Abstract
Gene therapy for hemophilia A would be facilitated by development of smaller expression cassettes encoding factor VIII (FVIII), which demonstrate improved biosynthesis and/or enhanced biologic properties. B domain deleted (BDD) FVIII retains full procoagulant function and is expressed at higher levels than wild-type FVIII. However, a partial BDD FVIII, leaving an N-terminal 226 amino acid stretch (N6), increases in vitro secretion of FVIII tenfold compared with BDD-FVIII. In this study, we tested various BDD constructs in the context of either wild-type or codon-optimized cDNA sequences expressed under control of the strong, ubiquitous Spleen Focus Forming Virus promoter within a self-inactivating HIV-based lentiviral vector. Transduced 293T cells in vitro demonstrated detectable FVIII activity. Hemophilic mice treated with lentiviral vectors showed expression of FVIII activity and phenotypic correction sustained over 250 days. Importantly, codon-optimized constructs achieved an unprecedented 29- to 44-fold increase in expression, yielding more than 200% normal human FVIII levels. Addition of B domain sequences to BDD-FVIII did not significantly increase in vivo expression. These significant findings demonstrate that shorter FVIII constructs that can be more easily accommodated in viral vectors can result in increased therapeutic efficacy and may deliver effective gene therapy for hemophilia A.
Introduction
Hemophilia A is a serious bleeding disorder caused by a deficiency in, or complete absence of, the blood coagulation factor VIII (FVIII). It is the most common hereditary coagulation disorder with an incidence approaching approximately 1 in 5000 males.1 The disorder is an attractive candidate for gene therapy because only a modest increase in FVIII plasma concentration is needed for therapeutic benefit, with levels of more than 1% able to achieve markedly reduced rates of spontaneous bleeding and long-term arthropathy.2 However, although preclinical results using gene therapy in animal models of hemophilia A have been encouraging, no approach as yet has been translated to clinical success where insufficient FVIII expression has been observed.3
Low FVIII expression is principally caused by inefficient expression of the mRNA,4-6 a significant proportion of protein misfolding with subsequent intracellular degradation, and inefficient transport of the primary translation product from the endoplasmic reticulum (ER) to the Golgi.7,8 This results in expression levels of FVIII approximately 2 to 3 orders of magnitude lower than those of other comparably sized secreted proteins.4 Insights over the past 2 decades into the secretion pathway, FVIII protein structure and function, and mechanisms of inhibitor development have led to the incorporation of bioengineered forms of FVIII in gene transfer systems. Bioengineering aims to improve properties, such as biosynthesis, secretion efficiency, functional activity, and plasma half-life, and to reduce antigenicity/immunogenicity.9 FVIII is produced as a large 330-kDa glycoprotein with the domain structure A1-A2-B-A3-C1-C2,10,11 where both the A and C domains have internal sequence homology and approximately 40% sequence identity to the A and C domains of factor V (FV), which shares the same domain structure.12,13 The B domain, which constitutes 38% of the total sequence, shares no amino acid sequence identity with other known proteins, including the B domain of FV. It is, however, extensively glycosylated and contains 19 of the 26 asparagine (N)-linked glycosylation sites on the whole FVIII molecule.14 The FVIII B domain is dispensable for procoagulant activity. FVIII in which the B domain is deleted (BDD) and replaced by a short 14 amino acid linker (FVIII SQ; Figure 1B) is in clinical use as a replacement recombinant FVIII product (Refacto, Wyeth Pharma).15
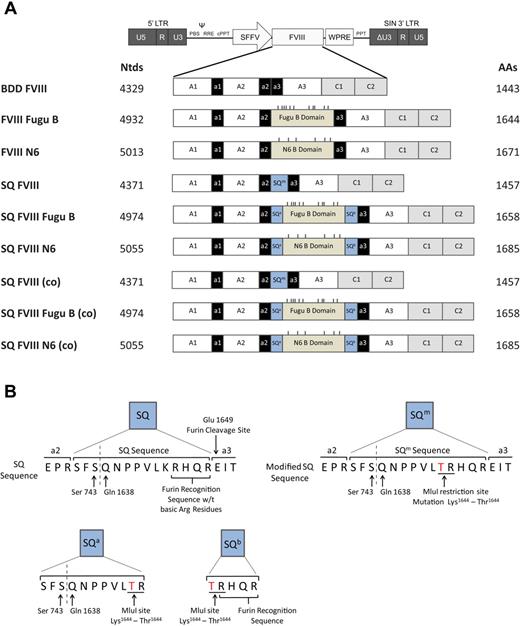
Schematic representation of human FVIII variants designed and cloned into a SIN LV backbone. (A) Nine different human FVIII variants were designed and cloned into a LV backbone plasmid: BDD FVIII, B domain deleted human FVIII; FVIII Fugu B, BDD FVIII containing the Fugu B domain; FVIII N6, BDD FVIII containing the human N6 B domain; SQ FVIII, BDD FVIII containing a modified version of the SQ amino acid sequence SQm; SQ FVIII Fugu B, SQ FVIII containing the Fugu B domain between the SQm sequence to create the N terminal SQa and C terminal SQb sequences; and SQ FVIII N6, SQ FVIII containing the human N6 B domain. And constructs SQ FVIII (co), SQ FVIII Fugu B (co), and SQ FVIII N6 (co) are the same amino acid structure as constructs SQ FVIII, SQ FVIII Fugu B, and SQ FVIII N6, respectively, but are produced from a codon-optimized cDNA sequence. Dashes on constructs indicate asparagine (N)-linked glycosylation sites within the B domain only. (B) Schematics of SQ and modified SQ sequences; SQm, SQa, and SQb. The SQ sequence is a 14-amino acid bridge between the a2 and a3 domains of FVIII created by fusing Ser743 and Gln1638 in the B domain. The sequence promotes efficient intracellular cleavage by containing the 4 amino acid protease recognition site RHQR. A modified SQ sequence (SQm) was created containing a missense mutation from Lys1644 to Thr1644 caused by the creation of an MluI restriction enzyme site within the cDNA sequence for insertion of the Fugu and N6 B domains. SQa is the 11-amino acid sequence created at the N-terminal of the B domain after insertion of the N6 or Fugu B domain sequences into the SQ FVIII construct. SQb is the 5 amino acid sequence created at the C-terminal of the B domain after insertion of the N6 or Fugu B domain sequences into the SQ FVIII construct; this sequence retains the 4 amino acid protease recognition site. MluI restriction sites are shown underlined, and the K to T missense mutation is shown in red.
Schematic representation of human FVIII variants designed and cloned into a SIN LV backbone. (A) Nine different human FVIII variants were designed and cloned into a LV backbone plasmid: BDD FVIII, B domain deleted human FVIII; FVIII Fugu B, BDD FVIII containing the Fugu B domain; FVIII N6, BDD FVIII containing the human N6 B domain; SQ FVIII, BDD FVIII containing a modified version of the SQ amino acid sequence SQm; SQ FVIII Fugu B, SQ FVIII containing the Fugu B domain between the SQm sequence to create the N terminal SQa and C terminal SQb sequences; and SQ FVIII N6, SQ FVIII containing the human N6 B domain. And constructs SQ FVIII (co), SQ FVIII Fugu B (co), and SQ FVIII N6 (co) are the same amino acid structure as constructs SQ FVIII, SQ FVIII Fugu B, and SQ FVIII N6, respectively, but are produced from a codon-optimized cDNA sequence. Dashes on constructs indicate asparagine (N)-linked glycosylation sites within the B domain only. (B) Schematics of SQ and modified SQ sequences; SQm, SQa, and SQb. The SQ sequence is a 14-amino acid bridge between the a2 and a3 domains of FVIII created by fusing Ser743 and Gln1638 in the B domain. The sequence promotes efficient intracellular cleavage by containing the 4 amino acid protease recognition site RHQR. A modified SQ sequence (SQm) was created containing a missense mutation from Lys1644 to Thr1644 caused by the creation of an MluI restriction enzyme site within the cDNA sequence for insertion of the Fugu and N6 B domains. SQa is the 11-amino acid sequence created at the N-terminal of the B domain after insertion of the N6 or Fugu B domain sequences into the SQ FVIII construct. SQb is the 5 amino acid sequence created at the C-terminal of the B domain after insertion of the N6 or Fugu B domain sequences into the SQ FVIII construct; this sequence retains the 4 amino acid protease recognition site. MluI restriction sites are shown underlined, and the K to T missense mutation is shown in red.
It has been shown that deletion of the entire B domain leads to a 17-fold increase in mRNA and primary translation product; however, only a 30% increase in the levels of secreted protein, suggesting that the rate of ER-Golgi transport is actually reduced.16 Efficient FVIII secretion requires carbohydrate-facilitated transport by lectin mannose binding-1 (LMAN1) mediated by mannose residues of N-linked oligosaccharides post-translationally attached to the B domain. To build on the advantages of BDD-FVIII while aiding LMAN1-mediated transport, Miao et al17 added back a short B domain sequence to the BDD-FVIII, optimally 226 amino acids and retaining 6 sites for N-linked glycosylation (N6). This resulted in a 10-fold increase in secretion in vitro from transfected COS-1 cells and a 5-fold increase in vivo following hydrodynamic hepatic gene delivery.17
The teleost puffer fish Fugu rubripes is a commonly used organism for investigation of genetics. Fugu has a basic vertebrate genome and contains a similar repertoire of genes to humans; however, in 1993, it was shown that the Fugu genome is only 390 Mb, approximately one-eighth the size of the human genome.18 This makes Fugu an extremely useful model for annotating the human genome and a valuable “reference” genome for identifying genes and other functional elements. Sequence analysis of genes in the blood coagulation system showed that Fugu amino acid sequences are highly conserved relative to their human orthologs. For FVIII cDNA sequences, the Fugu A1, A2, A3, C1, and C2 domains show 46%, 43%, 47%, 52%, and 50% sequence identity to human orthologs, respectively. Conversely, the Fugu factor VIII B domain shares only 6% sequence identity to its human counterpart. However, although there is no apparent sequence conservation between B domains, the Fugu B domain is also highly glycosylated with 10 asparagine (N)-linked glycosylation attachment sites across its 224 amino acid length.19
In this study, we examined the expression of human BDD FVIII constructs containing the previously described “SQ” B domain element, the N6 B domain fragment, and the Fugu B domain. Constructs were tested under the control of the Spleen Focus Forming Virus (SFFV) promoter in the context of a self-inactivating (SIN) HIV-1 based lentiviral vector (LV). Furthermore, constructs were expressed from either a codon-optimized or non–codon-optimized cDNA sequence. Multiple transcriptional silencers and inhibitory motifs are widely distributed throughout the FVIII cDNA,4,6,20-22 and these sequences act as potent inhibitors of RNA production and protein formation, which can hamper expression in vivo. FVIII expression for all constructs was compared in vitro by transduction of 293T cells and in vivo by intravenous injection of vector into neonatal hemophilia A mice. The most significant observation was the effect of codon optimization on FVIII expression both in vitro and in vivo. Direct comparison of bioengineered FVIII constructs showed that significantly greater levels (up to a 44-fold increase and in excess of 200% normal human levels) of active FVIII protein were detected in the plasma of mice transduced with vector expressing FVIII from a codon-optimized cDNA sequence, successfully correcting the disease model. To date, this is the highest relative increase in FVIII expression after bioengineering of BDD FVIII, resulting in unprecedented, stable FVIII expression in vivo using a LV-based approach.
Methods
FVIII transgene and LV construction
The expression plasmid pMT2-FVIII was obtained as a kind gift from Dr Steven W. Pipe (University of Michigan); this plasmid contained the human FVIII gene with a Fugu B domain. The hFVIII gene had a B domain deletion from amino acids 740 to 1649 and an MluI restriction site (ACG-CGT) engineered by site-directed mutagenesis at amino acid positions 739 to 740, causing the missense mutation Pro739 to Thr739 in the a2 domain. The Fugu B domain had been cloned in using flanking MluI restriction sites on 5′ and 3′, creating a 4935-bp hFVIII Fugu B gene. The FVIII Fugu B gene was removed in 3 parts using a digest with XhoI and KpnI to remove a 1.83-kb fragment, a partial digest with KpnI and MluI to remove a 1.06-kb fragment, and polymerase chain reaction (PCR) amplification of the last 2.066-kb section using primers that created MluI and SbfI sites on the 5′ and 3′ ends, respectively. Each section was sequentially cloned into pLNT/SFFV-MCS using the same enzymes to create pLNT/SFFV-FVIII Fugu B. The construct was fully sequenced on completion. pLNT/SFFV-BDD FVIII was produced by digest of pLNT/SFFV-FVIII Fugu B with MluI to remove the Fugu B domain and religation. The 226/N6 B domain sequence was manufactured by GeneArt to produce a standard GeneArt plasmid containing 226/N6; pGA_N6_nonopt, the sequence was obtained by taking the first 678 bp of the human FVIII B domain (cDNA found at GenBank: A05328); 5′ and 3′ flanking MluI sites were then added. N6 was then removed from pGA_N6_nonopt and ligated into pLNT/SFFV-BDD FVIII using MluI to create pLNT/SFFV-FVIII N6. The SQ cDNA sequence was obtained23 and was modified to contain an MluI site (underlined) to give the SQm cDNA sequence: 5′-AGC-TTC-AGC-CAG-AAC-CCC-CCC-GTG-CTG-ACG-CGT-CAC-CAG-CGG-3′ (Figure 1B). LNT/SFFV-SQ FVIII Fugu B was produced by site-directed mutagenesis performed by Eurofins MWG Operon to add the flanking SQa and SQb (Figure 1B) sequences into the plasmid pLNT/SFFV-FVIII Fugu B to produce pLNT/SFFV-SQ FVIII Fugu B. pLNT/SFFV-SQ FVIII was then produced by removal of the Fugu B domain from pLNT/SFFV-SQ FVIII Fugu B by digest with MluI and religation. pLNT/SFFV-SQ FVIII N6 was produced by removal of the 226/N6 B domain from pGA_N6_nonopt by digestion with MluI and ligation into pLNT/SFFV-SQ FVIII. In this construct, there is a repeat of the 11 amino acid SQa sequence caused by the insertion of the N6 B domain into the SQm sequence. Codon-optimized sequences were created by analysis of the SQ FVIII Fugu B cDNA and adaption of the codon usage to the bias of Homo sapiens using codon adaptation index performed by GeneArt (GeneArt AG) using their in-house proprietary software GeneOptimizer. Optimization also removed cis-acting sequence motifs, including internal TATA-boxes, chi-sites and ribosomal entry sites, AT- or GC-rich sequence stretches, AU-rich elements, inhibitory and cis-acting repressor sequence elements, repeat sequences, RNA secondary structures, and all cryptic splice sites. The optimized gene was cloned into pLNT/SFFV-MCS to give the plasmid pLNT/SFFV-SQ FVIII Fugu B (co). The plasmid pLNT/SFFV-SQ FVIII (co) was created by digestion of pLNT/SFFV-SQ FVIII Fugu B (co) with MluI and religation. The 226/N6 B domain sequence from pGA_N6_nonopt was codon-optimized and manufactured by GeneArt. It was received in the plasmid pGA_N6_opt; and as the MluI restriction sites were maintained cloned directly into the pLNT/SFFV-SQ FVIII (co) plasmid to obtain the construct pLNT/SFFV-SQ FVIII N6 (co), again, this construct will contain an 11 amino acid SQa repeat sequence caused by the insertion of the B domain into the SQm sequence. Each construct was fully sequenced before testing.
Quantitative real-time PCR
As described previously,24 quantitative PCR was performed using an ABI 7700 Sequence Detection System (ABI, Applied Biosystems) using the following oligonucleotide primers and TaqMan probes: Total viral DNA was quantified using primers specific for the viral woodchuck hepatitis regulatory element (WPRE) sequence contained in the vector 5′-TGTGTGCCCGTCTGTTGTGT-3′ and 5′-GAGTCCTGCGTCGAGAGAGC-3′ and TaqMan probe (FAM) 5′-CGCCCGAACAGGGACTTGAA-3′ (TAMRA). The mouse titin gene (Ttn) was used as an endogenous 2-copy gene control for mouse cells and was quantified using primers 5′-AAAACGAGCAGTGACCTGAGG-3′ and 5′-TTCAGTCATGCTGCTAGCGC-3′ and TaqMan probe (FAM) 5′-TGCACGGAATCTCGTCTCAGTC-3′ (TAMRA). The human β-actin gene (ACTB) was used as an endogenous 2-copy gene control for 293T cells and was quantified using primers 5′-TCACCCACAAGTTGCCCATCTACGA-3′ and 5′-CAGCGGAACCGCTCATTGCCAATGG-3′ and TaqMan probe (FAM) 5′-ATGCCCTCCCCCATGCCATCCTGCGT-3′ (TAMRA).
LV production
Described previously,25,26 LVs were produced by transient cotransfection of HEK293T (293T) cells. A total of 107 cells were seeded in one 175-cm2 tissue culture flask overnight before transfection in Dulbecco modified Eagle medium (Invitrogen) containing GlutaMAX supplemented with 10% (vol/vol) heat-inactivated fetal calf serum (Invitrogen) and 1% (vol/vol) penicillin/streptomycin (herein referred to as “complete Dulbecco modified Eagle medium”). A total of 100 μg plasmid was used for transfection of one flask: 50 μg LV plasmid, 17.5 μg pMD.G2 (vesicular stomatitis virus glycoprotein envelope plasmid), and 32.5 μg pCMVΔ8.74 (gag-pol packaging plasmid, Plasmid Factory). Plasmid was precomplexed with 2.5nM polyethylenimine (Sigma-Aldrich) in OptiMEM for 20 minutes at room temperature. Transfection media was added to the cells for 3 to 4 hours and then replaced by 15 mL complete Dulbecco modified Eagle medium. Viral supernatant was harvested 48 and 72 hours after transfection and concentrated using ultracentrifugation (100 000g for 2 hours at 4°C). Aliquots of viruses were stored at −80°C.
Titration of LV
The physical titer in nanograms per microliter of all LVs was determined using a colorimetric reverse transcriptase (RT) enzyme-linked immunosorbent assay (ELISA) kit (Roche) according to the manufacturer's instructions. This value was used to determine an estimated vector titer in transducing units (TU) per milliliter by assuming that one vector particle contains 9.88 × 10−18 g of RT protein. Vector titers were also determined by quantitative PCR on genomic DNA from transduced 293T cells. Briefly, 1 × 105 293T cells were transduced with serial dilutions of vector, genomic DNA was extracted from transduced cells 72 hours after transduction using the ZR-96 Quick-gDNA (Zymo Research) as per the manufacturer's instructions, and quantitative PCR was carried out in triplicate on samples to determine both total viral DNA and human β-actin (ACTB). The number of vector genomes (vg) per transduced cell was normalized assuming 2 alleles ACTB per cell and titers in vector genomes per milliliter calculated as vector genome value × number of transduced cells/volume vector (milliliter).
Quantitative RT-PCR to determine FVIII mRNA expression
Quantitative RT-PCR was carried out on transduced 293T cells using the CellsDirect One-Step quantitative RT-PCR Kit with ROX (Invitrogen), including DNase I treatment, as per the manufacturer's instructions. Assays were run in triplicate using an ABI 7700 Sequence Detection System (ABI, Applied Biosystems) with oligonucleotides primer and TaqMan probe sets for WPRE and the human housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (both obtained from ABI, Applied Biosystems).
By directly comparing the cycle threshold (Ct) value obtained for the WPRE sequence (contained in all FVIII mRNA transcripts) relative to that for the housekeeping gene the expression levels of FVIII can be compared across samples using the ΔΔ Ct method.
Measurement of FVIII
The cofactor activity of blood plasma samples and in vitro cell culture media samples was assessed using the Biophen Factor VIII:C Chromagenic Assay (Biophen, Quadratech Diagnostics) as per the manufacturer's instructions. Samples were diluted 1:20 to 1:40 in sample diluent provided and analyzed in duplicate. A standard curve was constructed by diluting normal control plasma (Biophen, Quadratech Diagnostics) 1:20, carrying out 4 1:2 serial dilutions, and running in duplicate. Abnormal control plasma (Biophen, Quadratech Diagnostics) was also used as a further quality control for the assay. The FVIII antigen levels of cell culture media samples were determined using the Asserachrom VIII:Ag kit (Diagnostica Stago) as per the manufacturer's instructions. Samples were diluted 1:5 to 1:40 in sample diluent provided and analyzed in duplicate. Standard curves in percentage FVIII antigen activity were constructed by diluting normal control plasmas (Diagnostica Stago & Biophen) and Refacto (Wyeth), a commercially available SQ FVIII. FVIII activity was determined by a one-stage coagulation assay using FVIII deficient plasma on an ACL Futura Advance instrument (Instrumentation Laboratory) according to the manufacturer's protocol.
In vivo methods
All mice were handled according to procedures approved by the United Kingdom Home Office and the Imperial College London Research Ethics Committee. Hemophilia A mice (F8tm2Kaz) generated by deletion of exon 1727 were maintained on a 129SV background; 0- to 1-day-old neonatal mice were subject to brief (< 5 minutes) hypothermic anesthesia and 40 μL of concentrated LV (equivalent to 4 × 107 to 1 × 108 transducing units per mouse) injected into the superficial temporal vein. For coagulation factor assays, 100 μL of peripheral blood was collected from anesthetized mice by tail vein bleed. Blood was mixed immediately in a ratio of 1:9 with sodium citrate, centrifuged at 13 000g in a microcentrifuge for 5 minutes and plasma transferred to fresh microcentrifuge tube and stored at −20°C before assaying.
Statistical analysis
Data are expressed as mean values plus or minus SD. Statistical analyses were performed using a general linear model based on one-way analysis of variance with individual pairwise comparisons performed using Bonferroni simultaneous tests (Minitab 15 software; Minitab Inc).
Results
Generation of bioengineered FVIII variants and production of FVIII-expressing SIN LVs
To overcome low protein expression associated with hemophilia A gene transfer applications, we investigated the expression from bioengineered FVIII transgenes containing various B domain elements from codon-optimized or non–codon-optimized cDNA sequences. The following 9 FVIII variants were generated (Figure 1A; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article): BDD FVIII, encoding a human FVIII with a B domain deletion between amino acids 740 to 1649 and a missense mutation Pro739 to Thr739 in the a2 domain previously described by Miao et al17 ; FVIII Fugu B, encoding a human BDD FVIII in which the entire B domain from Fugu replaces the human B domain between amino acids 740 and 1649; FVIII N6, encoding a human BDD FVIII, which retains the N6 human B domain fragment containing 6 (N)-linked glycosylation sites between amino acids 740 and 164917 ; SQ FVIII, containing a modified 14-amino acid SQ activation peptide SQm between amino acids 740 and 1649 (SFSQNPPVLTRHQR; missense mutation Lys to Thr italicized), which contains the RHQR furin recognition sequence to increase intracellular cleavage, the original SQ activation peptide sequence (SFSQNPPVLKRHQR) was described by Sandberg et al23 ; SQ FVIII Fugu B, encoding a SQ FVIII with the Fugu B domain inserted into the SQm sequence. This causes the SQm sequence to be split either side of the B domain insert with the N-terminal sequence (SFSQNPPVLTR) is referred to as SQa, and the C-terminal sequence containing the furin recognition site RHQR as SQb (TRHQR); SQ FVIII N6, encoding the SQ FVIII with the 226 amino acid/N6 B domain inserted into the SQm sequence creating SQa and SQb sequences on the N- and C-terminal sides of the B domain, respectively. In this construct, there is a repeat of the 11 amino acid SQa sequence caused by the insertion of the N6 B domain into the SQm sequence. The effect that this repeat will have on FVIII secretion and function is unknown. Representation of SQ, SQm, SQa, and SQb is shown in Figure 1B.
Constructs SQ FVIII (co), SQ FVIII Fugu B (co), and SQ FVIII N6 (co) are identical in amino acid structure as constructs SQ FVIII, SQ FVIII Fugu B, and SQ FVIII N6, respectively, but are translated from codon-optimized cDNA sequences (Figure 1A). All 3 constructs share the identical codon-optimized SQ FVIII sequence. Codon optimization of SQ FVIII included the removal of 14 cryptic splice sites, an increase in GC-content from approximately 45% to approximately 60%, and an increase in codon adaptation index from 0.74 to 0.97. The optimized cDNA has 75.8% sequence identity at the nucleotide level to the original nonoptimized sequence and 907 of the 1457 amino acid codons were altered. In addition, a Kozak sequence was introduced to increase translation initiation, and 2 stop codons were added to ensure efficient termination. The Fugu B domain and the human FVIII N6 B domain sequences were codon-optimized in an identical fashion.
All constructs were cloned into a SIN lentiviral backbone under control of the SFFV promoter, and transgene sequences were confirmed by automated DNA sequencing. Vectors were produced for all 9 factor VIII constructs and tested for physical titer using the RT protein assay. They were then tested using quantitative PCR to determine an approximate titer in vector genomes per milliliter (Table 1). There was no substantial difference in titer between constructs.
Physical titer of FVIII vectors as determined by reverse transcriptase assay and quantitative PCR
| Virus . | Average reverse transcriptase, ng/μL . | Estimated titer, TU/mL . | Titer, vector genomes/mL . |
|---|---|---|---|
| BDD FVIII | 10.9 | 3.71 × 109 | 1.14 × 108 |
| FVIII Fugu B | 46.5 | 1.58 × 1010 | 1.58 × 109 |
| FVIII N6 | 30.7 | 1.04 × 1010 | 1.07 × 109 |
| SQ FVIII | 68.3 | 2.32 × 1010 | 2.91 × 109 |
| SQ FVIII Fugu B | 44.8 | 1.52 × 1010 | 1.18 × 109 |
| SQ FVIII N6 | 78.0 | 2.65 × 1010 | 2.0 × 109 |
| SQ FVIII (co) | 69.6 | 2.37 × 1010 | 4.45 × 109 |
| SQ FVIII Fugu B (co) | 71.8 | 2.40 × 1010 | 2.65 × 109 |
| SQ FVIII N6 (co) | 87.9 | 2.99 × 1010 | 3.39 × 109 |
| Virus . | Average reverse transcriptase, ng/μL . | Estimated titer, TU/mL . | Titer, vector genomes/mL . |
|---|---|---|---|
| BDD FVIII | 10.9 | 3.71 × 109 | 1.14 × 108 |
| FVIII Fugu B | 46.5 | 1.58 × 1010 | 1.58 × 109 |
| FVIII N6 | 30.7 | 1.04 × 1010 | 1.07 × 109 |
| SQ FVIII | 68.3 | 2.32 × 1010 | 2.91 × 109 |
| SQ FVIII Fugu B | 44.8 | 1.52 × 1010 | 1.18 × 109 |
| SQ FVIII N6 | 78.0 | 2.65 × 1010 | 2.0 × 109 |
| SQ FVIII (co) | 69.6 | 2.37 × 1010 | 4.45 × 109 |
| SQ FVIII Fugu B (co) | 71.8 | 2.40 × 1010 | 2.65 × 109 |
| SQ FVIII N6 (co) | 87.9 | 2.99 × 1010 | 3.39 × 109 |
Quantification of RT protein concentration in viral stocks, measured by performing an RT colorimetric assay, quantified in nanograms per microliter and estimated titer calculated from this. Data are mean values; n = 3. Quantification of titer was determined using quantitative PCR. A total of 1 × 105 293T cells were transduced with a serial dilution of vector; after 72 hours, genomic DNA was extracted from cells and quantitative PCR carried out for both WPRE and the human housekeeping gene β-actin. Data are mean values; n = 5.
Expression of FVIII in vitro
Relative FVIII protein expression was measured for each construct in the human embryonic kidney cell line 293T. Cells were transduced with an equal volume of vector and cultured for 48 hours, after which cells were washed and fresh serum-free media added for a further 24 hours. FVIII activity was assayed by one-stage clotting and chromogenic assays, and FVIII antigen was assayed by ELISA. Genomic DNA was then extracted from cells to determine viral copy number per cell by quantitative PCR and expression values normalized against copy number allowing accurate values for FVIII protein activity per gene copy to be determined (Figure 2). Total cellular RNA was also prepared and FVIII transgene expression quantified by RT real-time quantitative PCR analysis of the WPRE element present in all transcripts.
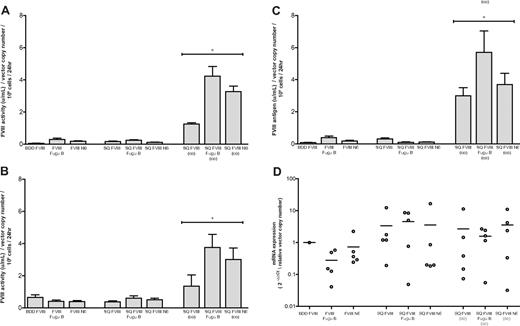
Relative human FVIII activity of FVIII constructs in vitro as determined by chromogenic assay. A total of 1 × 105 293T cells were transduced with 3 μL BDD FVIII, FVIII Fugu B, FVIII N6, SQ FVIII, SQ FVIII Fugu B, SQ FVIII N6, SQ FVIII (co), SQ FVIII Fugu B (co), or SQ FVIII N6 (co). At 48 hours, cell media was changed for 500 μL serum-free media. After a further 24 hours, incubation media was collected from all wells and assayed for factor VIII expression using (A) chromogenic-based assay, (B) a one-stage clotting assay to measure factor VIII cofactor activity, and (C) ELISA to measure FVIII antigen. Results were normalized on virus copy number per cell determined by quantitative PCR. Data are mean plus or minus SD; n = 5. *Statistical analyses were performed using general linear model based on 2-way analysis of variance with individual pairwise comparisons performed using Bonferroni simultaneous tests (Minitab 15 software). Results show a highly significant increase for SQ FVIII (co), SQ FVIII Fugu B (co), and SQ FVIII N6 (co) compared with their non–codon-optimized equivalents SQ FVIII, SQ FVIII Fugu B, and SQ FVIII N6, respectively (P < .0001). In addition, results for codon-optimized vectors also show a significant increase for SQ FVIII N6 (co) compared with SQ FVIII (co) (P < .0001) and a significant increase for SQ FVIII Fugu B (co) compared with both SQ FVIII (co) and SQ FVIII N6 (co) (P < .0001). (D) Transgene mRNA expression was quantified by quantitative RT-PCR, normalized to glyceraldehyde 3-phosphate dehydrogenase and compared with expression levels in cells transduced with BDD FVIII using the ΔΔ Ct method.
Relative human FVIII activity of FVIII constructs in vitro as determined by chromogenic assay. A total of 1 × 105 293T cells were transduced with 3 μL BDD FVIII, FVIII Fugu B, FVIII N6, SQ FVIII, SQ FVIII Fugu B, SQ FVIII N6, SQ FVIII (co), SQ FVIII Fugu B (co), or SQ FVIII N6 (co). At 48 hours, cell media was changed for 500 μL serum-free media. After a further 24 hours, incubation media was collected from all wells and assayed for factor VIII expression using (A) chromogenic-based assay, (B) a one-stage clotting assay to measure factor VIII cofactor activity, and (C) ELISA to measure FVIII antigen. Results were normalized on virus copy number per cell determined by quantitative PCR. Data are mean plus or minus SD; n = 5. *Statistical analyses were performed using general linear model based on 2-way analysis of variance with individual pairwise comparisons performed using Bonferroni simultaneous tests (Minitab 15 software). Results show a highly significant increase for SQ FVIII (co), SQ FVIII Fugu B (co), and SQ FVIII N6 (co) compared with their non–codon-optimized equivalents SQ FVIII, SQ FVIII Fugu B, and SQ FVIII N6, respectively (P < .0001). In addition, results for codon-optimized vectors also show a significant increase for SQ FVIII N6 (co) compared with SQ FVIII (co) (P < .0001) and a significant increase for SQ FVIII Fugu B (co) compared with both SQ FVIII (co) and SQ FVIII N6 (co) (P < .0001). (D) Transgene mRNA expression was quantified by quantitative RT-PCR, normalized to glyceraldehyde 3-phosphate dehydrogenase and compared with expression levels in cells transduced with BDD FVIII using the ΔΔ Ct method.
All constructs produced detectable FVIII activity using chromogenic and one-stage clotting assays (Figure 2A-B) and FVIII antigen by ELISA (Figure 2C). Cells transduced with constructs expressed from non–codon-optimized cDNA sequences produced on average 0.05 to 0.29, 0.37 to 0.65, and 0.08 to 0.38 U/mL/vector copy number/106 cells/24 hours as determined by chromogenic, one-stage clotting and antigen assays. There was no significant difference in expression of FVIII between equivalent constructs where the SQm, SQa, and SQb activation peptide sequences were present (BDD FVIII vs SQ FVIII, FVIII Fugu B vs SQ FVIII Fugu B, and FVIII N6 vs SQ FVIII N6; P > .05, Bonferroni simultaneous test). In addition, there was no significant increase in expression where the Fugu B or N6 B domains were present (FVIII Fugu B, SQ FVIII Fugu B, FVIII N6, and SQ FVIII N6) compared with SQ FVIII or BDD FVIII constructs (P > .05).
However, a highly significant increase in expression was observed with constructs expressed from codon-optimized cDNA sequences. Cells expressing SQ FVIII (co), SQ FVIII Fugu B (co), and SQ FVIII N6 (co) produced, 1.26 plus or minus 0.07, 4.22 plus or minus 0.61, and 3.27 plus or minus 0.34 U/mL/vector copy number/106 cells/24 hours, respectively, as assayed by the chromogenic assay, a 7- to 30-fold increase compared with expression from equivalent non–codon-optimized cDNA sequences (P < .0001). Equivalent FVIII antigen levels were produced at 1.64 plus or minus 0.28, 3.12 plus or minus 0.73, and 2.04 plus or minus 0.39 U/mL/vector copy number/106 cells/24 hours, for cells expressing SQ FVIII (co), SQ FVIII Fugu B (co), and SQ FVIII N6 (co), respectively, a 9- to 37-fold increase compared with expression from equivalent non–codon-optimized cDNA sequences (P < .0001; Figure 2C). A significant increase in expression was also observed from constructs SQ FVIII Fugu B (co) and SQ FVIII N6 (co) compared with SQ FVIII (co) (P < .0001); furthermore, the SQ FVIII Fugu B (co) had expression significantly higher than both SQ FVIII (co) and SQ FVIII N6 (co) (P < .0001; Figure 2A-C). The ratio of FVIII antigen to activity was 1.27 plus or minus 0.30 for all the constructs with the exception of SQ FVIII Fugu B (0.39 ± 0.08) and SQ FVIII (co) (2.35 ± 0.49). The discrepancy in antigen/activity ratio of SQ FVIII Fugu B is probably the result of an underestimate of FVIII antigen resulting from low expression levels and limiting sample. In contrast, the FVIII antigen and activity (both chromogenic and one-stage clotting) measurements of SQ FVIII (co) are extremely robust and suggest that the absence of a B domain in this variant may make it more susceptible to C-terminal proteolysis, thus leading to reduced activity relative to antigen.
Relative FVIII mRNA levels were also determined in vitro to establish whether B domain variation or codon optimization of cDNA sequences affects steady-state mRNA levels. Quantitative RT-PCR was used to evaluate FVIII mRNA transgene levels in cells transduced with the various constructs relative to those transduced with BDD FVIII. All constructs had detectable FVIII mRNA, with Ct values ranging from 24.41 to 31.59. After normalization to glyceraldehyde 3-phosphate dehydrogenase mRNA levels, no significant difference (P > .05, Bonferroni simultaneous test) in mRNA expression was observed from constructs expressed from a non–codon-optimized or codon-optimized cDNA sequence. Furthermore, there was no significant difference in FVIII mRNA levels per gene copy number between equivalent constructs where the SQm, SQa, and SQb activation peptide sequences were present (BDD FVIII vs SQ FVIII, FVIII Fugu B vs SQ FVIII Fugu B, and FVIII N6 vs SQ FVIII N6; P > .05).
Comparison of FVIII expression in vivo after intravenous delivery of vector into neonatal hemophilia A mice
SQ-containing FVIII expression cassettes were tested in vivo. Six constructs, SQ FVIII, SQ FVIII Fugu B, SQ FVIII N6, SQ FVIII (co), SQ FVIII Fugu B (co), and SQ FVIII N6 (co), were tested by direct intravenous injection of LV into neonatal (0- to 1-day-old) hemophiliac mice. All mice received between 4.72 × 107 and 1.78 × 108 vg with 6 to 10 mice injected per vector group. Blood plasma samples were collected via tail vein bleed approximately every 30 days for a total of approximately 250 days. FVIII activity was assessed using a functional chromogenic assay.
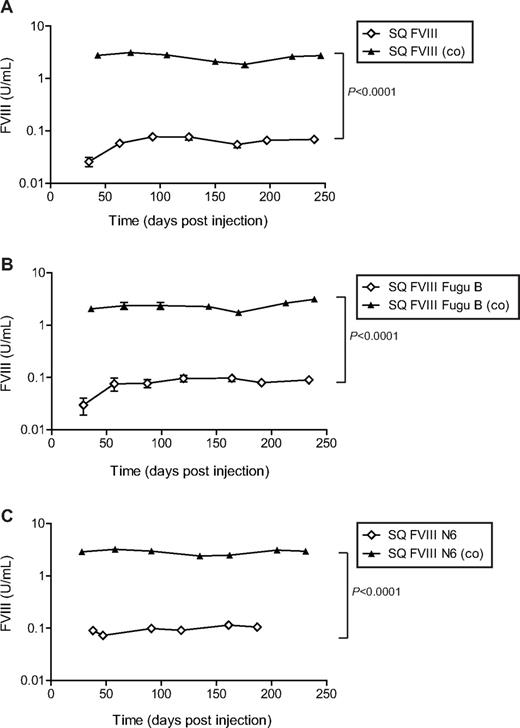
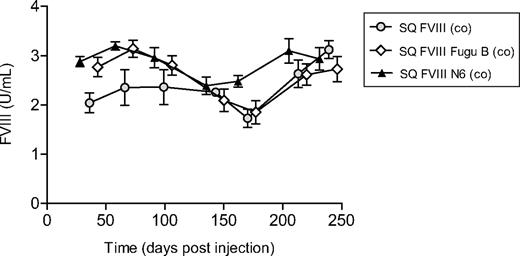
Functional FVIII was detected in the plasma of all transduced mice at all time points (Figure 3). Plasma from mice transduced with vector containing non–codon-optimized FVIII sequences; SQ FVIII, SQ FVIII Fugu B, or SQ FVIII N6 contained on average 0.05 plus or minus 0.02, 0.08 plus or minus 0.04, and 0.10 plus or minus 0.02 U/mL human FVIII activity, respectively, for the duration of the experiment. The ability to clot rapidly after tail vein bleeds indicated that the mice treated with sequences SQ FVIII Fugu B, or SQ FVIII N6 were able to achieve adequate hemostasis; however, 4 of the 6 mice injected in the SQ FVIII vector group did not survive, indicating that the levels of FVIII were insufficient to correct the murine hemophilia A phenotype. None of the other vector groups showed morbidity associated with low FVIII expression. For mice transduced with vector containing codon-optimized FVIII cDNA sequences, SQ FVIII (co), SQ FVIII Fugu B (co), or SQ FVIII N6 (co), average FVIII levels were detected at 2.56 plus or minus 0.63, 2.32 plus or minus 0.74, and 2.83 plus or minus 0.56 U/mL human FVIII activity, respectively, for the duration of the experiment. This is a 44-, 29-, and 29-fold increase in expression for SQ FVIII (co), SQ FVIII Fugu B (co), and SQ FVIII N6 (co), respectively, compared with expression from equivalent non–codon-optimized sequences (P < .0001, Bonferroni simultaneous test). Furthermore, no substantial loss in FVIII expression was observed in any vector groups. Importantly, no significant difference in expression was observed for constructs containing different B domain elements for vectors containing codon-optimized or non–codon-optimized cDNA sequences (Figure 4).
Expression of human FVIII activity in vivo in blood plasma of hemophiliac mice after intravenous injection of SIN LVs expressing bioengineered FVIII constructs. A total of 6 to 10 F8tm2Kaz hemophilic neonatal mice were injected intravenously via the superficial temporal vein with SIN LVs expressing bioengineered human FVIII constructs. Mice were bled at various time points over approximately 250 days, and a chromogenic assay was used to calculate the activity of human FVIII in blood plasma taken from each mouse. (A) SQ FVIII (◇) versus SQ FVIII (co) (▴). (B) SQ FVIII Fugu B (◇) versus SQ FVIII Fugu B (co) (▴). (C) SQ FVIII N6 (◇) versus SQ FVIII N6 (co) (▴). Points on graphs represent the mean; error bars represent SD. Statistical analyses were performed using general linear model based on 2-way analysis of variance with individual pairwise comparisons performed using Bonferroni simultaneous tests (Minitab software).
Expression of human FVIII activity in vivo in blood plasma of hemophiliac mice after intravenous injection of SIN LVs expressing bioengineered FVIII constructs. A total of 6 to 10 F8tm2Kaz hemophilic neonatal mice were injected intravenously via the superficial temporal vein with SIN LVs expressing bioengineered human FVIII constructs. Mice were bled at various time points over approximately 250 days, and a chromogenic assay was used to calculate the activity of human FVIII in blood plasma taken from each mouse. (A) SQ FVIII (◇) versus SQ FVIII (co) (▴). (B) SQ FVIII Fugu B (◇) versus SQ FVIII Fugu B (co) (▴). (C) SQ FVIII N6 (◇) versus SQ FVIII N6 (co) (▴). Points on graphs represent the mean; error bars represent SD. Statistical analyses were performed using general linear model based on 2-way analysis of variance with individual pairwise comparisons performed using Bonferroni simultaneous tests (Minitab software).
FVIII activity levels in vivo in plasma taken from mice injected with vector expressing FVIII from codon-optimized cDNA sequences. Activity of human FVIII in blood plasma taken from mice injected with LV expressing SQ FVIII (co) (gray circles), SQ FVIII Fugu B (co) (◇), and SQ FVIII N6 (co) (▴) collated. Points on graphs represent the mean; error bars represent the SD. No significant difference in expression is noted between constructs expressing different B domains (P > .5, Bonferroni simultaneous test).
FVIII activity levels in vivo in plasma taken from mice injected with vector expressing FVIII from codon-optimized cDNA sequences. Activity of human FVIII in blood plasma taken from mice injected with LV expressing SQ FVIII (co) (gray circles), SQ FVIII Fugu B (co) (◇), and SQ FVIII N6 (co) (▴) collated. Points on graphs represent the mean; error bars represent the SD. No significant difference in expression is noted between constructs expressing different B domains (P > .5, Bonferroni simultaneous test).
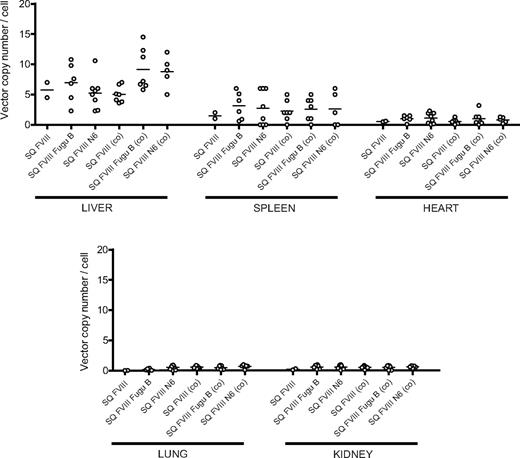
Analysis of viral copy number in the organs of transduced mice
From 187 and 246 days after injection, mice were killed to determine vector copy number in liver, spleen, heart, lung, and kidney tissue by real-time quantitative PCR (Figure 5). Vector genomes were detected predominantly in the liver and spleen tissue with negligible copies in heart, lung, and kidney tissues for all mice in all vector groups. Liver tissue taken from mice transduced with vector containing non–codon-optimized cDNA sequences contained an average of 5.75, 6.97, and 5.25 vector copies per cell for SQ FVIII, SQ FVIII Fugu B, and SQ FVIII N6, respectively. In spleen tissue, average copy number was 1.50, 3.13, and 2.75 copies per cell for SQ FVIII, SQ FVIII Fugu B, and SQ FVIII N6, respectively. There was no significant difference in the vector copy number detected in liver tissues of animals injected with vector containing codon-optimized sequences (P > .1, Bonferroni simultaneous test). Average copy number in liver tissue was detected at 5.04, 9.17, and 8.80 copies per cell, and in spleen tissue copy was 2.28, 2.57, and 2.60 copies per cell, for SQ FVIII (co), SQ FVIII Fugu B (co), and SQ FVIII N6 (co), respectively. In all cases, similar copy number was found in all tissues for all animals regardless of vector group.
Quantification of vector copy number in tissues of hemophiliac mice after intravenous injection of SIN LVs expressing bioengineered FVIII constructs. Liver, spleen, heart, lung, and kidney tissue were taken from mice killed at approximately 250 days after neonatal injection of LVs expressing SQ FVIII, SQ FVIII Fugu B, SQ FVIII N6, SQ FVIII (co), SQ FVIII Fugu B (co), and SQ FVIII N6 (co). Genomic DNA was extracted and viral copy number determined using quantitative PCR. Line represents the mean of all points. No significant difference in copy number was observed between any vector group (P > .1, Bonferroni simultaneous test).
Quantification of vector copy number in tissues of hemophiliac mice after intravenous injection of SIN LVs expressing bioengineered FVIII constructs. Liver, spleen, heart, lung, and kidney tissue were taken from mice killed at approximately 250 days after neonatal injection of LVs expressing SQ FVIII, SQ FVIII Fugu B, SQ FVIII N6, SQ FVIII (co), SQ FVIII Fugu B (co), and SQ FVIII N6 (co). Genomic DNA was extracted and viral copy number determined using quantitative PCR. Line represents the mean of all points. No significant difference in copy number was observed between any vector group (P > .1, Bonferroni simultaneous test).
Discussion
One of the significant limitations in the generation of efficient viral gene delivery systems for the treatment of hemophilia A by gene therapy is the large size of the FVIII cDNA (7053 nucleotides encoding 2351 amino acids). A number of bioengineered forms of human FVIII have been incorporated into gene transfer systems and have been shown to have enhanced expression both in vitro and in vivo.28 BDD factor VIII constructs are used widely in gene transfer experiments as there is no loss of FVIII procoagulant function and its smaller size (1443 vs 2351 amino acids) is more easily incorporated into vectors. A variation of this construct is a BDD FVIII containing the 14-amino acid link SQ between the A2 and A3 domains, currently produced as a recombinant product and marketed as Refacto (Wyeth).23 The SQ link has previously been shown to promote efficient intracellular cleavage of the primary single chain translation product of FVIII as it contains the intracellular furin recognition and cleavage site.23,29 This construct has been incorporated into plasmid vectors where it has conferred therapeutic levels of expression.30-32 A human BDD FVIII construct containing the first 226 amino acids of the B domain, including 6 N-linked asparagine glycosylation sites has been incorporated into many gene transfer vectors, including plasmid,33 LVs,34 and γ-retroviral vectors35 and is more efficiently secreted both in vitro17,35-37 and in vivo.17,37
The goal of this study was to investigate the effect of FVIII expression cassettes with various B domain constructs. These consist of SQ (14 amino acids), N6 (228 amino acids), and Fugu B (201 amino acids), a BDD FVIII construct containing the entire B domain from the puffer fish Fugu rubripes, which contains 10 N-linked asparagine glycosylation sites, which potentially would promote more efficient trafficking from the ER to the Golgi and therefore be more efficiently secreted. We also investigated the expression of these constructs from cDNA sequences, which had been codon-optimized for expression in Homo sapiens. Our study found that in vitro incorporation of B domain regions into constructs did not cause a significant increase in expression for non–codon-optimized constructs compared with their B domain deleted equivalents. However, a 7- to 30-fold increase in expression of functional factor VIII per integrated gene copy was observed from codon-optimized sequences. For codon-optimized sequences, significantly higher expression of both SQ FVIII Fugu B (co) and SQ FVIII N6 (co) was observed compared with SQ FVIII (co).
In vivo, after neonatal injection of a similar number of LV genomes the presence of a B domain did not significantly affect the steady-state levels of circulating FVIII activity for either codon-optimized or non–codon-optimized constructs. However, we observed a 29- to 44-fold increase in steady-state plasma levels of functional FVIII in hemophilia A mice to levels more than 200% of normal human FVIII expression from codon-optimized constructs compared with non–codon-optimized equivalents. Importantly, these levels of circulating FVIII were associated with a correction of the bleeding diatheses. In contrast, the levels of FVIII activity observed in mice treated with non–codon-optimized FVIII expression cassettes were associated with fatal hemorrhage after tail bleeds.
Multiple transcriptional silencers and inhibitory sequences are widely distributed throughout the FVIII cDNA,4,6,21,22 and the increased expression after codon optimization may be in part the result of the elimination of such sequences. However, deletion of the entire B domain, which led to a 17-fold increase in mRNA and primary translation product only resulted in a 30% increase in the levels of secreted protein, suggesting that the rate of ER-Golgi transport was reduced16 and that levels of FVIII mRNA were not limiting expression. The introduction of multiple N-linked glycosylation sites known to be important in ER-Golgi transport of FVIII increased levels of secreted FVIII, suggesting that the rate of ER-Golgi transport may be a rate-limiting step.17 However, a significant amount of FVIII within the ER never transits to the Golgi compartment resulting from a failure to fold correctly and misfolded FVIII accumulation in the ER can result in oxidative damage and apoptosis, perhaps suggesting that FVIII folding is the rate-limiting step in FVIII expression.36 Although protein secondary structure is determined primarily by the amino acid sequence, protein folding within the cell is affected by a range of factors: these include interaction with other proteins (chaperones) and ligands, translocation through the ER membrane and redox conditions. The rate of translation can also affect protein folding, and it has been suggested that codon usage may be a mechanism to regulate translation speed and thus allow stepwise folding of individual protein domains.38,39 FVIII is a complex multidomain protein in which nonsequential segments of the nascent polypeptide chain may interact in the three-dimensional fold. Ribosome stalling at “rare” codons may therefore lead to alternative folding pathways generating altered conformations and potentially misfolded protein. A potential explanation for the observed effect of codon-optimized sequences used in this study may be that they allow effcient translation and transport across the ER membrane, allowing the nascent FVIII polypeptide chain to fold correctly leading to the increased levels of secreted FVIII observed in vitro and in vivo.
Expression of more than 200% is not required in hemophilia patients, and production of such high levels of FVIII may be detrimental to producer cells.4,36 However, a major advantage of the optimized sequence is the ability to minimize the number of genetically modified cells needed to produce therapeutic levels, thereby reducing the risk of insertional mutagenesis and insertion site-dependent positional effects. In addition, the use of strong, ubiquitous promoter elements, such as SFFV, which were previously required to drive high expression of FVIII constructs, could be replaced by weaker, tissue-specific promoters, which are less prone to transcriptional silencing.33 Furthermore, these expression cassettes are only approximately 5 kb in size and are applicable for any viral (including AAV) or nonviral gene delivery system.
Previous in vivo studies have demonstrated expression of therapeutic levels of FVIII in vivo in adult hemophilia A mice after systemic injection of vector,34,40-42 transplantation of transduced bone marrow cells,33,35 transplantation of transduced bone marrow cells with targeted platelet-specific expression,43,44 and transplantation of transduced blood outgrowth endothelial cells.45 However, FVIII expression levels mediated from many of these approaches have been low (1%–5% normal human) and expression transient because of formation of neutralizing antibodies. In this study, we used a LV system to deliver the transgene to neonatal mice by an intravascular route where because of immune tolerance the formation of inhibitory antibodies or host immune responses against transduced cells would be extremely unlikely.46-48 This allowed us to directly compare FVIII expression from FVIII constructs containing various B domains from non–codon-optimized and codon-optimized cDNA sequences without the confounding effect of variable immune responses against human FVIII, neo epitopes and the Fugu B domain. The dramatic increase in the observed level of secreted FVIII from codon optimized cDNA sequences in this model system will now need to be validated in immune competent animals but open the possibility of development of safer, more efficacious vectors for gene therapy of hemophilia A.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Wellcome Trust (A.J.T.) and Medical Research Council (N.J.W.).
Wellcome Trust
Authorship
Contribution: N.J.W. designed and performed the research and wrote the paper; S.M.K.B. performed the research; S.N.W., A.C.N., and J.H.M. designed the research and wrote the paper; T.V.D., M.K.L.C., and J.M. contributed vital reagents; and E.G.D.T., C.K., and A.J.T. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John H. McVey, Molecular Medicine, Thrombosis Research Institute, Manresa Rd, London, SW3 6LR, United Kingdom; e-mail: jmcvey@tri-london.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal