independent from the BCR-ABL kinase activity.25 Consequently, TKI therapy seems unlikely to be fully curative. We report here in patients with sustained UMRD that LSCs could indeed persist after the cessation of therapy. These results raise several questions about the mechanisms of long-term LSC dormancy as well as the role of an immunomodulatory effect of the treatments. In addition, our data show that some patients who appear clinically “cured” have persistent LSCs, suggesting that the definition of a “clinical cure” should take into account the investigation of the stem cell compartment. The analysis of BCR-ABL–expressing stem cells in patients with CML with sustained UMRD induced by second-generation TKIs as a first-line therapy could also be of great interest, because these drugs generate molecular responses more rapidly than IM. Finally, future studies are required to determine whether, before TKI discontinuation, the evaluation of LSCs could be of clinical value to assess the burden of dormant tumor cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Alison Rocher for her assistance in the preparation of this manuscript.
This work was supported by grants from INCa (Institut National du Cancer), Cancéropôle Grand Ouest, Université de Poitiers, Université Paris-Sud 11, Inserm; from the association Laurette Fugain (Project ALF 09-04); and a supplementary contribution from Novartis Pharma SAS France.
Authorship
Contribution: J.-C.C., A.B.-G., F.G., and A.G.T designed the study; M.-L.B., A.B., M.-C.M., M.M., S.F., and N.S. performed experiments; F.G. collected clinical data; and J.-C.C. and A.G.T analyzed the data and wrote the manuscript. All authors read and approved the manuscript in its final version.
Conflict-of-interest disclosure: F.G. has received consulting fees and research funding from Novartis Oncology and Bristol Myers Squibb. A.G.T has received lecture fees and research funding from Novartis Oncology and Bristol Myers Squibb. The remaining authors declare no competing financial interests.
Correspondence: Ali G. Turhan, Service d'Hématologie et Oncologie Biologique – Inserm U935, CHU de Poitiers, 2 rue de la Milétrie, 86021 Poitiers Cedex, France; e-mail: a.turhan@chu-poitiers.fr.
References
Author notes
A.B.-G. and F.G. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal