Abstract
The differentiation of natural killer (NK) cells and a subpopulation of NK cells which requires an intact thymus, that is, thymic NK cells, is poorly understood. Previous in vitro studies indicate that double negative (CD4−CD8−, DN) thymocytes can develop into cells with NK cell markers, but these cells have not been well characterized. Herein, we generated and characterized NK cells differentiating from thymic DN precursors. Sorted DN1 (CD44+CD25−) CD122−NK1.1− thymocytes from Rag1−/− mice were adoptively transferred into Rag1−/−Ly5.1 congenic mice. After intrathymic injection, donor-derived cells phenotypically resembling thymic NK cells were found. To further study their differentiation, we seeded sorted DN1 CD122−NK1.1− thymocytes on irradiated OP9 bone marrow stromal cells with IL-15, IL-7, Flt3L, and stem cell factor. NK1.1+ cells emerged after 7 days. In vitro differentiated NK cells acquired markers associated with immature bone marrow–derived NK cells, but also expressed CD127, which is typically found on thymic NK cells. Furthermore, we found that in vitro cells generated from thymic precursors secreted cytokines when stimulated and degranulated on target exposure. Together, these data indicate that functional thymic NK cells can develop from a DN1 progenitor cell population.
Introduction
NK cells are critical innate immune components that do not require prior sensitization to be activated on exposure to tumor or infected cells.1 On activation, NK cells lyse target cells through degranulation and also produce cytokines such as IFNγ and TNFα.2 While there has been much progress in defining the specificity and function of NK cells, their differentiation process has not been fully elucidated.
NK cells are generally thought to be bone marrow (BM)–derived, and differentiate through the sequential acquisition of markers and functional receptors.3,4 Unlike other lymphocytes, NK cells do not require somatic gene rearrangements for expression of their receptors, and can be found normally in mice defective in antigen receptor gene rearrangement, such as Rag1−/− mice. Current models suggest that NK cell development can be divided into several major steps.5-7 First, hematopoietic stem cells (HSCs) commit to the NK lineage, becoming natural killer cell precursors (NKPs). Second, the NKPs acquire receptors and molecules involved in target detection, making them immature NK cells (iNKs). Finally, the iNKs terminally differentiate into mature NK cells (mNK) that have appropriate effector functions. Thus, NK cells undergo several developmental steps to become phenotypically and functionally mature.3,4
In greater detail, the earliest step of murine NK cell differentiation involves the commitment of HSCs to the lymphoid lineage and become early lymphoid progenitors (ELP) (Lin−cKithighSca1+Flt3+).5 Further differentiation generates common lymphoid progenitors (CLP; Lin−cKitlowSca1lowIL7Rα+).6 The acquisition of CD122 (IL2/IL15Rβ) defines the transition from a CLP to a committed NKP.7 The maturation of NKP to iNK and later mNK cells can be further characterized by several putative intermediate stages, based on marker expression.8 Expression of CD122 on NKPs marks the beginning of intermediate stage I while NK1.1, CD94/NKG2 and NKG2D expression defines stage II. During stages III and IV, immature NK cells acquire CD117, Ly49 receptors, and integrin α2 (DX5). At stage IV, NK cells undergo expansion. After proliferation, iNK cells become mNK cells characterized by high levels of CD11b (Mac1) and CD43 and acquire the capacity to kill targets and produce IFNγ. However, it is important to note that these putative developmental stages are based on correlating marker expression in vivo and developmental progression has not been directly observed.
While conventional splenic NK cells appear to differentiate in this manner within the BM, CLPs also have the ability to become bipotential T/NK progenitors (T/NKP) and are found in various fetal organs.9-14 The T/NKP found in fetal thymus can generate either TCRαβ T cells or NK cells when transferred into a thymic environment.15 In addition, a small thymic DN1 (CD4−CD8−CD44+CD25−, double negative 1) population in adult mice also harbors T/NK potential,16-18 although these cells were not further characterized. Later studies described an unusual, phenotypically distinct thymic NK cell.19 These cells were CD127+CD69highLy49lowCD11blow, failed to lyse target cells as well as their splenic counterparts, yet were more efficient at producing cytokines. These cells are absent in athymic nude mice, indicating that a functional thymus is required for their development. However, their relationship to previously described T/NKP cells has not been elucidated. Furthermore, it is not known if their progenitors seed the thymus as cells already expressing NK cell markers or if they differentiate from uncommitted thymic precursors. Improved understanding of mouse thymic NK cell development should aid knowledge of human CD56bright NK cells which resemble murine thymic NK cells.19,20
Meanwhile, NK cells can be generated in vitro from BM ELPs with stem cell factor (SCF), IL-7, Flt3L, and IL-15,21-23 and require direct contact with BM stromal cells, such as OP9 cells, to acquire a mature phenotype.22,24 However, the stages through which NK cells differentiate in vitro were not described. In addition, DN1 thymocytes cultured with OP9 cells develop into B and NK1.1+ cells, whereas T cells develop when exposed to OP9 cells transfected with a Notch receptor-ligand.16,17 Recent in vivo experiments further showed that thymic NK cells do not express Rag2 and do not rearrange TCRγ locus, suggesting that thymic NK cells are not derived from a committed T cell progenitor.25 In either case, whether these NK1.1+ cells were functional is unknown. Therefore, prior studies provide a framework for considering NK cell differentiation, but further analysis is required to understand the acquisition of markers during development and the relationship of DN1 cells to thymic and conventional NK cells.
In this study, we addressed the hypothesis that DN1 thymocytes contain precursors with the potential to develop into NK cells. We isolated DN1 CD122−NK1.1− thymocytes from Rag1−/− mice and performed in vivo adoptive transfer and in vitro culture studies. Taken together, our data suggest that thymic NK cells differentiate from thymocyte precursors.
Methods
Mice
B6.129S7-Rag1tm1Mom/J (Rag1−/−) Ly5.2 were purchased from The Jackson Laboratory. Rag1−/− Ly5.1 congenic mice on the C57BL/6 background were generated by crossing B6.SJL-Ptprca Pep3b/BoyJ (Ly5.1) mice (The Jackson Laboratory) with Rag1−/− mice. All mice were studied at 4-16 weeks of age, housed in specific pathogen free conditions, and used in accordance with the animal protocol approved by the Animal Studies committee at Washington University.
Antibodies, flow cytometry, and cytokines
Antibodies purchased from BD Biosciences: anti-CD44 (clone IM7); anti-CD25 (PC61); anti-NK1.1 (PK136); anti-CD122 (TM-β1); anti-CD45.1 (Ly5.1, clone A20); anti-CD45.2 (Ly5.2, clone 104); anti-CD43 (Ly48); anti-CD117 (2B8); anti-CD62L (MEL-14); anti-B220 (RA3-62); and streptavidin PerCP-Cy5.5 (SA-PerCP-Cy5.5). Antibodies purchased from eBioscience: anti-Ly49A/D (12A8); anti-Ly49C/I/F/H (14B11); anti-CD49 (DX5); anti-CD11b (M1/70); anti-NKG2D (CX5); anti-CD127 (A7R34); anti-CD69 (H1.2F3); anti-CD94 (18d3); anti-CD16/32 (clone 93); anti-CD103 (2E7); CD86 (GL1); anti-CD45.1 (A20); SA-PerCP-Cy5.5; and anti-CD107a (eBio1D4B). Surface staining was performed on ice in staining buffer (3% FBS, 0.1% NaN3 in PBS). Nonspecific antibody binding was blocked with 2.4G2 (anti-FcγRII/III) from American Type Culture Collection (ATCC). Samples were collected on a FACSCanto (BD Biosciences) using FACSDiva software (BD Biosciences), and data were analyzed using FlowJo Version 6.4.7 software for Macintox (TreeStar Inc). IL-15, IL-7, Flt3L, and IL-12 were purchased from PeproTech; we used SCF (Fitzgerald Industries) and IL-18 (R&D Systems).
Cell sorting
DN1 CD122−NK1.1− cells were sorted from Rag1−/− thymic single cell suspensions stained with FITC-conjugated anti-CD44, phycoerythrin (PE)-conjugated anti-CD25, allophycocyanin (APC)–conjugated anti-NK1.1, biotinylated anti-CD122 and SA-PerCP-Cy5.5. Pilot studies showed that Rag1−/− thymus contains only DN cells so routine CD4 and CD8 staining was not required. Cells were stained in MTHc buffer which contained MTH (308 mOsm solution of Hanks Balanced Salt Solution, 1M HEPES), 5% FBS and 0.5% 1mg/mL DNase I stock solution. Cells were sorted in MTHs buffer which contained MTH, 0.5% FBS and 0.5% 1 mg/mL DNase I stock solution. Thymic and splenic NK cells were enriched by generation of single cell suspensions from Rag1−/− mice and stained with APC-conjugated anti-NK1.1 in sorting buffer (1% FBS, 0.2% NaN3 in PBS). All cells were sorted by flow cytometry to > 98% purity on a Dako MoFlo (Beckman Coulter) in the Siteman Cancer Center Flow Cytometry Core.
Adoptive transfer
DN1 CD122−NK1.1− cells were sorted from 50-80 Rag1−/−Ly5.2 mice and washed several times with PBS to avoid contaminating recipient mice with remaining antibodies or FBS. Approximately 2 × 105 cells were injected into age- and sex-matched sublethally γ-irradiated (700cGy) Rag1−/−Ly5.1 congenic mice, either via lateral tail vein (intravenously) or intrathymically (IT). IT recipient mice were given buprenorphine hydorchloride for pain every 24 hours for 72 hours and sulfamethoxazole and trimethoprim antibiotics for the duration of the experiment.
Coculture and resting cells
OP9 stromal cells (ATCC) were resuspended in fresh OP9 media (DMEM, 20% FBS and 1% HEPES), γ-irradiated at 2500 rad, seeded at 1.5 × 104 cell per well in 96-well U-bottom plates and incubated for 20-24 hours at 37°C in 5% CO2. DN1 CD122−NK1.1− cells were sorted from 10-20 Rag1−/−Ly5.2 mice and seeded in wells containing the OP9 monolayer in R10 media (RPMI, 10% FBS, l-glutamine and pen/strep) supplemented with IL-7 (5 ng/mL), Flt3L (5 ng/mL), SCF (5 ng/mL), and IL-15 (20 ng/mL). Cells received fresh media every 3 days and cultured from 4-22 days. To rest in vitro generated NK cells, we pooled wells after 19 days, washed cells twice with PBS, resuspended them in R10 with low dose IL-15 (10 ng/mL) and seeded at 1-2 × 106 cells per well in 12 well plates for 36 hours. To rest freshly isolated and sorted splenic and thymic NK cells, we cultured them in R10 supplemented with low dose IL-15 for 16-20 hours. These cells were not cultured longer because of lower yield at later time points. There was no change in phenotypic markers or activation during this incubation period. All cells were incubated at 37°C in 5% CO2.
In vitro stimulation assays, intracellular cytokine staining and cytometric bead array
Briefly, 1 × 105 cells were cultured in either a 96-well plate with R10 media alone or R10 media with IL-12 (10 ng/mL) and IL-18 (50 ng/mL). Cells were incubated at 37°C and 5% CO2 for 1 hour then with brefeldin A (GolgiPlug; BD Biosciences) for another 7 hours. Cells were fixed and permeabilized (Cytofix/Cytoperm; BD Biosciences) and IFNγ was detected by intracellular cytokine staining and flow cytometry as described previously.8 For cytometric bead arrays, cells were stimulated for 8 hours without brefeldin A. Then we collected supernatants and followed the manufacturer's protocol for the Mouse Inflammation Kit (BD Biosciences). Cytokine secretion was measured with the FACSCalibur using CellQuest Pro Version 4.0 (Mac; BD Biosciences) and BD CBA Version 1.4 (Mac) software.
Degranulation assay
Cells were cocultured with YAC-1 target cells at various ratios in 96-well V-bottom plates. Anti-CD107a antibody and monensin (eBioscience) were added to each well. Plates were incubated for 2 hours at 37°C, after which surface staining for flow cytometry was performed.
Results
In vivo differentiation of NK1.1+ cells from thymic DN1 progenitors
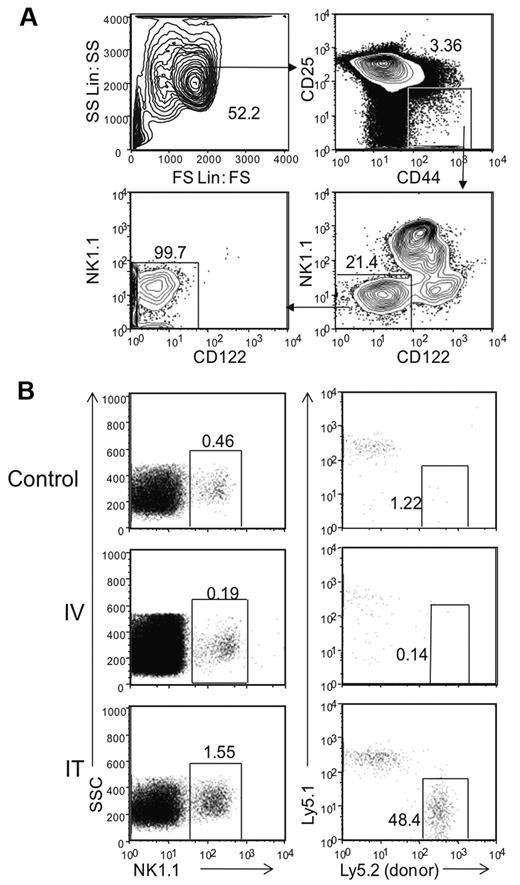
We enriched potential progenitors by FACS sorting a subset of DN1 thymocytes based on expression of CD44 and low or absent CD25 from Rag1−/− mice (Figure 1A). Presort analysis showed that < 4% of thymocytes were in the DN1 (CD44+CD25−) stage of development as most were arrested in the DN3 stage (CD44−CD25+). While other studies have shown that DN1 can be further subdivided by the expression of CD117,17 we did not find this initially feasible because < 1.5% of DN1 cells in Rag1−/− mice express this marker. In addition, within the DN1 population, we found CD122−NK1.1−, CD122+NK1.1−, and NK1.1+CD122+ cells, constituting ∼ 21%, 11%, and 65% of the cells, respectively. To study the NK cell developmental potential of DN1 cells, we further enriched the DN1 population by collecting only CD122−NK1.1− cells which represent cells that are not already committed to the NK cell lineage, as determined for developing NK cells in BM7 (Figure 1A and data not shown). Postsort analysis showed the DN1 CD122−NK1.1− thymic population had a 99% purity level, indicating there were little to no contaminating mature NK cells.
Donor derived DN1 CD122−NK1.1− thymocytes differentiate into NK cells in the thymus. (A) Thymic DN1 CD122−NK1.1− cells from Rag1−/− mice were sorted and collected for post sort analysis with the gating parameters shown. (B) Thymi from Rag1−/− Ly5.2 mice were harvested and sorted based on DN1CD122−NK1.1− phenotype as shown in panel A. Cells were transferred into irradiated Rag1−/− Ly5.1 mice either intravenously or intrathymically. Irradiated littermates were used as controls. Cells were harvested from the thymus 32 days after transfer. NK1.1+ cells from the lymphocyte population were then examined for host (Ly5.1+) and donor (Ly5.2+) cells. Data are representative of 2 experiments.
Donor derived DN1 CD122−NK1.1− thymocytes differentiate into NK cells in the thymus. (A) Thymic DN1 CD122−NK1.1− cells from Rag1−/− mice were sorted and collected for post sort analysis with the gating parameters shown. (B) Thymi from Rag1−/− Ly5.2 mice were harvested and sorted based on DN1CD122−NK1.1− phenotype as shown in panel A. Cells were transferred into irradiated Rag1−/− Ly5.1 mice either intravenously or intrathymically. Irradiated littermates were used as controls. Cells were harvested from the thymus 32 days after transfer. NK1.1+ cells from the lymphocyte population were then examined for host (Ly5.1+) and donor (Ly5.2+) cells. Data are representative of 2 experiments.
To assess whether this population had the potential to generate NK1.1+ cells, we adoptively transferred sorted DN1 CD122−NK1.1− cells from Rag1−/−Ly5.2 mice into sub-lethally irradiated Rag1−/−Ly5.1 recipients either intravenously or intrathymically. Thirty-two days after transfer, we analyzed recipient thymus for donor-derived NK1.1+ cells (Figure 1B). In mice receiving intravenously transferred cells, the thymus was completely devoid of donor-derived NK1.1+ cells. However, donor-derived cells were present in mice receiving IT transferred cells, where ∼ 50% of the thymic NK cells were derived from Ly5.2+ donor cells. Repeated experiments with fewer (4 × 103 to 40 × 103) transferred DN1 CD122−NK1.1− cells or transfer into nonirradiated hosts did not yield enough detectable donor-derived NK1.1+ cells for analysis (data not shown). Nonetheless, these results suggest that, on intrathymic adoptive transfer, the DN1 CD122−NK1.1− thymic subpopulation can differentiate into NK1.1+ cells in the thymus.
In vivo differentiation of thymic DN1 cells generates nonconventional NK cells
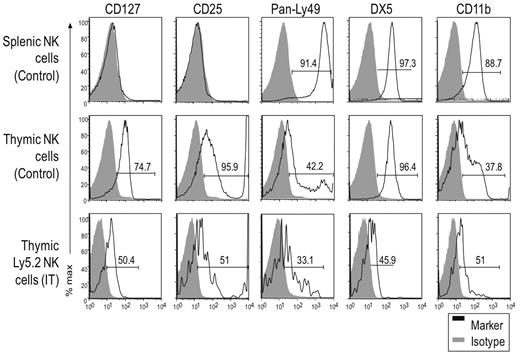
While the number of donor cells in recipient mice receiving IV transferred cells was too few for analysis, we could further characterize the NK1.1+ cells generated by IT transfer by analyzing informative markers associated with thymic and splenic NK cells (Figure 2). We found that, unlike control splenic NK cells, thymic NK cells from unmanipulated mice uniformly expressed the thymic marker CD127, as did IT transferred donor (Ly5.2+) cells, albeit at somewhat lower levels for unclear reasons. In addition, CD25 was selectively expressed by thymic NK cells in control Rag1−/− mice, and also by donor-derived IT cells. This was a surprising finding because previous studies did not note CD25 expression on thymic NK cells in C57BL/6 mice.19,26 This expression may be related to our use of Rag1−/− mice because we also did not find CD25 expressed on thymic NK cells from C57BL/6 mice (data not shown). Regardless, unmanipulated splenic NK1.1+ cells did not express CD25, while thymic NK1.1+ cells from both unmanipulated and IT mice had CD25low and CD25high populations, the latter usually falling outside the range of detection. The CD25high staining was unlikely due to nonspecific Fc receptor staining or dead cell contamination because splenic Rag1−/− cells which contain abundant FcR-bearing cells did not stain and there were few dead cells in our preparations. Because of the low yield of Ly5.2+ NK cells in the IT mice, we were unable to assess individual Ly49 expression; Ly49 antibodies with broader specificities were used to detect Ly49C/I/F/H and Ly49A/D. A sizable fraction of donor Ly5.2+ NK cells were Ly49low, similar to thymic NK cells from unmanipulated Rag1−/− mice, whereas a smaller fraction expressed Ly49 receptors at higher levels. However, essentially all splenic NK cells from control mice were Ly49high. Finally, DX5 and CD11b were detected on donor-derived cells, although their expression level was lower than on unmanipulated thymic NK cells. Taken together, particularly based on differential staining of CD127, CD25, and Ly49 receptors on thymic versus splenic NK cells, these data suggest that IT transferred DN1 CD122−NK1.1− cells can differentiate into cells that resemble thymic NK cells.
DN1 CD122−NK1.1− thymocytes differentiate into NK1.1+ cells with a unique phenotype. DN1 CD122−NK1.1− thymocytes from Rag1−/− Ly5.2 mice were transferred into irradiated Rag1−/− Ly5.1 mice intrathymically. Donor (Ly5.2+) NK cells from IT mice were compared with NK cells from unmanipulated (nonirradiated) littermates 32 days after transfer. Gray-filled histograms represent cells stained with an isotype control and black-line histograms represent cells stained with the indicated antibody. Although the staining patterns for each marker were somewhat different, even for the same marker on different cell populations, ie, some markers were expressed on all cells while others were expressed on subsets, we used the percent of positive cells (above isotype control staining) as a convenient (although technically imprecise) means to compare and describe the staining profiles for a large number of markers. Data are representative of 2 experiments.
DN1 CD122−NK1.1− thymocytes differentiate into NK1.1+ cells with a unique phenotype. DN1 CD122−NK1.1− thymocytes from Rag1−/− Ly5.2 mice were transferred into irradiated Rag1−/− Ly5.1 mice intrathymically. Donor (Ly5.2+) NK cells from IT mice were compared with NK cells from unmanipulated (nonirradiated) littermates 32 days after transfer. Gray-filled histograms represent cells stained with an isotype control and black-line histograms represent cells stained with the indicated antibody. Although the staining patterns for each marker were somewhat different, even for the same marker on different cell populations, ie, some markers were expressed on all cells while others were expressed on subsets, we used the percent of positive cells (above isotype control staining) as a convenient (although technically imprecise) means to compare and describe the staining profiles for a large number of markers. Data are representative of 2 experiments.
In vitro differentiation of NK1.1+ cells from thymic DN1 CD122−NK1.1− progenitors
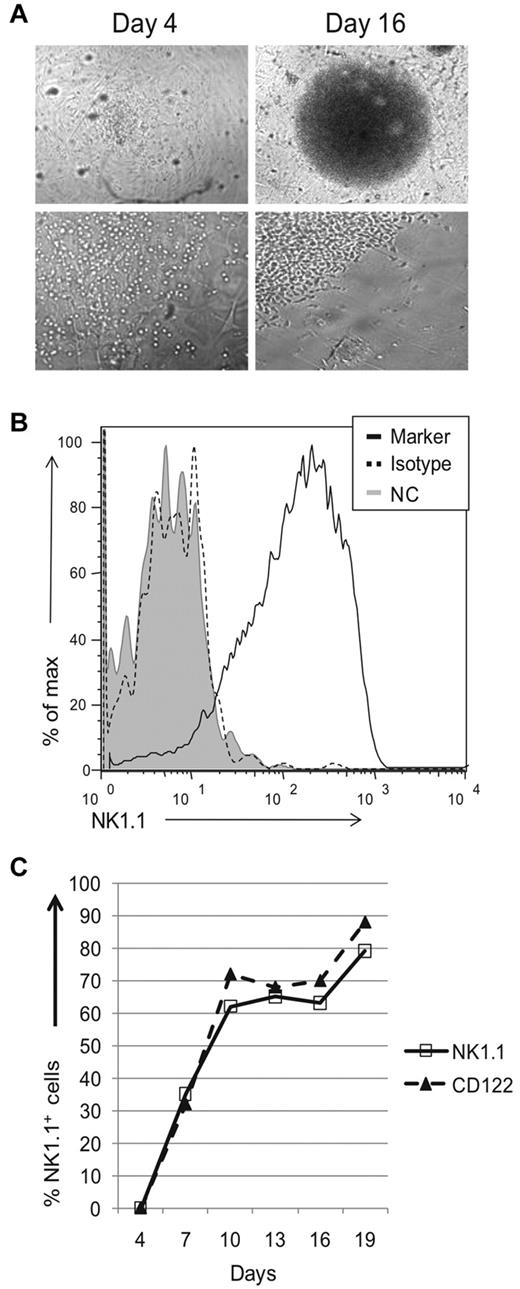
Because of low donor-derived NK cell numbers, we were unable to perform more detailed experiments with adoptive transfer experiments. Instead we used an in vitro system in which thymic progenitors were cocultured with stromal cells in the presence of cytokines. Sorted DN1 CD122−NK1.1− thymocytes from Rag1−/− mice were seeded onto a confluent monolayer of irradiated OP9 stromal cells and cultured in IL7, Flt3L, SCF, and IL15. After 10 days of culture, we visualized a colony of cells at the center of most wells. During early stages of development, the progenitors made contact with stromal cells, but by day 16 cells appeared in areas devoid of OP9 cells (Figure 3A and data not shown). To assess whether these cells had differentiated, we pooled growth-positive wells identified by visual inspection and analyzed surface expression of NK1.1 via flow cytometry. By day 16, most cells were NK1.1+ (Figure 3B). Kinetic analysis revealed no NK1.1+ cells on day 4 but NK1.1+ cells were readily detected on day 7, and continued to increase on day 10 (Figure 3C). Thereafter, the percentage of NK1.1+ cells was relatively stable. CD122 expression paralleled that of NK1.1 expression. OP9 cells and all cytokines were necessary for growth of NK1.1+ wells, because few to no NK1.1+ wells were obtained if any component was excluded (data not shown). Together, these results suggest that DN1 CD122−NK1.1− thymocytes contain progenitors capable of developing into NK1.1+ cells in vitro with OP9 stromal cells and cytokines.
Thymic progenitors can differentiate into NK 1.1+ cells in vitro. Sorted DN1 CD122−NK1.1− thymocytes from Rag1−/− mice were cocultured with OP9 cells and cytokines. (A) Wells were visually and microscopically examined for growth on different days. Images were acquired using a Nikon Diaphot 200 microscope (Nikon) with a Hamamatsu digital camera (Hamamatsu Photonic) and processed using MetaVue imaging software (Molecular Devices Corp). Top panels are at 4× magnification, while bottom panels are at 20× magnification. (B) Growth positive wells by visual inspection were pooled, stained, and analyzed for expression of NK1.1. For the negative controls, cells were either left unstained (gray-filled histograms) or stained using the appropriate isotype antibody (black dotted histogram). Cells were gated based on lymphocyte population by scatter parameters. Data are representative of at least 3 experiments. (C) Kinetic analysis of NK1.1 and CD122 expression on lymphocyte population, gated by scatter parameters. At various culture periods, wells were examined for the indicated markers as described in panel B. Data are representative of 3-5 experiments
Thymic progenitors can differentiate into NK 1.1+ cells in vitro. Sorted DN1 CD122−NK1.1− thymocytes from Rag1−/− mice were cocultured with OP9 cells and cytokines. (A) Wells were visually and microscopically examined for growth on different days. Images were acquired using a Nikon Diaphot 200 microscope (Nikon) with a Hamamatsu digital camera (Hamamatsu Photonic) and processed using MetaVue imaging software (Molecular Devices Corp). Top panels are at 4× magnification, while bottom panels are at 20× magnification. (B) Growth positive wells by visual inspection were pooled, stained, and analyzed for expression of NK1.1. For the negative controls, cells were either left unstained (gray-filled histograms) or stained using the appropriate isotype antibody (black dotted histogram). Cells were gated based on lymphocyte population by scatter parameters. Data are representative of at least 3 experiments. (C) Kinetic analysis of NK1.1 and CD122 expression on lymphocyte population, gated by scatter parameters. At various culture periods, wells were examined for the indicated markers as described in panel B. Data are representative of 3-5 experiments
In vitro generated NK1.1+ cells have a unique phenotype
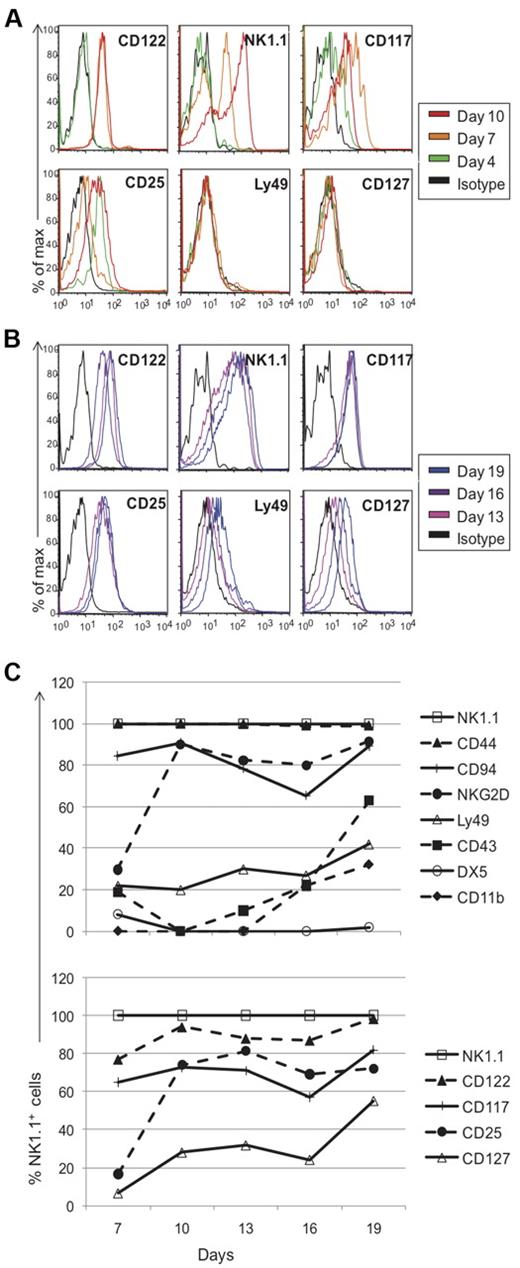
To directly follow the kinetics of NK-cell development from progenitor stage to NK1.1-expressing cells, we performed experiments informed by a limiting dilution analysis (supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Because of the small number of cells available, we used a seeding concentration of 1000 cells per well to insure that each well will have differentiating NK1.1+ cells (from an average of 1-2 progenitors) and chose to pool wells for analysis. After 4 days of culture, most markers were absent, including CD122 and NK1.1 (Figures 3C, 4A), except CD117 and CD25 which were expressed at low levels (Figure 4A). While ∼ 3% of sorted DN1 CD122−NK1.1− cells expressed CD117 before culture, none expressed CD25 (Figure 1A and data not shown). By day 7, all cells expressed CD122 and > 30% were NK1.1+, with NK1.1+ cells increasing to > 85% by day 19 (Figures 3C, 4B). Taken together, these data strongly suggest that the cells were differentiating into NK1.1+ cells rather than representing outgrowth of contaminating mature NK cells because NK1.1 was not initially expressed and markers associated with more mature NK cells were primarily expressed only after 1 week.
Phenotypic profile of cells generated in vitro from thymic progenitors. Cells cultured in vitro were pooled on different days and assessed for marker expression via flow cytometry. Black histograms represent cells stained with appropriate isotype controls and colored-line histograms correspond to different days of analysis. Early (A) and late (B) marker acquisition is shown. Expression on gated NK1.1+ cells is shown, except for all markers on day 4 and NK1.1 expression on different days, which were gated on the lymphocyte population. (C) Developmental kinetics of sorted thymocytes as a function of days versus the percentage of NK1.1+ cells expressing the given marker. Again, as described for Figure 2, the staining patterns for each marker were somewhat different but we used the percent of positive cells as a convenient means to compare and describe the staining profiles for numerous antibodies used. Data are representative of 3-5 experiments.
Phenotypic profile of cells generated in vitro from thymic progenitors. Cells cultured in vitro were pooled on different days and assessed for marker expression via flow cytometry. Black histograms represent cells stained with appropriate isotype controls and colored-line histograms correspond to different days of analysis. Early (A) and late (B) marker acquisition is shown. Expression on gated NK1.1+ cells is shown, except for all markers on day 4 and NK1.1 expression on different days, which were gated on the lymphocyte population. (C) Developmental kinetics of sorted thymocytes as a function of days versus the percentage of NK1.1+ cells expressing the given marker. Again, as described for Figure 2, the staining patterns for each marker were somewhat different but we used the percent of positive cells as a convenient means to compare and describe the staining profiles for numerous antibodies used. Data are representative of 3-5 experiments.
To further analyze the kinetics of marker expression, we compared markers on NK1.1+ cells from different culture time points to each other, expressed as a percentage of NK1.1+ cells (Figure 4C). Both CD122 and NK1.1 were expressed relatively early with ready detection on day 7 (Figure 4A). Interestingly, > 99% of cells were CD44+ throughout the entire culture time (Figure 4C). Developing cells failed to express CD94 on day 4, but > 80% of NK1.1+ cells expressed it by day 7. By contrast, NKG2D was poorly expressed on day 7, and were expressed by essentially all NK1.1+ cells on day 10 (Figure 4C), supporting the previously described acquisition of CD94 then NKG2D expression on developing NK cells after NK1.1 and CD122 are expressed.3,8,27 With the subsequent expression of Ly49 on day 13, it appeared that the in vitro generated NK1.1+ cells had progressed from stage II of development to stage III during days 10-13. Limited by the number of available cells, we were unable to assess specific Ly49 expression with mono-specific anti-Ly49 antibodies. Nonetheless, only a fraction of the cells expressed Ly49s. However, unlike conventional splenic NK cells, the in vitro generated cells lacked DX5 expression, and CD43 and CD11b were detected only after approximately 16 days in culture. Finally, low levels of CD127 was detected on the in vitro cultured cells, although predominantly late in culture period. While 10%-25% of the cells were CD127low on day 10, there were > 50% NK1.1+CD127+ cells by day 19 (Figure 4). Overall, these studies allowed us to directly follow the sequential development of NK1.1+ cells from DN1 thymocytes, which generally recapitulates developmental expression of most markers on developing conventional NK cells in the BM as previously determined by correlation analysis.
Comparison between in vitro generated cells and freshly isolated NK cells
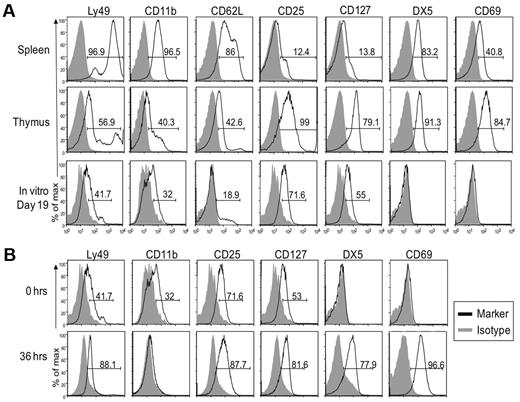
Our results suggested that in vitro generated NK1.1+ cells resembled thymic NK cells, as also suggested by our adoptive transfer studies. Here, we compared expression of surface markers on in vitro derived NK cells after 19 days in culture with that of freshly isolated thymic and splenic NK cells (Figure 5A). Similar to thymic CD127+ NK cells, less than half of the in vitro generated cells had low expression of Ly49 receptors, CD11b, and CD62L. Further detailed analysis of the Ly49 receptors confirmed these data because splenic NK cells readily expressed Ly49A, Ly49C, Ly49D, Ly49F, Ly49G2/I, and Ly49H, some of which are expressed on > 50% of cells, while < 20% of thymic NK cells expressed any one of these receptors (data not shown). In vitro generated cells lacked Ly49G2/I and < 10% expressed low levels of Ly49A, Ly49D, Ly49F, or Ly49H (data not shown). Kinetic analysis showed that CD25 was up-regulated during the first 4 days of culture and expression was then constant through day 19 (Figure 4). Surprisingly, as noted in Figure 2, CD25 was also detected in thymic NK cells from Rag1−/− mice with essentially all cells expressing it at intermediate to high levels. Most importantly, in vitro generated NK cells uniformly expressed CD127, a unique marker of thymic NK cells,19 albeit at lower levels. Overall, these phenotypic comparisons suggest that in vitro generated NK1.1+ cells more closely resemble thymic, and not conventional splenic NK cells.
In vitro developed NK cells resemble those found in the thymus and not conventional splenic NK cells. Cells from indicated tissues of Rag1−/− mice, along with in vitro differentiated NK cells were analyzed by cytometry. Gray-filled histograms represent cells stained with an isotype control and black line histograms represent cells stained with the indicated antibody. (A) In vitro generated NK cells were compared with freshly isolated splenic and thymic NK cells. Gated NK1.1+ cells are shown. (B) The phenotype of in vitro generated NK cells in culture for 19 days (top panel) was compared with that of cells that were removed from culture after 19 days and rested in LD IL15 for 36 hours (bottom panel). Gated NK1.1+ cells are shown. As described for Figure 2 and here, the staining patterns for each marker were somewhat different but we used the percentage of positive cells as a convenient means to compare and describe the staining profiles for numerous antibodies used. Data are representative of 2-5 experiments.
In vitro developed NK cells resemble those found in the thymus and not conventional splenic NK cells. Cells from indicated tissues of Rag1−/− mice, along with in vitro differentiated NK cells were analyzed by cytometry. Gray-filled histograms represent cells stained with an isotype control and black line histograms represent cells stained with the indicated antibody. (A) In vitro generated NK cells were compared with freshly isolated splenic and thymic NK cells. Gated NK1.1+ cells are shown. (B) The phenotype of in vitro generated NK cells in culture for 19 days (top panel) was compared with that of cells that were removed from culture after 19 days and rested in LD IL15 for 36 hours (bottom panel). Gated NK1.1+ cells are shown. As described for Figure 2 and here, the staining patterns for each marker were somewhat different but we used the percentage of positive cells as a convenient means to compare and describe the staining profiles for numerous antibodies used. Data are representative of 2-5 experiments.
Unlike thymic NK cells, the in vitro NK1.1+ cells did not express the integrin DX5 or CD69. However, a slight modification in the culture protocol was informative. After 19 days, in vitro generated NK1.1+ cells were harvested, washed, and incubated in low-dose (LD) IL15 alone for 36 hours. Results showed the cells up-regulated DX5 and CD69 and down-regulated CD11b (Figure 5B), resulting in a phenotype strikingly similar to thymic NK cells (Figure 5A).
In vitro generated cells are functional
We further sought to characterize the in vitro generated NK1.1+ cells by comparing their function to thymic NK cells. For these studies, we used in vitro differentiated NK cells cultured under standard conditions with OP9 cells and cytokines for 19 days. At this point, the cells were secreting several cytokines and chemokines, including monocyte chemotactic protein-1 (MCP-1), IFNγ, TNF, IL-6, and IL-10, without further stimulation (data not shown). However, after washing and culturing the cells in LD IL-15 alone, we found that the unstimulated cells no longer secreted MCP-1 or the other cytokines (Figure 6B). Despite up-regulation of CD69 (Figure 5B), a classic activation marker, these data suggested that the in vitro differentiated NK cells functionally achieved a “resting” state when cultured in LD IL15 alone.
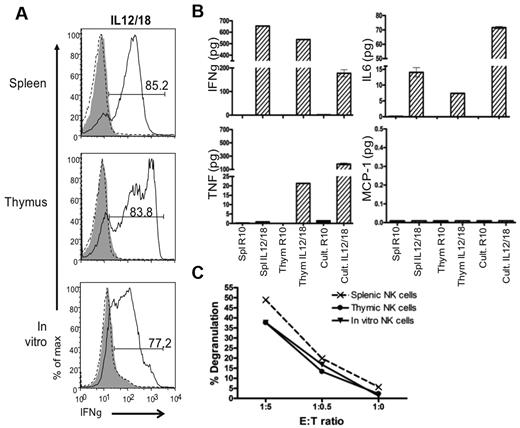
In vitro generated NK cells are functional. The function of in vitro differentiated NK cells grown in culture (Cult) for 19 days then rested in LD IL15 was compared with the function of Rag1−/− splenic and thymic NK cells cultured in LD IL15. Sorted NK1.1+ cells were stimulated with IL-12/IL-18 or target YAC-1 cells. (A) IFNγ production was measured by intracellular flow cytometry. For negative controls, cells were either not stained (gray-filled histograms) or not stimulated (black-dotted histograms). All histograms are gated on NK1.1+ cells. (B) Cytokines secreted were assessed by cytometric bead array. (C) CD107 degranulation on target encounter was measured by flow cytometry. Data are representative of 3 experiments.
In vitro generated NK cells are functional. The function of in vitro differentiated NK cells grown in culture (Cult) for 19 days then rested in LD IL15 was compared with the function of Rag1−/− splenic and thymic NK cells cultured in LD IL15. Sorted NK1.1+ cells were stimulated with IL-12/IL-18 or target YAC-1 cells. (A) IFNγ production was measured by intracellular flow cytometry. For negative controls, cells were either not stained (gray-filled histograms) or not stimulated (black-dotted histograms). All histograms are gated on NK1.1+ cells. (B) Cytokines secreted were assessed by cytometric bead array. (C) CD107 degranulation on target encounter was measured by flow cytometry. Data are representative of 3 experiments.
To assess whether the “rested” in vitro generated NK cells could be activated, we performed a target cell-free stimulation assay with IL-12 plus IL-18 for 8 hours (Figure 6A). The majority of these cells produced a large amount of IFNγ. Similar results were obtained with splenic and thymic NK cells also pre-cultured in LD IL15 (rested). Taken together, these data indicate that the in vitro generated NK cells can be stimulated to produce IFNγ.
To more broadly compare the function of in vitro differentiated NK cells to thymic and splenic NK cells, we used a cytometric bead array (CBA). As expected from the intracellular staining assay, rested splenic and thymic cells and in vitro differentiated cells secreted a large amount of IFNγ when stimulated with IL-12/IL-18. Interestingly, TNFα was produced by thymic and in vitro generated NK cells on stimulation by IL-12/IL-18, whereas splenic NK cells failed to produced this cytokine. IL-6 was detected after IL-12/IL-18 stimulation but MCP-1 and IL-10 were not detected at any point (Figure 6B and data not shown). These results indicate that the in vitro generated NK cells are comparable with thymic NK cells in cytokine production.
Finally, to determine whether the in vitro generated NK cells were capable of responding to target cells, rested in vitro generated and rested splenic and thymic NK cells were coincubated with YAC-1 target cells. In a similar manner, in vitro differentiated, thymic, and splenic NK1.1+ cells degranulated on target encounter (Figure 6C). Overall, the phenotypic and functional analyses show more similarities between thymic and in vitro generated NK cells, and together they share differences from their splenic counterparts.
Discussion
Here we show that DN1 CD122−NK1.1− thymocytes can differentiate into NK cells. We used highly purified DN1 CD122−NK1.1− cells from Rag1−/− mice that were unlikely to have contaminating mature NK cells that grew out in vitro for several reasons: (1) Purified, congenically marked, DN1 CD122−NK1.1− cells gave rise to cells with NK cell markers when adoptively transferred into sub-lethally irradiated recipients in vivo. (2) During in vitro culture, the cells gradually acquired markers associated with developing NK cell subsets. If they represented outgrowth of mature NK cells, they should have expressed markers characteristic of later developmental stages early in the culture period unlike what we observed. (3) Although NK1.1 and CD122 are among the earliest markers on developing NK cells, they were not expressed at the earliest times in culture, indicating that the cells acquired these markers during differentiation. (4) The number of cells expressing NK cell markers during early times in culture cannot be accounted for by proliferation of a small contaminating pool of mature NK cells which typically have a 24-hour doubling time in vitro. (5) Finally, acquisition of the NK cell markers did not occur unless the cultures had all cytokines and OP9 cells, whereas mature NK cells can proliferate with IL15 alone. Taken together, these considerations strongly support that DN1 CD122−NK1.1− thymocytes can differentiate into cells with NK markers in vivo and in vitro.
Early studies examining the heterogeneity of the thymic DN1 population showed that T cells or NK1.1+ cells could be generated in vitro using either OP9 cells with or without the expression of the Notch ligand Delta-like 1 (OP9-DL1), respectively. These studies showed that after 6 days of DN1/OP9 coculture, NK1.1-expressing cells could be detected.17 A recent study indicates that BM-derived precursors can also give rise to NK1.1+ cells with a similar in vitro system.28 However, in either case, the NK cells generated were not further characterized. Our studies show that these in vitro differentiated cells have a phenotype that differs from conventional splenic NK cells. Moreover, when we modified the in vitro culture conditions and incubated the cells in LD IL15 alone, the cells very closely resembled thymic NK cells. Another interesting point is that CD127 is a relatively late marker on these cells, appearing around day 13 of in vitro differentiation. Thus, the DN1 CD122−NK1.1− cells differentiated in vitro into NK1.1+ cells more closely resembling thymic NK cells.
The differentiation of thymic NK cells in vitro allowed us to directly examine the acquisition of markers on developing NK cells. This acquisition seem to follow the stages proposed previously on the basis of correlating marker expression with each other on immature BM NK cells.8 In such studies, CD94, NKG2D, Ly49, CD43, and CD11b are sequentially up-regulated after CD122 and NK1.1 expression. Regardless of the percentage of cells that expressed each marker, we were able to detect the acquisition and the incremental expression, compared with isotype controls, of the positive population as time progressed. These studies suggest that conventional splenic and thymic NK cells undergo similar stages during development.
Unlike their conventional counterparts in the spleen, little is known about thymic NK cells. While our in vitro differentiated cells more closely resembled thymic NK cells than conventional NK cells, there were some subtle differences between the in vitro differentiated cells and thymic NK cells. For example, it was only after we “rested” the day 19 cultured cells in LD IL-15 alone did we find functional profiles that more closely resembled thymic NK cells from naive animals, perhaps reflecting a dynamically changing in vivo cytokine environment during normal development. In addition, subtle differences in the expression levels of Ly49 molecules in thymic, adoptively transferred, and in vitro generated NK cells may be because of stromal factors that regulate Ly49 expression that are difficult to recapitulate in vitro.23 For example, irradiation may alter stromal molecules or the MHC on OP9 cells could affect Ly49 expression or acquisition. Regardless, the studies described here provide future opportunities to dissect the influence of differing cytokine environments and stromal components on thymic NK cells and their development.
While the majority of splenic NK cells originate and develop in the BM, early studies have shown the existence of a T/NK bipotential progenitor population in the thymus.9,11,12 Recent in vitro differentiation studies suggest a similar T/NK bipotential progenitor population may be present in the BM.28 However, thymic NK cells did not develop on adoptive transfer, unlike our results, suggesting that these BM precursors lack the signals to develop into thymic NK cells. On the other hand, T/NK bipotential precursors are most commonly found within fetal thymus, suggesting that thymic NK cells also may develop in adult thymus.9 Furthermore, previous studies did not characterize the NK1.1+ cells that developed in vitro in the absence of Notch signaling, and thymic NK cells were described subsequently; their relationship to thymic NK cells was not investigated. Here, our adoptive transfer and in vitro differentiation studies strongly suggest that bipotential progenitor populations exist in the adult thymus which can differentiate into thymic NK cells rather than conventional NK cells.
While our studies demonstrate that thymic NK cells can differentiate from DN1 CD122− NK1.1− cells, more work is needed to determine whether a single progenitor cell in the thymus could give rise to a T cell or NK cell on appropriate maturation signals. While our studies suggest that a small population of DN1 thymic progenitors are destined to become thymic NK cells, rather than conventional NK cells, we do not know if they acquire this commitment once they reside in the thymus or if experimental constraints allowed preferential development of thymic NK cells. To address these issues, further dissection of the DN1 progenitor pool will be necessary.
Taking into account that NK cells do not require gene rearrangement for their receptors, unlike T and B cell receptors, we opted to perform all of our experiments using Rag1−/− mice on a C57BL/6 background. During thymic development, RAG genes are first expressed in committed T cell precursors in the DN2 stage and T-cell development is thus blocked after the DN1 stage used in our studies.29,30 While NK cell differentiation as described here should therefore exist in wild type mice, development of DN1 CD122−NK1.1− thymocytes from B6 mice need confirmation in future experiments. Regardless, our studies yielded an unexpected difference with respect to CD25 that we and others did not find expressed on wild type thymic NK cells.19,26 The basis for this difference is not clear at the moment but at minimum, it appears to be another marker selectively expressed on thymic NK cells in Rag1−/− mice. Furthermore, our choice to use Rag1−/− mice was further supported by a recent publication indicating that thymic NK cells are not derived from thymocytes having undergone antigen receptor rearrangement.25 While earlier in vitro studies showed that NK1.1+ cells are generated from CD117+ cells within the DN1 thymic population,17 our studies indicate that in Rag1−/− mice, there is a DN1 CD122+NK1.1+ population that may have skewed our results if not sorted out. Future experiments to enrich for the CD117+ population may be informative but additional markers will still be needed to markedly increase precursor frequency. Nonetheless, the studies reported here show that this approach is feasible.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Howard Petrie (Scripps Research Institute, Florida campus) for initial help with isolation and culture of DN1 cells. They also thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St Louis, MO, for the use of the High Speed Cell Sorter Core, which provided the cell-sorting service. The Siteman Cancer Center is supported in part by NCI Cancer Center Support Grant #P30 CA91842. They also thank Megan Cooper and Takashi Ebihara for their critical review of this manuscript.
This work was supported by the Howard Hughes Medical Institute and grants RO1-AI33903 and R37-AI34385 from the National Institutes of Health. C.L.V. is supported by the Training Program in Immunology and Immunogenetics Grant AI007163.
National Institutes of Health
Authorship
Contribution: C.L.V., J.P.L., and W.M.Y. designed the research; C.L.V. and L.Y. performed in vivo studies; C.L.V. performed in vitro studies; W.M.Y. contributed resources; C.L.V. and W.M.Y. analyzed all results and C.L.V. made the figures; and C.L.V. and W.M.Y. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wayne M. Yokoyama, MD, Rheumatology Division Box 8045, Washington University School of Medicine, 660 S Euclid Ave, St Louis, MO 63110; e-mail: yokoyama@dom.wustl.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal