Abstract
HIV-1 depends on host-cell resources for replication, access to which may be limited to a particular phase of the cell cycle. The HIV-encoded proteins Vpr (viral protein R) and Vif (viral infectivity factor) arrest cells in the G2 phase; however, alteration of other cell-cycle phases has not been reported. We show that Vif drives cells out of G1 and into the S phase. The effect of Vif on the G1-to-S transition is distinct from its effect on G2, because G2 arrest is Cullin5-dependent, whereas the G1-to-S progression is Cullin5-independent. Using mass spectrometry, we identified 2 novel cellular partners of Vif, Brd4 and Cdk9, both of which are known to regulate cell-cycle progression. We confirmed the interaction of Vif and Cdk9 by immunoprecipitation and Western blot, and showed that small interfering RNAs (siRNAs) specific for Cdk9 inhibit the Vif-mediated G1-to-S transition. These data suggest that Vif regulates early cell-cycle progression, with implications for infection and latency.
Introduction
HIV-1 is the retrovirus that causes AIDS. It carries 6 accessory genes (tat, rev, vif, vpr, vpu, and nef), and 3 structural genes (gag, pol, and env).1 Vif (viral infectivity factor) of HIV-1 is a 23-kDa phosphoprotein that is expressed late in the retroviral life cycle. Vif is conserved among all of the primate lentiviruses except the equine infectious anemia virus. It is required for HIV-1 infection in primary CD4 T cells, macrophages, and certain T-cell lines (eg, H9 and CEM).2-6 Viruses lacking a functional vif gene (Δvif) fail to mount a spreading infection in the above “nonpermissive” cell types. In contrast, many “permissive” T-cell lines (eg, Jurkat and CEMss) and nonhematopoietic cell lines (eg, HeLa and 293T) fully support HIV-1 spreading infection in the absence of Vif. Results from heterokaryon analyses, in which permissive and nonpermissive cell lines have been fused, suggest that the nonpermissive cells express a host-restriction factor that inhibits the replication of HIV-1 virus lacking the vif gene.7,8 Using a subtractive hybridization approach for analysis of 2 closely related cell lines differing in permissivity, Sheehy et al demonstrated that this host antiviral restriction factor is APOBEC3G (A3G).9 Vif suppresses the functions of A3G and increases the infectivity of virus produced from infected cells by preventing the packaging of A3G into viral particles. Therefore, Vif plays a vital role in viral replication in primary CD4 T cells both in vitro and in vivo.
HIV-1, like all retroviruses, depends on host-cell resources for replication. Access to those resources may be limited to a particular phase of the cell cycle. In human T cells, infection with HIV-1 causes cell-cycle arrest or delay in the G2 phase of the cell cycle, conferring some advantage to the virus,10 but leading ultimately to cell death.11,12 The only viral protein implicated in G2 arrest of infected cells was Vpr (viral protein R),11,13-17 until we and others demonstrated that a second viral protein, Vif, also induces G2 arrest.18-21 Vpr has been shown to alter the cell cycle by inhibiting the activation of Cdc2/Cdk1, a kinase controlling the G2/M checkpoint, to prevent or delay entry of infected cells into mitosis.13-15 In contrast, how Vif induces G2 arrest is not yet completely known. Previously published data21 and our own observations have shown that Vif does not induce G2 arrest by interacting with its primary known target, A3G; therefore, it must target other cellular protein(s) to mediate cell-cycle alterations during HIV-1 infection.
To identify the possible mechanisms by which Vif induces G2 arrest, and the cellular partner(s) with which Vif interacts to regulate the cell cycle in HIV-1 infection, we synchronized HeLa cells transfected with wild-type Vif (derived from HIV-1 NL4-3 or HXB2) or various Vif mutants at the G1 phase of the cell cycle and analyzed the cell-cycle profiles of transfected cells after release from cell-cycle synchronization. Surprisingly, we found that, in addition to arresting cells at phase G2, Vif drove cells out of G1 and into the S phase. This latter effect, which we reproduced in HIV-1–infected HeLa and CEMss T cells, appeared to be mediated by a different set of cellular interaction partners than those that contribute to Vif-mediated G2 arrest, because G2 arrest is Cullin5-dependent (consistent with a role for proteosomal degradation), while the G1-to-S progression was Cullin5-independent. Using mass spectrometry, we identified 2 cellular proteins, Brd4 and Cdk9, which interact with Vif to induce cell-cycle alterations, specifically to accelerate the transition from G1 to S. The work presented here sheds new light on the role of Vif in regulating the cell cycle during HIV-1 infection.
Methods
Cells and reagents
The human epithelial carcinoma cell line HeLa was maintained in Dulbecco modified Eagle medium (Mediatech) supplemented with 10% fetal bovine serum (Gemini Bioproducts), 100 U/mL of penicillin G + 100 μg/mL of streptomycin (GibcoBRL), and 2mM l-glutamine (Mediatech). CEMss T cells were cultured in 10% fetal bovine serum RPMI 1640 medium. All cultures were maintained at 37°C and 5% CO2.
The plasmid encoding for farnesylated enhanced green fluorescent protein (pEGFP-F) vector (BD Biosciences) was used for all transfection experiments. NL4-3 or NL4-3vif (referred to as NL4-3ΔVif) plasmids were the generous gift of Dr Una O'Doherty (University of Pennsylvania, Philadelphia, PA), and were derived from ligation of the viral DNAs p83-2 (5′ half of NL4-3) or p1971-1 (5′ NLΔVif) with p83-10 (3′ half of NL4-3), obtained from the National Institutes of Health AIDS Research and Reference Reagent Program.19 Sequencing of the NL4-3ΔVif clone confirmed all genes typical of HIV-1 with the exception of vif (data not shown). Viral stocks of NL4-3 and NL4-3ΔVif were prepared by transfecting plasmids into 293T cells, followed by virus amplification in CEMss cells. Vpr was expressed in both wild-type NL4-3 and NL4-3ΔVif–infected cells (data not shown). The Cullin5 mutant vectors Cullin5ΔN1 and Cullin5ΔNedd8 were generated as described previously.22 Cullin5ΔNedd8 is a dominant-negative mutant of Cullin5, while Cullin5ΔN1 is a negative-control mutant of Cullin5. The plasmid VR1012 was used as an empty vector control. The codon-optimized HIV-1 Vif expression plasmid (sometimes referred to below simply as “Vif”) derived from the Vif sequences of HIV-1 NL4-3 was the generous gift of Dr Warner Greene (University of California, San Francisco, CA) and was generated as described previously.19
Two mutants of Vif, SLQ/AAA and C114S/C133S, were generated using the QuikChange site-directed mutagenesis kit (Stratagene). These mutations render Vif unable to bind to Cullin5 (Cul5)22-24 or to mediate degradation of A3G (Figure 2A).22-24
In some experiments, short interfering RNAs (siRNAs) were also transfected using Fugene 6 transfection reagent (Roche Applied Science) according to the manufacturer's instructions. Thymidine, diethylamino ethyl-dextran hydrochloride, and siRNAs directed against Cdk9 were purchased from Sigma-Aldrich. siRNA directed against Brd4 was purchased from Dharmacon RNAi Technologies. Fluorescein isothiocyanate–labeled anti–Gag antibody (KC57) was purchased from Beckman Coulter.
Transfections and cell-cycle synchronization
HeLa cells (0.8-1 × 106 cells in 100mM culture plates) were cotransfected using Fugene 6 with 18 μg of pcDNA or with the Vif or Vif mutant plasmid SLQ and 2 μg of pEGFP-F. Two to 3 days after transfection, cells were harvested, fixed in ethanol, and stained with propidium iodide (PI; 50-100 μg/mL of PI, 50 μg/mL of RNase A, 0.1% saponin, and 5mM EDTA [ethylenediaminetetraacetic acid] in PBS) for cell-cycle analysis. In some experiments, cells that had been transfected using this protocol were synchronized with double-thymidine treatment. This was carried out using the following protocol. Briefly, 2-4 hours after transfection, cells were treated with 2mM thymidine for 18 hours at 37°C, followed by washout and continued culture for 6-9 hours without thymidine. To release cells from the G1 phase, cells were treated for a second time with 2mM thymidine for 17 hours at 37°C, followed by washout with PBS or Dulbecco modified Eagle medium. Cell-cycle profiles were analyzed by bromodeoxyuridine/7-amino-actinomycin D (BrdU/7AAD), BrdU/PI, or PI staining at different time points after thymidine washout. Western blot results suggested that exogenous Vif expression in transfected cells was at similar levels as Vif expression in infected cells (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
To study the effect of Cul5 in Vif-mediated G2 arrest, we transfected HeLa cells (0.8-1 × 106 cells in 100mM culture plates) with 18 μg of Vif and Cul5 mutant plasmids at different ratios of Vif and Cul5 mutants, along with 2 μg of pEGFP-F. We used the control vector VR1012 to normalize equal amounts of DNA in each transfection. In brief, we used 12 μg of Vif, 6 μg of Cul5 mutants, and 2 μg of pEGFP-F for cells transfected with the 2:1 ratio of Vif and Cul5 mutants; 6 μg of Vif, 6 μg of Cul5 mutants, 6 μg of pcDNA, and 2 μg of pEGFP-F for the 1:1 ratio of Vif and Cul5 mutants; and 6 μg of Vif, 12 μg of Cul5 mutants, and 2 μg of pEGFP-F for the 1:2 ratio of Vif and Cul5 mutants. Two to 3 days after transfection, cells were harvested, fixed in ethanol, and stained with PI for cell-cycle analysis as described in the previous paragraph.
Infection or transfection by HIV-1 NL4-3 or the HIV-1 mutant NL4-3ΔVif
Stocks of HIV-1 virus NL4-3 were obtained from 24- and 48-hour harvests of supernatants from infected CEMss cells. The HIV-1 mutant deficient in Vif, NL4-3ΔVif, was generated as described previously.19 CEMss T cells were infected with NL4-3 or NL4-3ΔVif, or mock-infected (as a control) in the presence of 20 μg/mL of diethylamino ethyl-dextran hydrochloride for 2 hours at 37°C. Infection was accomplished by incubating the cells for 2 hours with equal amounts of viruses by viral p24 assessed by enzyme-linked immunosorbent assay. Cells were washed with medium twice and resuspended in growth medium at a concentration of 3 × 105 cells/mL. At day 2 after infection, infected cells were harvested and stained with anti–Gag antibody for the presence of the viral antigen Gag and with PI for the DNA content, as described previously.18,19 The protein levels of Vpr in cells infected with NL4-3ΔVif were similar to those in NL4-3–infected cells (data not shown), which is consistent with our previous observations.19
To determine whether Vif-induced G1-to-S progression is also observed in HIV-infected cells, we transfected HeLa cells (3-5 × 105 cells per well in 6-well plates) with 5 μg of NL4-3 or NL4-3ΔVif plasmids, along with 1 μg of p-EGFP using Fugene 6 transfection reagent. At 2-4 hours after transfection, cells were synchronized at the G1 phase of the cell cycle with double-thymidine treatment as described in “Transfections and cell-cycle synchronization.” Cell-cycle profiles in green fluorescent protein–positive (GFP+) cells were analyzed by BrdU/7AAD at 1.5 and 3 hours after thymidine washout.
Analysis of DNA content for cell-cycle profiles
To analyze the percentage of GFP+ cells in the G1, S, and G2 phases of the cell cycle, as described previously,18,19 transfected or infected cells were fixed in 70% ethanol or 0.5% paraformaldehyde and stained with PI solution at day 2-3 after transfection (or, for some experiments, at different time points after release from thymidine blockade). To more precisely define the percentage of cells at the S phase of the cell cycle, cells were also analyzed with BrdU-allophycocyanin/7AAD staining following the manufacturer's instructions (BrdU-allophycocyanin BD Pharmingen). The cells with active DNA synthetic activities (defined as cells in the S phase of the cell cycle) can be identified by correlating total DNA expression with the level of BrdU incorporation. The ModFit LT software program (Verity Software House) was used to determine the distribution of cells in each phase of the cell cycle (G1, S, and G2/M) with PI staining. Acceptance criteria were based on Shankey et al.25 Fold increases were calculated relative to control siRNA and pcDNA cotransfected cells after normalization of the corresponding S/G1 ratio to 1.
Mass spectrometry
To identify the interaction partners of Vif regulating the alteration of the cell cycle in transfected HeLa cells, PCR was used to Flag tag the codon-optimized Vif at the C-terminus. HeLa cells were then transfected with the Vif-Flag plasmid or pcDNA in 100mM culture plates (0.8-1 × 106 cells per plate) using Fugene 6 and cultured for 3 days. Transfected cells were lysed in buffer containing 50mM Tris, pH 7.5, with 150mM NaCl, 1% Triton X-100, 1mM EDTA, complete protease inhibitor cocktail tablets, and phenylmethanesulfonyl fluoride. Lysates were applied to an anti–Flag M2 affinity gel (4%-12% Bis-Tris gel; Sigma-Aldrich) at 4°C overnight. After washing, the Flag-tagged proteins and their associated cofactors in transfected cell extracts were isolated with Flag peptide (Sigma-Aldrich) following the manufacturer's instructions. The bands of interest were cut and analyzed by mass spectrometry. The criteria for protein identification by mass spectrometry were more than 2 peptides matched and a mascot score greater than 35.
Western blotting
Transfected cells (5 to 10 × 106) were harvested and lysed in NP-40 buffer (0.5% NP-40, 75mM NaCl, 1mM EDTA, 0.5mM Na3VO4, 25mM Tris, pH 7.5, 5mM NaF, 5mM Na4P2O7, and 0.5mM phenylmethanesulfonyl fluoride) for Western blotting. Western blotting was carried out on a 4%-12% Bis-Tris Nupage Ready Gel (Invitrogen). Equal amounts of protein were loaded in each lane. Optical density of positive bands was measured with Bio-Rad Quantity One on a Geldoc 2000. Antibodies used for Western blotting were: rabbit anti–HIV-1 Vif Ab (National Institutes of Health AIDS Research and Reference Reagent Program); rabbit anti–Cdk9 antibody and rabbit anti–Cullin5 antibody (Santa Cruz Biotechnology); rabbit anti–Brd4 antibody (kindly provided by Dr J. You, University of Pennsylvania, Philadelphia, PA); and mouse mAbs against GFP and hemagglutinin (HA; Covance).
Statistical analyses
The Student t test was used to determine whether the averages of 2 groups were significantly different. The 2-tailed test P value is provided.
Results
Vif induces an early G1-to-S transition in Vif-transfected HeLa cells
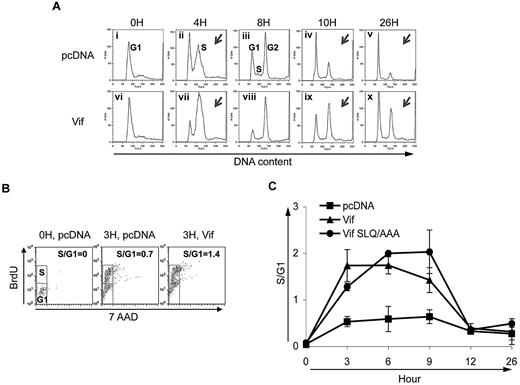
We and others have demonstrated that Vif causes G2 arrest.18-21 To further study the mechanisms of Vif-induced cell-cycle alterations, we synchronized HeLa cells transfected with control DNA (pcDNA), or Vif with double-thymidine treatment at the G1 phase of the cell cycle, and analyzed cell-cycle profiles at various time points after release from thymidine blockade. As expected, at 10 and 26 hours after release from G1, the percentage of cells in the G2 phase of the cell cycle was increased in cells transfected with wild-type Vif (Figure 1A panels iv and v compared with panels ix and x, and supplemental Figure 2 pcDNA compared with Vif), which is consistent with our published results. Intriguingly, these experiments also revealed a second interesting effect of Vif on the cell cycle: at 4 hours after release from G1, a larger proportion of cells that had been transfected with Vif had moved into S phase compared with cells that had been transfected with the control vector (compare the size of the “S” peak in Figure 1Avii to the size of the “S” peak in Figure 1Aii, and see supplemental Figure 2). This result suggests that, in addition to influencing cell-cycle phase G2, Vif drives cells out of G1 and into the S phase.
Vif induces an early G1-to-S transition in Vif-transfected HeLa cells. (A) Cell-cycle profiles of transfected HeLa cells at 5 different time points after release from cell-cycle synchronization. Cells were transfected with pcDNA (top) or wild-type Vif (bottom). Vertical axes denote relative cell number. Horizontal axes denote DNA content as measured by PI staining. Peaks in each plot denote different phases of the cell cycle as labeled in Ai-iii. Note that at 4 hours in Vif-transfected cells (Aix), the height of the S peak compared with the G1 peak is larger than in control cells (Aii). At 10 hours, the height of the G2 peak relative to the G1 peak is greater in Vif-transfected cells (Aviii) than in control cells (Aiv). (B) Proportion of cells in the S phase at time 0 and 3 hours after release from cell-cycle synchronization for the same types of cultures as in panel A after treatment with BrdU/7AAD. Vertical axes show the number of cells with incorporated BrdU. Horizontal axes show total DNA content by 7AAD staining. The top small box in each treatment condition denotes the cell number at the S phase of the cell cycle, while the lower small box denotes the cells at G1. The calculated S/G1 ratio is noted in the upper right of each of the 3 plots. (C) Combined results from multiple experiments of the types shown in panels A-B. Vertical axis denotes ratio of number of cells in S phase to number of cells in G1. Horizontal axis denotes time after release from synchronization in the S phase. The proportion of cells in S compared with G1 was obtained by analyzing the peaks of cell-cycle profiles such as those shown in (A). The proportion of cells in the S phase was increased in cells transfected with either wild-type Vif or the Vif mutant SLQ/AAA at 3, 6, and 9 hours after release from G1, compared with cells transfected with control vector. ■, control vector; ▲, wild-type Vif; and ●, Vif mutant SLQ/AAA.
Vif induces an early G1-to-S transition in Vif-transfected HeLa cells. (A) Cell-cycle profiles of transfected HeLa cells at 5 different time points after release from cell-cycle synchronization. Cells were transfected with pcDNA (top) or wild-type Vif (bottom). Vertical axes denote relative cell number. Horizontal axes denote DNA content as measured by PI staining. Peaks in each plot denote different phases of the cell cycle as labeled in Ai-iii. Note that at 4 hours in Vif-transfected cells (Aix), the height of the S peak compared with the G1 peak is larger than in control cells (Aii). At 10 hours, the height of the G2 peak relative to the G1 peak is greater in Vif-transfected cells (Aviii) than in control cells (Aiv). (B) Proportion of cells in the S phase at time 0 and 3 hours after release from cell-cycle synchronization for the same types of cultures as in panel A after treatment with BrdU/7AAD. Vertical axes show the number of cells with incorporated BrdU. Horizontal axes show total DNA content by 7AAD staining. The top small box in each treatment condition denotes the cell number at the S phase of the cell cycle, while the lower small box denotes the cells at G1. The calculated S/G1 ratio is noted in the upper right of each of the 3 plots. (C) Combined results from multiple experiments of the types shown in panels A-B. Vertical axis denotes ratio of number of cells in S phase to number of cells in G1. Horizontal axis denotes time after release from synchronization in the S phase. The proportion of cells in S compared with G1 was obtained by analyzing the peaks of cell-cycle profiles such as those shown in (A). The proportion of cells in the S phase was increased in cells transfected with either wild-type Vif or the Vif mutant SLQ/AAA at 3, 6, and 9 hours after release from G1, compared with cells transfected with control vector. ■, control vector; ▲, wild-type Vif; and ●, Vif mutant SLQ/AAA.
To corroborate and quantify the effect of Vif on the G1-to-S transition, we used BrdU/7AAD staining (Figure 1B) and BrdU/PI staining (Figure 1C). Figure 1B shows that at 3 hours after release from cell-cycle synchronization, a greater proportion of Vif-transfected than control cells had entered the S phase, indicated by the number of cells in the top box labeled “S” compared with number of cells in the lower box labeled “G1.“ Data from 3 such experiments are quantified and summarized in Figure 1C, demonstrating that at 3, 6, and 9 hours after release from G1, the cell-cycle progression from G1 to the S phase was increased in Vif-transfected cells. A Vif-induced G1-to-S transition was also observed with an alternate Vif expression vector derived from HIV-1 HXB222 (data not shown). These results indicate that Vif not only induces G2 arrest, but also promotes the cell-cycle transition out of G1 and into the S phase.
Vif-mediated G2 arrest is Cullin5-dependent
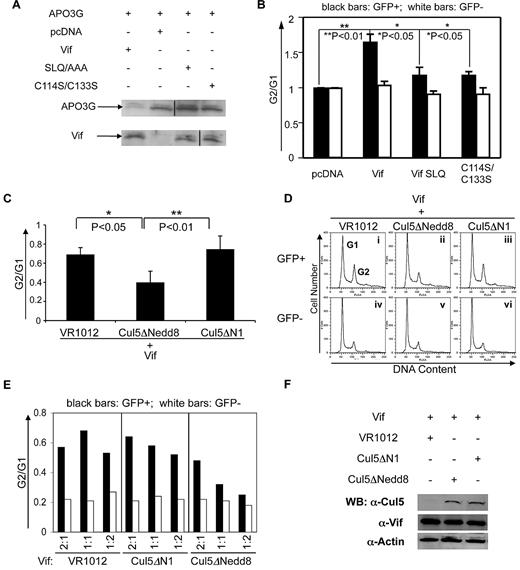
It is well established that in HIV-1–infected cells, one key role of Vif is to degrade the host antiviral factor A3G by forming a complex with the E3 ubiquitin ligase Cul5-ElonginB/C, thereby marking A3G for destruction by the ubiquitin/proteosome system.22-24 Based on this finding, we hypothesized that Vif might also induce cell-cycle alterations in HIV-1–infected cells by interacting with Cullin5 to promote the degradation of a protein that is critical for cell-cycle regulation. We therefore first determined whether the Cul5-ElonginB/C E3 ubiquitin ligase is required for Vif-mediated G2 arrest by making 2 mutants of Vif, SLQ/AAA and C114S/C133S, and testing their effects on Vif-induced cell-cycle alterations. Neither the SLQ/AAA nor the C114S/C133S mutant binds to the Cul5-ElongB/C E3 ligase complex. As anticipated, both Vif mutants failed to degrade A3G (Figure 2A), suggesting that an interaction between Vif and Cullin5 is needed for this process. Next, we analyzed cell-cycle profiles after transfection with wild-type Vif or the mutants SLQ or C114S/C133S. The degree of G2 arrest is indicated by the ratio of G2/G1. We found that the ratio of G2/G1 in cells transfected with either of the Vif mutants was reduced compared with cells transfected with wild-type Vif, suggesting that Vif-induced G2 arrest in transfected HeLa cells depends on an interaction with Cullin5 (Figure 2B).
Cullin5 is required for Vif-induced G2 arrest and its effect is dose dependent. (A) Western blot of HeLa cell lysates at day 3 after transfection with APO3G-HA + Vif vector (lane 1), control vector (lane 2), Vif mutant SLQ (lane 3), or Vif mutant C114S/C113S (lane 4). Expression of Vif and APO3G was measured in cell lysates run on parallel gels; blots were probed with anti–HA antibody (for APO3G expression, upper bands) or with anti–Vif antibody (lower bands). Vif expression was similar among cells transfected with Vif or Vif mutants. Vertical lines have been inserted to indicate a repositioned gel lane. (B) Vif-induced G2 arrest is dependent on an interaction between Cul5 and Vif. Vertical bars indicate the G2/G1 ratio obtained from cell-cycle profiles of HeLa cells transfected with control vector (pcDNA), Vif alone (Vif), or one of the Vif mutants (Vif SLQ or C114S/C113S) at 2-3 days after transfection. The SLQ and C114S/C133S mutants fail to interact with the Cul5-ElongB/C E3 ligase. The mean ± SE of 3 experiments is shown. (C-F) All experiments show results obtained from 3-day cultures of HeLa cells cotransfected with GFP and either VR1012 (control vector) or one of the Cul5 mutants Cul5ΔNedd8 (dominant-negative mutant) or Cul5ΔN1 (negative-control mutant). (C) Proportion of cells in G2 compared with G1 after cotransfection with Vif and VR1012, Cul5ΔNedd8, or Cul5ΔN1. The average ± standard error of 3 experiments is shown for each condition. (D) Cell-cycle profiles of Control (column 1) and Cul5 mutant–transfected HeLa cells (columns 2 and 3), marked by the presence (top row) versus the absence (bottom row) of GFP. Vertical axes denote cell number. Horizontal axes denote total DNA content by PI staining. Peaks in each plot denote cells at the G1 or G2 phase of the cell cycle as labeled. Each plot shows one representative experiment of 3 conducted for each condition. (E) Dose dependence of the proportion of cells in G2 compared with G1 after transfection with decreasing ratios of Vif + control vector or one of the Cul5 mutants. For each condition, 3 ratios were tested: 2:1, 1:1, and 1:2 (horizontal axes). Black bars, GFP+ cells; white bars, GPF− cells. Cell-cycle profiles were analyzed by PI staining at 3 days after transfection. The proportion of cells in G2 compared with G1 was obtained by analysis of peaks of cell-cycle profiles such as those shown in (B). (F) Western blot of cell lysates 3 days after transfection with control vector (lane 1), the Cullin5 dominant-negative mutant (lane 2), or the Cullin5 control mutant (lane 3). Blot was probed with anti–Cul5 antibody or anti–Vif. Anti–actin antibody confirms that equal amounts of cell extract were run in each lane.
Cullin5 is required for Vif-induced G2 arrest and its effect is dose dependent. (A) Western blot of HeLa cell lysates at day 3 after transfection with APO3G-HA + Vif vector (lane 1), control vector (lane 2), Vif mutant SLQ (lane 3), or Vif mutant C114S/C113S (lane 4). Expression of Vif and APO3G was measured in cell lysates run on parallel gels; blots were probed with anti–HA antibody (for APO3G expression, upper bands) or with anti–Vif antibody (lower bands). Vif expression was similar among cells transfected with Vif or Vif mutants. Vertical lines have been inserted to indicate a repositioned gel lane. (B) Vif-induced G2 arrest is dependent on an interaction between Cul5 and Vif. Vertical bars indicate the G2/G1 ratio obtained from cell-cycle profiles of HeLa cells transfected with control vector (pcDNA), Vif alone (Vif), or one of the Vif mutants (Vif SLQ or C114S/C113S) at 2-3 days after transfection. The SLQ and C114S/C133S mutants fail to interact with the Cul5-ElongB/C E3 ligase. The mean ± SE of 3 experiments is shown. (C-F) All experiments show results obtained from 3-day cultures of HeLa cells cotransfected with GFP and either VR1012 (control vector) or one of the Cul5 mutants Cul5ΔNedd8 (dominant-negative mutant) or Cul5ΔN1 (negative-control mutant). (C) Proportion of cells in G2 compared with G1 after cotransfection with Vif and VR1012, Cul5ΔNedd8, or Cul5ΔN1. The average ± standard error of 3 experiments is shown for each condition. (D) Cell-cycle profiles of Control (column 1) and Cul5 mutant–transfected HeLa cells (columns 2 and 3), marked by the presence (top row) versus the absence (bottom row) of GFP. Vertical axes denote cell number. Horizontal axes denote total DNA content by PI staining. Peaks in each plot denote cells at the G1 or G2 phase of the cell cycle as labeled. Each plot shows one representative experiment of 3 conducted for each condition. (E) Dose dependence of the proportion of cells in G2 compared with G1 after transfection with decreasing ratios of Vif + control vector or one of the Cul5 mutants. For each condition, 3 ratios were tested: 2:1, 1:1, and 1:2 (horizontal axes). Black bars, GFP+ cells; white bars, GPF− cells. Cell-cycle profiles were analyzed by PI staining at 3 days after transfection. The proportion of cells in G2 compared with G1 was obtained by analysis of peaks of cell-cycle profiles such as those shown in (B). (F) Western blot of cell lysates 3 days after transfection with control vector (lane 1), the Cullin5 dominant-negative mutant (lane 2), or the Cullin5 control mutant (lane 3). Blot was probed with anti–Cul5 antibody or anti–Vif. Anti–actin antibody confirms that equal amounts of cell extract were run in each lane.
To further confirm that Cullin5 is required for G2 arrest, we cotransfected HeLa cells with Vif and the Cul5 dominant-negative mutant Cul5ΔNedd8, the Cul5 control mutant Cul5ΔN1, or the empty vector VR1012, and analyzed cell-cycle profiles at 3 days. We found that G2 arrest in cells cotransfected with Vif and Cul5ΔNedd8 (Figure 2C-Dii) was decreased compared with cells transfected with Vif + Cul5ΔN1 (Figure 2C-Diii) or with Vif + the empty vector, VR1012 (Figure 2C-Di). This suggests that the dominant-negative Cul5ΔNedd8 blocked Vif-mediated G2 arrest, presumably by interfering with the interaction between Vif and Cullin5. In addition, when we transfected cells with Vif and Vif mutants at different ratios, we found that the inhibition of Vif-induced G2 arrest by Cul5ΔNedd8 was dose dependent (Figure 2E). These data show that either Cul5 E3 ligase activity or an interaction between Cul5 and Vif is required for Vif-induced G2 arrest.
To be certain that our results were not complicated by unequal expression of Cullin5 in transfections with Cul5ΔN1 or Cul5ΔNedd8, cell lysates from transfections such as those shown in Figure 2C were run on a Western blot and probed with anti–Cul5 antibody. Amounts of Cullin5 protein were found to be similar in cells transfected with either Cul5ΔN1 or Cul5ΔNedd8 (Figure 2F).
Vif-mediated early entry into the S phase does not require an interaction with Cullin5
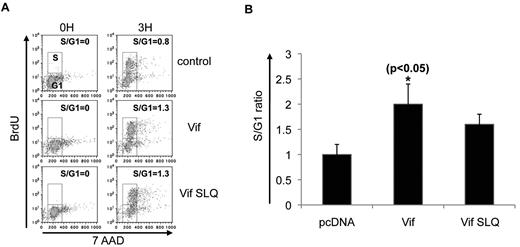
We then asked whether Vif-mediated early entry into the S phase also requires Cul5-ElongB/C E3 ligase, or if this cell-cycle transition relies on different intracellular mechanisms. To pursue this question, we transfected HeLa cells with the wild-type Vif or the Vif mutant SLQ (which fails to interact with the Cul5-ElongB/C E3 ligase complex), and then observed cell-cycle profiles at 3-4 hours after release from thymidine block. Surprisingly, in contrast to the Cullin5-dependent G2 arrest induced by Vif, the early transition from G1 to the S phase was still observed, indicating that the lack of interaction between Vif and Cullin5 did not prevent early entry into S-phase (Figure 3A and supplemental Figure 2). Data from multiple experiments of this type confirmed that Vif SLQ did not alter the Vif-mediated early entry into the S phase at 3, 6, and 9 hours after release from thymidine blockade (Figures 3B and 1C). Thus, because the Vif-mediated early G1-to-S transition appears to be independent of an interaction with Cullin5, the biologic effect of Vif on the G1-to-S transition appears to be distinct from the Vif effect on G2 arrest.
Vif induces an early G1-to-S transition that is Cul5 independent. (A) HeLa cells were transfected with pcDNA, wild-type Vif, or the Vif mutant SLQ, and the proportion of cells in S phase at time 0 and 3 hours after release from cell-cycle synchronization after treatment with BrdU/7AAD was determined. Vertical axes show the number of cells that incorporated BrdU. Horizontal axes show total DNA content by 7AAD staining. The top small box in each plot denotes the cell number at the S phase of the cell cycle, while the lower small box denotes the cells at G1. The calculated S/G1 ratio is noted in the upper right of each plot. (B) Combined results from multiple experiments of the type shown in panel A. Vertical axis denotes the ratio of the number of cells in the S phase to the number of cells in G1. The SLQ mutants fail to interact with the Cul5-ElongB/C E3 ligase complex. *P < .05 for bar 2 vs bar 1. The mean ± SE of 3 experiments is shown.
Vif induces an early G1-to-S transition that is Cul5 independent. (A) HeLa cells were transfected with pcDNA, wild-type Vif, or the Vif mutant SLQ, and the proportion of cells in S phase at time 0 and 3 hours after release from cell-cycle synchronization after treatment with BrdU/7AAD was determined. Vertical axes show the number of cells that incorporated BrdU. Horizontal axes show total DNA content by 7AAD staining. The top small box in each plot denotes the cell number at the S phase of the cell cycle, while the lower small box denotes the cells at G1. The calculated S/G1 ratio is noted in the upper right of each plot. (B) Combined results from multiple experiments of the type shown in panel A. Vertical axis denotes the ratio of the number of cells in the S phase to the number of cells in G1. The SLQ mutants fail to interact with the Cul5-ElongB/C E3 ligase complex. *P < .05 for bar 2 vs bar 1. The mean ± SE of 3 experiments is shown.
Vif induces an early G1-to-S transition in HIV-infected cells
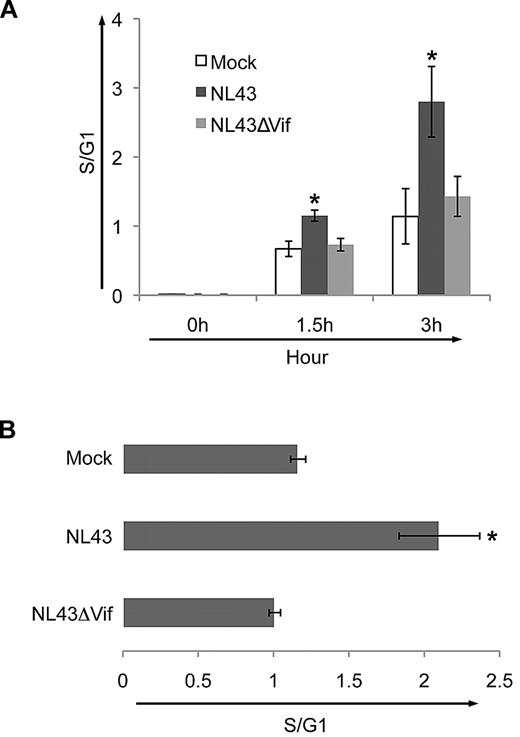
Next, we asked if the Vif-induced G1-to-S progression is also observed in cells infected with HIV. We transfected HeLa cells with wild-type NL4-3 or with mutant virus deleted in Vif (NL4-3ΔVif) along with GFP, stained them with BrdU/7AAD after release from G1, and calculated the S/G1 ratio for transfected (GFP+) cells. At 1.5 and 3 hours after release from thymidine blockade, the ratio of S/G1 in transfected cells was significantly increased in NL4-3 compared with mock-transfected cultures (Figure 4A white bars compared with black bars). Transfection with virus deficient in Vif (NL4-3ΔVif) did not promote the early G1-to-S transition (Figure 4A gray bars). These data support a role for Vif in driving cells into the S phase during infection with HIV-1.
Vif drives HIV-1–infected cells from G1 into the S phase of the cell cycle. (A) Relative proportion of HeLa cells in the S phase compared with G1 after cotransfection with GFP and pcDNA (white bars), wild-type NL4-3 (black bars), or NL4-3ΔVif (gray bars), along with GFP. Horizontal axis shows each transfection condition at 0, 1.5, and 3 hours after release from 24 hours of synchronization. The ratio of S/G1 in cells transfected with NL4-3 was significantly increased compared with cells transfected with NL4-3ΔVif. (B) Ratio of number of cells in S compared with G1 (S/G1) in CEMss T cells 2 days after infection with wild-type NL4-3 or NL4-3ΔVif or mock infection. The mean ± SE of 3 experiments is shown. Infection levels, as measured by the percentage of Gag+ cells, were 0.1% ± 0.1% for mock, 51% ± 5.4% for NL4-3, and 55% ± 8.4% for NL4-3ΔVif. Significantly more cells entered the S phase when infected with the wild-type virus NL4-3 than with the wild-type virus deleted in Vif, NL4-3ΔVif (P < .05).
Vif drives HIV-1–infected cells from G1 into the S phase of the cell cycle. (A) Relative proportion of HeLa cells in the S phase compared with G1 after cotransfection with GFP and pcDNA (white bars), wild-type NL4-3 (black bars), or NL4-3ΔVif (gray bars), along with GFP. Horizontal axis shows each transfection condition at 0, 1.5, and 3 hours after release from 24 hours of synchronization. The ratio of S/G1 in cells transfected with NL4-3 was significantly increased compared with cells transfected with NL4-3ΔVif. (B) Ratio of number of cells in S compared with G1 (S/G1) in CEMss T cells 2 days after infection with wild-type NL4-3 or NL4-3ΔVif or mock infection. The mean ± SE of 3 experiments is shown. Infection levels, as measured by the percentage of Gag+ cells, were 0.1% ± 0.1% for mock, 51% ± 5.4% for NL4-3, and 55% ± 8.4% for NL4-3ΔVif. Significantly more cells entered the S phase when infected with the wild-type virus NL4-3 than with the wild-type virus deleted in Vif, NL4-3ΔVif (P < .05).
To confirm these results in T cells, we infected CEMss T cells with wild-type NL4-3 or NL4-3ΔVif viruses. At day 2 after infection, cultures infected with NL4-3 had a significantly higher S/G1 ratio compared with cultures infected with wild-type NL4-3ΔVif (Figure 4B). These data support the hypothesis that Vif promotes the G1-to-S transition in T cells.
Vif interacts with Cdk9 in Vif-transfected HeLa cells
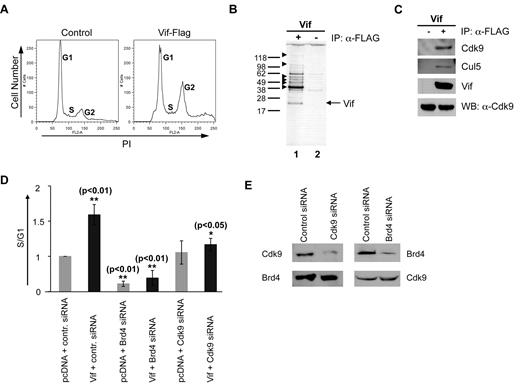
To discover other potential cellular partner(s) that may regulate the Vif-mediated cell-cycle alterations in HIV-1 infection, we used a mass spectrometry approach to screen for cellular proteins that interact with Vif to control the cell cycle. To efficiently precipitate Vif from transfected cells, we first generated a codon-optimized, Flag-tagged Vif. This Vif-Flag clone was as functional in inducing G2 arrest in transfected HeLa cells as was the pure Vif clone (compare Figure 1A at 10 and 26 hours with Figure 5A). When lysates of Vif-Flag–transfected HeLa cells were probed with the anti–Flag antibody, several potential Vif-interaction partners were observed (Figure 5B lane 1).
Mass spectrometry and a proteomic screen of Vif-Flag–transfected HeLa cells yield known and novel cellular proteins and RNP partners of Vif. (A) Cell-cycle profiles of HeLa cells transfected with control vector or with Vif-Flag at 3 days after transfection. Note that Vif-Flag, like untagged Vif, causes transfected cells to arrest in G2. Vertical axes denote cell number. Horizontal axes denote total DNA content by PI staining. Peaks in each plot denote different phases of the cell cycle as labeled. One representative experiment is shown. (B) Silver-stained Western blot of immunoprecipitates (with anti–Flag antibody) of cell lysates obtained from cells transfected with Vif-Flag vector (lane 1) or control vector (lane 2). One representative of 3 experiments is shown. The gel shown was silver stained. (C) Western blot of Vif-Flag–transfected HeLa cell lysates immunoprecipitated with anti–Flag antibody. Left lane, control; right lane, Vif-Flag–transfected cells. Equal amounts of cell extract were analyzed using antibodies against Cdk9 and Cul5 and Vif. Cdk9, Cyclin-dependent kinase; Cul5, Cullin5; Vif, viral infectivity factor; WB, Western blot; α, anti-. (D) Relative proportion of cells in S compared with G1 after transfection with Vif and siRNA against its potential binding partners Brd4 and Cdk9. The various vectors used to transfect synchronized HeLa cells are shown across the x-axis. **P < .01 for bar 2 vs bar 1 and for bar 4 vs bar 2; *P < .05 for bar 6 vs bar 2. The mean ± standard error of 3 experiments is shown. Inhibition of Brd4 or Cdk9 with siRNA reduces the Vif-mediated G1-to-S cell-cycle transition in transfected cells. Fold increases were calculated relative to control siRNA and pcDNA cotransfected cells after normalization of the corresponding S/G1 ratio to 1. (E) Western blot of lysates from HeLa cells transfected with control siRNA or siRNA against Brd4 or Cdk9. Lysates were probed with anti–Cdk9 or anti–Brd4 antibody and analyzed at 2 days after transfection. Note how transfection with Cdk9 siRNA or Brd4 siRNA reduced the expression of Cdk9 and Brd4 proteins, respectively.
Mass spectrometry and a proteomic screen of Vif-Flag–transfected HeLa cells yield known and novel cellular proteins and RNP partners of Vif. (A) Cell-cycle profiles of HeLa cells transfected with control vector or with Vif-Flag at 3 days after transfection. Note that Vif-Flag, like untagged Vif, causes transfected cells to arrest in G2. Vertical axes denote cell number. Horizontal axes denote total DNA content by PI staining. Peaks in each plot denote different phases of the cell cycle as labeled. One representative experiment is shown. (B) Silver-stained Western blot of immunoprecipitates (with anti–Flag antibody) of cell lysates obtained from cells transfected with Vif-Flag vector (lane 1) or control vector (lane 2). One representative of 3 experiments is shown. The gel shown was silver stained. (C) Western blot of Vif-Flag–transfected HeLa cell lysates immunoprecipitated with anti–Flag antibody. Left lane, control; right lane, Vif-Flag–transfected cells. Equal amounts of cell extract were analyzed using antibodies against Cdk9 and Cul5 and Vif. Cdk9, Cyclin-dependent kinase; Cul5, Cullin5; Vif, viral infectivity factor; WB, Western blot; α, anti-. (D) Relative proportion of cells in S compared with G1 after transfection with Vif and siRNA against its potential binding partners Brd4 and Cdk9. The various vectors used to transfect synchronized HeLa cells are shown across the x-axis. **P < .01 for bar 2 vs bar 1 and for bar 4 vs bar 2; *P < .05 for bar 6 vs bar 2. The mean ± standard error of 3 experiments is shown. Inhibition of Brd4 or Cdk9 with siRNA reduces the Vif-mediated G1-to-S cell-cycle transition in transfected cells. Fold increases were calculated relative to control siRNA and pcDNA cotransfected cells after normalization of the corresponding S/G1 ratio to 1. (E) Western blot of lysates from HeLa cells transfected with control siRNA or siRNA against Brd4 or Cdk9. Lysates were probed with anti–Cdk9 or anti–Brd4 antibody and analyzed at 2 days after transfection. Note how transfection with Cdk9 siRNA or Brd4 siRNA reduced the expression of Cdk9 and Brd4 proteins, respectively.
Using mass spectrometry, we identified 2 different groups of proteins with which Vif interacts (Table 1): the first group contains several RNA-associated proteins (eg, ribonucleoproteins [RNPs] hnRNP U and hnRNP L), RNA helicases (DHX9 and Gu), and some pre-mRNA splicing factors such as PRP9 and spliceosomal protein. These proteins have been demonstrated previously in A3G-associated RNP complexes.26-33 Cullin5 has also been shown to interact with Vif (22 and results shown in Table 1), and we confirmed this interaction by coimmunoprecipitation of Vif with Cullin5 (Figure 5C). More intriguingly, the mass spectrometry scan identified 2 Vif-interacting proteins, Cdk9 and Brd4, which are known to function in normal cell-cycle regulation. Cdk9 is a component of the heterodimer p-TEFb (positive transcription elongation factor b), which is crucial for efficient transcriptional elongation of HIV-1. Brd4 binds to the Cdk9/CyclinT (p-TEFb) complex34,35 and is required for the proper progression of the cell cycle: it regulates the G2-to-M transition36,37 and stimulates cell-cycle progression from G1 into S through recruitment of p-TEFb to chromosomes and stimulation of G1 gene expression during late mitosis.38,39 As shown in Figure 5C, interaction of Vif with Cdk9 was detected by coimmunoprecipitation of Vif with Cdk9. These results suggest that Vif interacts with Cdk9.
Mass spectrometry and a proteomic screen of Vif-Flag–transfected cells
| Common name . | No. of peptides matched . | Mascot score . |
|---|---|---|
| hnRNP L | 127 | 2317 |
| hnRNP U | 115 | 2005 |
| Spliceosomal protein | 4 | 132 |
| PRP19 | 8 | 123 |
| Cullin-associated protein | 13 | 247 |
| Heat-shock protein | 20 | 429 |
| DHX9 protein | 5 | 44 |
| Gu protein | 26 | 437 |
| Brd4 | 15 | 196 |
| Cdk9 | 4 | 93 |
| Common name . | No. of peptides matched . | Mascot score . |
|---|---|---|
| hnRNP L | 127 | 2317 |
| hnRNP U | 115 | 2005 |
| Spliceosomal protein | 4 | 132 |
| PRP19 | 8 | 123 |
| Cullin-associated protein | 13 | 247 |
| Heat-shock protein | 20 | 429 |
| DHX9 protein | 5 | 44 |
| Gu protein | 26 | 437 |
| Brd4 | 15 | 196 |
| Cdk9 | 4 | 93 |
Known and novel cellular proteins and RNP partners of Vif were identified by mass spectrometry in Vif-Flag–transfected HeLa cells. Using mass spectrometry, we identified two different groups of proteins with which Vif interacts: the first group contained a number of RNA-associated proteins (eg, ribonucleoproteins hnRNP U and hnRNP L), RNA helicases (DHX9 and Gu), and some pre-mRNA splicing factors (PRP9, spliceosomal protein). The second group included two cell-cycle regulatory proteins, Cdk9 and Brd4. The criteria for protein identification were more than 2 peptides matched and a mascot score > 35.
siRNA against Cdk9 counteracts the Vif-mediated G1-to-S transition
To confirm that Cdk9 and Brd4 play a role in the Vif-mediated G1-to-S transition, we used siRNAs against Cdk9 or Brd4 and examined the resulting cell-cycle profiles of Vif-transfected HeLa cells. We hypothesized that when cells were transfected with Vif + siRNA against either Cdk9 or Brd4, an early G1-to-S transition would not be observed.
First, we determined that siRNA against Brd4 or Cdk9 reduced the endogenous expression of Brd4 or Cdk9 in transfected HeLa cells (Figure 5E). Then, we synchronized HeLa cells transfected with Vif + siRNA against either Cdk9 or Brd4, and observed the G1-to-S transition after cells were released from G1. The effect on the cell cycle of 6 different combinations of control DNA, siRNA, and Vif at ∼48 hours after transfection is shown in Figure 5D. First, transfection with Vif + control siRNA significantly increased the proportion of cells in the S phase (Figure 5D bar 2 compared with bar 1). This increase was not observed when cells were transfected with Vif + siRNA against Cdk9 (Figure 5D bar 6 compared with bar 2), which is consistent with our postulated role for Cdk9 in the Vif-mediated early G1-to-S phase transition. When cells were transfected with Vif + siRNA against Brd4 (Figure 5D bar 4 compared with bar 2), the increase in the proportion of cells in the S phase also was not observed, although this result is complicated by the fact that Brd4 itself stimulates cell-cycle progression38,39 (Figure 5D bar 1 compared with bar 3). These data demonstrate that siRNA against Cdk9 counteracts the Vif-mediated G1-to-S transition, and suggest that the p-TEFb (Cdk9/CyclinT)/Brd4 complex interacts with Vif to drive cell-cycle progression in HIV-1 infection.
Discussion
We have previously demonstrated that HIV-1 Vif causes G2 arrest of HIV-infected cells.18,19 In the present study, we show that Vif also drives cells out of G1 and into the S phase of the cell cycle. The effect of Vif on the G1-to-S phase transition appears to be a biologic effect distinct from its effect on G2 arrest, because G2 arrest is Cullin5-dependent and the G1-to-S progression is Cullin5-independent. Published data21 and our own observations suggest that Vif does not induce G2 arrest by interacting with its primary known target, A3G; therefore, it must target other cellular protein(s) to mediate cell-cycle alterations in HIV-1 infection. Our mass spectrometry screen for candidate interaction partners of Vif yielded 2 novel targets of Vif, Cdk9 and Brd4, which are known to regulate cell-cycle progression. We further confirmed the interaction of Vif and Cdk9 by immunoprecipitation and Western blot of transfected HeLa cells. In addition, we used siRNAs to assay the functional effect of Cdk9 and Brd4 on the Vif-mediated G1-to-S transition. Our data suggest that Vif regulates cell-cycle progression from G1 to S through interactions involving Cdk9 and Brd4.
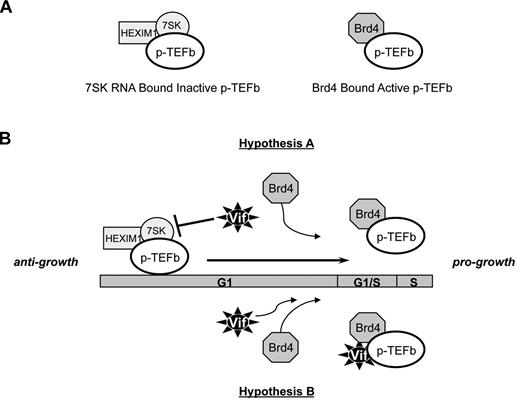
To explore the cellular mechanisms of the Vif-mediated transition from G1 to S, we tagged codon-optimized Vif with Flag and used mass spectrometry to identify cellular interacting partners. We identified 2 proteins, Cdk9 and Brd4, and confirmed their interaction by coimmunoprecipitation (Figure 4 and data not shown). siRNA specific for Cdk9 and Brd4 completely inhibited the G1-to-S transition, suggesting that Vif interacts with the cell-cycle–regulatory proteins Cdk9 and Brd4 to promote cell-cycle progression. Cdk9 is known to be a component of p-TEFb heterodimers. p-TEFb exists in vivo in either a kinase-active form (p-TEFb-Brd4 complex) or a kinase-inactive form (p-TEFb-HEXIM1-7SK RNA complex), as shown in Figure 6A.34,35 Brd4 and HEXIM1 act as positive and negative regulators, respectively, of the complex to reciprocally affect the functional equilibrium of p-TEFb. The appropriate balance of p-TEFb is known to be important for growth and differentiation.34,35 Intriguingly, both Brd4 and Cdk9 have been reported to be required for the proper progression of the cell cycle: Brd4 regulates the G2-to-M transition36,37 and stimulates cell-cycle progression from G1 into the S phase via recruitment of p-TEFb to chromosomes and stimulation of G1 gene expression during late mitosis.38,39 These data support our hypothesis that Brd4 and Cdk9 are part of a Vif interactome controlling cell-cycle progression. Exactly how Vif might affect the equilibrium of p-TEFb is not clear. We propose 2 possible mechanisms (as shown in Figure 6B): (1) Vif shifts the equilibrium of p-TEFb away from the inactive, anti-growth state and toward the active, pro-growth state by releasing p-TEFb from negative regulation by the HEXIM1-7SK RNA complex; or (2) Vif acts as a chaperone to recruit the certain host cofactors, such as Brd4 and Cdk9, into the HIV-1–pre-integration complex by forming the RNP complex. The inclusion of these transcriptional regulatory factors in the pre-integration complex may contribute to promoting the reactivation of the HIV-1 provirus on reactivation signaling. We believe that hypothesis B is more likely, given our data showing a strong interaction between Vif and Cdk9 by immunoprecipitation (Figure 5C) and the fact that siRNA against Cdk9 decreases the effect of Vif on the G1-to-S cell-cycle progression (Figure 5D). These mechanistic questions are currently under investigation.
Possible mechanisms by which Vif may modulate the in vivo equilibrium of p-TEFb. (A) p-TEFb exists in vivo in either a kinase-active form (p-TEFb-Brd4 complex) or a kinase-inactive form (p-TEFb-HEXIM1-7SK RNA complex). Brd4 and HEXIM1 act as positive and negative regulators, respectively, of the complex to reciprocally affect the functional equilibrium of p-TEFb. The appropriate equilibrium of p-TEFb is known to be important for growth and differentiation. (B) Postulated mechanisms by which Vif could alter the equilibrium of p-TEFb. Hypothesis A: Vif shifts the equilibrium of p-TEFb away from the inactive, anti-growth state and toward the active, pro-growth state by releasing p-TEFb from negative regulation by the HEXIM1-7SK RNA complex. Hypothesis B: Vif interacts directly with Brd4 and Cdk9 to promote the formation of the active, pro-growth state of p-TEFb.
Possible mechanisms by which Vif may modulate the in vivo equilibrium of p-TEFb. (A) p-TEFb exists in vivo in either a kinase-active form (p-TEFb-Brd4 complex) or a kinase-inactive form (p-TEFb-HEXIM1-7SK RNA complex). Brd4 and HEXIM1 act as positive and negative regulators, respectively, of the complex to reciprocally affect the functional equilibrium of p-TEFb. The appropriate equilibrium of p-TEFb is known to be important for growth and differentiation. (B) Postulated mechanisms by which Vif could alter the equilibrium of p-TEFb. Hypothesis A: Vif shifts the equilibrium of p-TEFb away from the inactive, anti-growth state and toward the active, pro-growth state by releasing p-TEFb from negative regulation by the HEXIM1-7SK RNA complex. Hypothesis B: Vif interacts directly with Brd4 and Cdk9 to promote the formation of the active, pro-growth state of p-TEFb.
To our knowledge, our data represent the first demonstration of an HIV-1–encoded protein that modulates the G1-to-S transition. This is of considerable interest, because virally encoded proteins that drive the cell from G1 to the S phase are prominent in infections by other latency-inducing viruses, such as Kaposi sarcoma-associated herpesvirus and Epstein-Barr virus.40-46 One longstanding obstacle to treating HIV-1 infection has been the ability of integrated virus to enter into a latent state and to remain latent until reactivated. How the virus integrates, remains latent, and reactivates is only partially understood. It is known, however, that reactivation of latent HIV-1 provirus occurs in association with T-cell activation and induced cycling of infected cells—that is, a transition from the resting state to cell-cycle entry. We speculate that the Vif-mediated G1-to-S transition regulates HIV-1 latency, for example, by promoting an active, pro-growth state of pTEFb, as described in the previous paragraph and Figure 6. Indeed, using an in vitro primary CD4 T-cell latency model,47,48 our preliminary data suggest that Vif promotes the integration or reactivation of latent provirus (data not shown). Alternatively, it is possible that the Vif-mediated G1-to-S transition may promote virus expression after reactivation of a latently infected cell.
Intriguingly, the G1-to-S phase transition is not the only point in the cell cycle where Vif has an effect. In human T cells, infection with HIV-1 causes cell-cycle arrest in the G2 phase of the cell cycle, where transcription of the viral genome is optimal. The 2 viral proteins implicated in G2 arrest of infected cells are Vpr11,13-17,49 and Vif.18-21 The full significance of HIV-1 induction of G2 arrest is not understood, nor is it clear why HIV-1 uses 2 different viral proteins to arrest cells in G2 or whether the ability of Vif to drive cells into the S phase contributes to G2 arrest. However, it seems reasonable to postulate that this is a critical viral function, because HIV-1 devotes 2 of its 9 genes to this effort. Viral regulation of cell-cycle proteins is a common method by which viruses (eg, hepatitis virus, Epstein-Barr virus, herpesvirus, etc) control their infectivity.44-46 We believe that an understanding of the molecular mechanisms of Vif function in HIV-1 pathogenesis, especially the way it works to affect the cell cycle, will lead to the identification of novel therapeutic targets to eliminate viral reservoirs and restore immune function in HIV-infected individuals.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grants R01 AI40003 and R01 AI35513 (T.H.F.), 1T32 AI007632 (E.L.R.), R01 AI062644 (X.-F.Y.), and R01 CA093237 (J.A.D.); by the Bender Foundation (T.H.F.); by the Joseph L. Hollander Chair of Pediatric Rheumatology (T.H.F.); and by the Children's Hospital of Philadelphia Research Institute and the University of Pennsylvania Center for AIDS Research (T.H.F.).
National Institutes of Health
Authorship
Contribution: J.F.W., J.A.D., X.-F.Y., and T.H.F. designed the research; J.F.W., E.L.R., J.M.S., L.J., and D.K.S. performed the research; X.-F.Y. and J.A.D. contributed vital new reagents; J.F.W., E.L.R., J.M.S., J.A.D., X.-F.Y., and T.H.F. analyzed and interpreted the data; and J.F.W. and T.H.F. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Terri H. Finkel, MD, PhD, Division of Rheumatology, The Children's Hospital of Philadelphia, Department of Pediatrics, University of Pennsylvania School of Medicine, 1102 Abramson Research Center, 3615 Civic Center Blvd, Philadelphia, PA 19104; e-mail: finkelt@email.chop.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal