Abstract
An important pathogenic event in Epstein-Barr virus (EBV)-associated lymphomas is the suppression of virus replication, which would otherwise lead to cell death. Because virus replication in B cells is intimately linked to their differentiation toward plasma cells, we asked whether the physiologic signals that drive normal B-cell differentiation are absent in EBV-transformed cells. We focused on BLIMP1α, a transcription factor that is required for plasma cell differentiation and that is inactivated in diffuse large B-cell lymphomas. We show that BLIMP1α expression is down-regulated after EBV infection of primary germinal center B cells and that the EBV oncogene, latent membrane protein-1 (LMP-1), is alone capable of inducing this down-regulation in these cells. Furthermore, the down-regulation of BLIMP1α by LMP-1 was accompanied by a partial disruption of the BLIMP1α transcriptional program, including the aberrant induction of MYC, the repression of which is required for terminal differentiation. Finally, we show that the ectopic expression of BLIMP1α in EBV-transformed cells can induce the viral lytic cycle. Our results suggest that LMP-1 expression in progenitor germinal center B cells could contribute to the pathogenesis of EBV-associated lymphomas by down-regulating BLIMP1α, in turn preventing plasma cell differentiation and induction of the viral lytic cycle.
Introduction
The Epstein-Barr virus (EBV) is a γ-herpesvirus that infects the majority of the world's adult population. In most persons, the virus is carried life-long as an asymptomatic infection where it establishes persistence by latently infecting memory B lymphocytes. However, in a minority of persons, EBV can contribute to the development of one of several B-cell malignancies, including Burkitt lymphoma (BL).1
In vitro infection of normal B cells with EBV gives rise to lymphoblastoid cell lines (LCLs) in which there is expression of a limited subset of latent virus genes that include the Epstein-Barr nuclear antigens (2, 3A, 3B, 3C, and LP) and the latent membrane proteins (LMP-1, and LMP-2).2 In contrast, most EBV-associated lymphomas show a more restricted pattern of latency; for example, in the majority of EBV+ BL, Epstein-Barr nuclear antigen-1 is the only viral protein expressed.3,4
As well as maintaining latency in B lymphocytes, the virus can also induce its replicative cycle in these cells. Thus, at any one time, a small proportion of cells in an LCL may spontaneously enter the lytic cycle or be induced to do so by treatment with chemical agents, such as phorbol esters, or by ligation of surface immunoglobulin (Ig).5
The replicative cycle of EBV is induced by expression of the immediate-early gene, BZLF1, which alone is sufficient to activate downstream lytic genes and complete viral replication in a permissive cell type.6,7 A number of studies suggest that EBV replicates in terminally differentiated plasma cells.8-12 These findings are supported by in vitro studies, which show that the BZLF1 promoter is active in memory cells only after they have been differentiated into plasma cells.12 The intimate association between terminal differentiation and EBV replication in B cells suggests that the switch from latency to the lytic cycle is controlled by factors that normally regulate plasma cell differentiation. We speculated that the absence of such factors could be important in the pathogenesis of EBV-associated lymphomas because virus replication would otherwise result in tumor cell death.
We have focused on BLIMP1, a transcription factor encoded by the PRDM1 gene, which orchestrates plasma cell differentiation by repressing genetic programs associated with the germinal center (GC) stages, while at the same time activating those programs associated with plasma cell functions.13,14 For example, the BLIMP1-mediated silencing of MYC, BCL6, and PAX5 expression has been shown to be required for plasma cell differentiation.15-19 BLIMP1 also induces IRF4 expression, which is an additional requirement of terminal B-cell differentiation.15,20 By turning off the expression of genes associated with cell-cycle progression and DNA synthesis (eg, MYC), BLIMP1 can promote the cell cycle exit that is characteristic of terminal differentiation.15 BLIMP1 can also apparently prime plasma cells for apoptosis by down-regulating the expression of antiapoptotic genes (eg, BCL2A1).15
The PRDM1 gene, which encodes BLIMP1, is inactivated in a subset of diffuse large B-cell lymphomas of the activated B-cell type where its loss is thought to contribute to lymphomagenesis by blocking post-GC B-cell differentiation.21,22 PRDM1 is expressed as 2 major isoforms, α and β; the latter lacks the amino-terminal acidic domain and part of the PR domain and is functionally impaired.23 In this study, we have investigated the possibility that the loss of BLIMP1α expression contributes to the pathogenesis of EBV+ GC-derived lymphomas.
Methods
Cell lines
LCLs were established by infection of GC B cells isolated from 3 separate donors with 2089 wild-type EBV and are referred to throughout as SL1-LCL, SL2-LCL, and SL3-LCL. OKU-LCL and SAL-LCL were generated by infection of B lymphocytes from an adult EBV-seronegative donor with virus rescued from atypical group I BL lines OKU and SAL lines (gift of Dr Gemma Kelly, University of Birmingham, Birmingham, United Kingdom).24 PER213 and HK-LCL were established using the reference EBV strain B95.8 and were a gift of Dr Heather Long (University of Birmingham, Birmingham, United Kingdom). BL2 and DG75 are EBV-negative BL cell lines. Akata is an EBV+ BL line. The B95.8 cell line was established from the peripheral blood lymphocytes of a cotton-top tamarin (Saguinus oedipus). All cell lines were cultured at 37°C in 5% CO2 in RPMI 1640 supplemented with 10% fetal calf serum, 2mM l-glutamine, and 1% penicillin-streptomycin solution (Sigma-Aldrich).
Isolation of tonsillar B cells
Fresh tonsils were obtained with informed consent from pediatric patients under local ethics committee approval (reference no. 06/Q2702/50) and transported in phosphate-buffered saline (PBS), pH 7.6, on ice. Tonsils were minced and mononuclear cells isolated by Ficoll-Isopaque centrifugation. CD10+ GC B cells were isolated by magnetic separation at 4°C with anti-CD10− phycoerythrin (eBioscience), antiphycoerythrin microbeads, and LS columns (both Miltenyi Biotec) following the protocol of the manufacturer. CD10+ cells were further separated into CD77+ and CD77− subpopulations using anti-CD77-FITC (BD Biosciences PharMingen). The purity of the isolated cells was investigated by FACS and shown to be > 95% with little or no contamination by plasma cells (data not shown).
Transfection
CD10+ GC B cells were cotransfected with either pcDNA3.1-LMP-1 (or in some cases, pSG5-LMP-1), pcDNA3.1-MYC (kindly provided by Georg Bornkamm), pcDNA3.1-PRDM1α, or corresponding empty vector together with pMACS LNGFR vector (Miltenyi Biotec) at a ratio of 7:3. Trans-fection was performed by nucleofection using the B Cell Nucleofector Kit B (Amaxa) for the Nucleofector II Device (Amaxa) using the U-15 program. Cells were cultivated overnight and stained with α-LNGFR-allophycocyanin (Miltenyi Biotec) and propidium iodide. Allophycocyanin-labeled and propidium iodide-negative cells were collected by FACS on a MoFlo sorter (Dako North America).
Cell lines were cotransfected with either pcDNA3.1-PRDM1α or empty pcDNA3.1 vector together with pMACS CD4.1 vector (Miltenyi Biotec) at ratios up to 9:1 by electroporation using the Bio-Rad Gene Pulser II electroporator (Bio-Rad). Cells were incubated for up to 72 hours at 37°C before they were pooled, and enriched as described previously.25
MYC knockdown
For knockdown experiments, DG75 and BL2 cells were transfected twice with smartpool siRNA targeting MYC or scrambled control siRNA by nucleofection with Cell Line Nucleofector Kit V (Amaxa). The siRNA were purchased from Dharmacon RNA Technologies. MYC smartpool sequences were: 5′-UCCAAGACGUUGUGUGUUCUU-3′, 5′-UUACGCACAAGAGUUCCGUUU-3′, 5′-UGUUGGUGAAGCUAACGUUUU-3′, and 5′-PUUCCACAGAAACAACAUCGUU-3′. After transfection, cells were cultivated in RPMI containing 10% fetal calf serum (Biochrom), 1% penicillin-streptomycin and glutamine (Sigma-Aldrich), and 25mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid at a concentration of 1 × 106 cells/mL (37°C in 5% CO2) for 3 hours, then diluted to a concentration of 0.5 × 106 cells/mL and cultivated for another 19 hours. Twenty-four hours after the first and second transfections, 1 × 106 cells were taken to monitor MYC knockdown by protein analysis and by transcript quantification.
Lytic cycle induction
For measurement of viral replication, Akata-BL cells were cotransfected with either pcDNA3.1-PRDM1α or empty pcDNA3.1 vector together with pMACS CD4.1 vector as described above and enriched. In control experiments, viral reactivation in Akata-BL cells was induced by cross-linking surface Ig. For cross-linking, cells were washed in complete media, resuspended at a concentration of 2 × 106 cells/mL, and exposed to α-IgG (10 μg/mL; Cappel) or α-IgM F(ab′)2 fragment (2 μg/mL; Jackson ImmunoResearch Laboratories). Cells were diluted to a concentration of 5 × 105 cells/mL after 24 hours and cultivated for a further 24 hours before harvesting. Immunohistochemistry was performed on treated Akata cells for detection of BZLF1 expression. A rabbit α-BLIMP1 (C14A4; Cell Signaling) was used to confirm expression of BLIMP1 in transfected cells. For detection of released EBV virions, an aliquot of culture supernatant was first treated with DNAse I (Turbo DNA-free, Ambion). Samples were then mixed with an equal volume of 2× lysis buffer (100mM KCl, 20mM Tris-HCl, pH 8.3, 3mM MgCl2, 0.5% Tween 20, and 400 μg/mL proteinase K) and digested for 1 hour at 55°C and finally heat inactivated for 10 minutes at 95°C. EBV DNA was quantified by quantitative PCR using primer/probe combinations to amplify EBV DNA polymerase (BALF5) and the cellular β2-microglobulin sequences.
Quantitative RT-PCR
RNA was extracted using RNeasy Mini Kit or Micro Kit, including removal of genomic DNA with RNase-Free DNase Set (QIAGEN). cDNA was generated in a reaction consisting of 400 ng (or less) of RNA, 250 ng of random primers (Promega United Kingdom), 10mM dNTP Mix (Roche Diagnostics), and SuperScript III Reverse Transcriptase (Invitrogen).
EBV gene expression was assayed by quantitative reverse-transcribed (RT)-PCR using the primer and probe combinations given in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) in a final reaction volume of 25 μL containing 1 × TaqMan universal PCR mastermix (Applied Biosystems), 7.5 pmol primers, 5 pmol probe, 0.5 μL of housekeeping assay (glyceraldehyde-3-phosphate dehydrogenase or β2-microglobulin; Applied Biosystems), and 5 μL cDNA (equivalent to required nanograms of input RNA). Thermal cycling conditions were: 2 minutes at 50°C, 12 minutes at 95°C, and 40 to 50 rounds of 15 seconds at 95°C and 1 minute at 60°C. Cellular gene transcripts were detected using the commercial assays (Applied Biosystems) detailed in supplemental Table 2. All test samples were run in triplicate, including template-negative reactions. The 2−ΔΔ CT method for quantifying expression relative to the housekeeping control was used; normalized values were expressed relative to one sample set to a value of 1. Results are reported as the mean of the 3 replicates. Unless otherwise stated, figures show the results of at least one of 3 independent experiments.
Immunohistochemistry
Cell lines were fixed in 10% formal saline and cytocentrifuged onto X-tra Adhesive slides (Surgipath Europe). Before staining, endogenous peroxidase activity was blocked in 0.3% hydrogen peroxide. After antigen retrieval (1mM EDTA in 0.1% Tween-20, pH 8.0; on a hot plate stirrer at 65°C for 16 hours), slides were incubated in primary antibody diluted in PBS-Tween-20 (0.1%) for 1 hour. Detection was performed with horseradish peroxidase-conjugated goat antirabbit or goat antimouse secondary IgG (Dako North America) for 30 minutes, followed by visualization in Vector NovaRED Substrate kit or diaminobenzidine (Vector Laboratories). Double immunohistochemistry was performed as previously described.26 Briefly, diluted primary antibody specific for second antigen was applied for 1 hour, followed by detection with horseradish peroxidase-conjugated goat antirabbit or goat antimouse secondary IgG (30 minutes) and visualization with Vector SG Substrate kit (Vector Laboratories). An antibody that recognizes all BLIMP1 isoforms was used in staining.26,27 Other antibodies used were directed to: BZLF1 (BZ-1, Santa Cruz Biotechnology28 ; 1:50); viral capsid antigen (VCA; clone L2, Chemicon; 1:1000); gp350 (72a.1, Millipore 29 ; 1:500), and HA (Y-11; 1:50, Santa Cruz Biotechnology). Photomicrographs were produced, using a Nikon Eclipse E400 microscope under immersion oil, captured using a Nikon DS-Fi1 digital camera, and a DS-5M-Ll Digital Sight system. Images were processed using PSPv7.
Immunoblotting
Cells were lysed in buffer consisting of 50mM Tris-HCl (pH 8.0), 150mM NaCl, 1mM EDTA, 1% Nonidet P40 (Roche Diagnostics) and protease inhibitors (Complete Protease Inhibitor Cocktail Tablets, Roche Diagnostics). Protein was quantified by Bio-Rad DC Protein Assay Kit (Bio-Rad). Laemmli sample buffer was added before SDS-PAGE, transfer on 0.45 μm nitrocellulose transfer membrane (Protan BA85 membrane; Schleicher & Schuell), and incubation at 4°C overnight with primary antibody diluted in 5% nonfat milk powder in PBS-Tween-20. Antibodies used were as follows: anti-LMP-1 (CS1–4; 1:1000, Dako North America), anti-c-MYC (9E10; 1:500, Santa Cruz Biotechnology), and anti-β-actin (c-2; 1:1000, Santa Cruz Biotechnology, Sc-8432). After a PBS-Tween-20 (0.1%) wash, blots were incubated for 30 minutes with HRP conjugated goat antimouse secondary IgG (Dako North America). Detection was with enhanced chemiluminescence (GE Healthcare).
Microarray analysis
CD10+ GC B cells were isolated from 2 donors and transfected as described in “Transfection”. RNA isolated from transfected cells was amplified with ExpressArt mRNA Nano Version and Nano plus Version Amplification Kits (AmpTec), biotin-labeled with an IVT labeling kit (Affymetrix), subjected to random fragmentation, and hybridized to Affymetrix HG-U133 Plus2 microarrays. Scanned images of microarray chips were analyzed using Affymetrix GeneChip Operating Software. The gene expression signal was calculated using the MAS5 algorithm of GeneChip Operating Software with the default settings, except that the target signal was set to 100. Gene expression profiles of BLIMP1α-transfected or LMP-1–transfected GC B cells were compared with that of the control pcDNA3.1–transfected GC B cells. Differentially expressed genes were identified using the GeneChip Operating Software pairwise analysis with the default settings. A gene was considered up-regulated or down-regulated if it was called “increased” or “decreased” in both replicates.
Identification of differential gene expression in B-cell subsets using a published dataset
A list of genes differentially expressed between plasma cells, memory cells, or both, compared with centrocytes, was derived from the GEO database a published dataset in which global gene expression was examined in different isolated human B-cell subsets (GSE12453).30
Results
Loss of BLIMP1α expression in EBV-transformed GC B cells
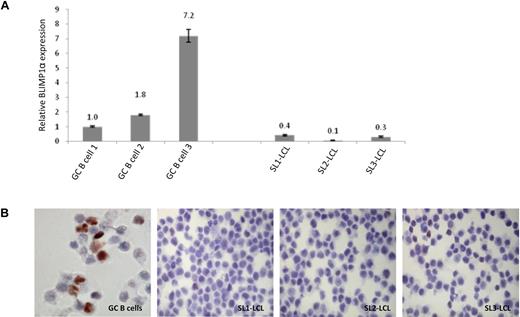
We first compared the expression of BLIMP1α in uninfected GC B cells with that in 3 EBV-transformed lymphoblastoid cell lines derived from GC B cells, which we had shown to be polyclonal in nature (data not shown). Figure 1A shows that, compared with GC B cells, BLIMP1α mRNA was lower in all 3 GC-derived LCLs; this down-regulation was confirmed at the protein level by immunohistochemistry (Figure 1B).
Differential expression of BLIMP1α in normal and transformed GC B cells. (A) Quantitative RT-PCR analysis shows that EBV infection of GC B cells was associated with the down-regulation of BLIMP1α transcription. Shown here are 3 LCLs (SL1-LCL, SL2-LCL, and SL3-LCL) established after the infection of GC B cells isolated from 3 separate donors. (B) Immunohistochemical analysis revealed strong BLIMP1 expression in a subpopulation of the uninfected GC B cells. In contrast, the LCLs showed homogeneously weaker BLIMP1 expression. Magnification ×450. Images acquired with 60×/1.40 NA oil immersion objective lens (Nikon Ltd).
Differential expression of BLIMP1α in normal and transformed GC B cells. (A) Quantitative RT-PCR analysis shows that EBV infection of GC B cells was associated with the down-regulation of BLIMP1α transcription. Shown here are 3 LCLs (SL1-LCL, SL2-LCL, and SL3-LCL) established after the infection of GC B cells isolated from 3 separate donors. (B) Immunohistochemical analysis revealed strong BLIMP1 expression in a subpopulation of the uninfected GC B cells. In contrast, the LCLs showed homogeneously weaker BLIMP1 expression. Magnification ×450. Images acquired with 60×/1.40 NA oil immersion objective lens (Nikon Ltd).
The EBV-encoded LMP-1 suppresses BLIMP1α expression in GC B cells
Having shown that EBV infection of GC B cells is followed by the down-regulation of BLIMP1α, we next studied the influence of the EBV oncogene LMP-1 on BLIMP1α expression in primary human GC B cells. To do this, we transfected GC B cells with an LMP-1 expression vector together with a truncated nerve growth factor receptor (LNGFR). Transfected cells were sorted on the basis of LNGFR expression as we have previously described.31 Figure 2B shows that, in GC B cells derived from 5 separate donors, BLIMP1α mRNA expression was decreased after the expression of LMP-1. Because approximately 15% to 20% of CD10+ GC B cells are BLIMP1α+, we repeated these experiments in the centrocyte-rich CD77− GC B cells.32,33 We showed that LMP-1 also down-regulated BLIMP1α expression in both the CD77− and CD77+ GC B-cell fractions (Figure 2C). Additional evidence for the repression of BLIMP1α by LMP-1 was provided by our immunohistochemical analysis of the B95.8 cell line, an LCL derived from the B-lymphocytes of a cotton-top tamarin. Although B95.8 cells are unusual insofar as they express both BLIMP1α and LMP-1, we found that the expression of these 2 proteins was mutually exclusive in these cells (Figure 2D).
LMP-1 down-regulates BLIMP1α expression in primary GC B cells and transformed B cells. (A) Ectopic expression of LMP-1 in GC B cells was confirmed by Western blotting or was published before.31 (B) Quantitative RT-PCR analysis of BLIMP1α mRNA in CD10+ GC B cells from 5 separate donors after transfection with LMP-1 (pSG5-LMP-1 or pcDNA3.1-LMP-1) or vector control only (pSG5 or pcDNA3.1); LMP-1 down-regulated BLIMP1α mRNA in all samples. (C) LMP-1 also down-regulated BLIMP1α mRNA in both CD77− (centrocytes [CCs]) and CD77+ (centroblasts [CBs]) subsets of CD10+ cells. (D) Immunohistochemistry revealed that the expression of BLIMP1α (gray) and LMP-1 (red) were mutually exclusive in B95.8 cells. Magnification ×800. Images acquired with 60×/1.40 NA oil immersion objective lens (Nikon Ltd).
LMP-1 down-regulates BLIMP1α expression in primary GC B cells and transformed B cells. (A) Ectopic expression of LMP-1 in GC B cells was confirmed by Western blotting or was published before.31 (B) Quantitative RT-PCR analysis of BLIMP1α mRNA in CD10+ GC B cells from 5 separate donors after transfection with LMP-1 (pSG5-LMP-1 or pcDNA3.1-LMP-1) or vector control only (pSG5 or pcDNA3.1); LMP-1 down-regulated BLIMP1α mRNA in all samples. (C) LMP-1 also down-regulated BLIMP1α mRNA in both CD77− (centrocytes [CCs]) and CD77+ (centroblasts [CBs]) subsets of CD10+ cells. (D) Immunohistochemistry revealed that the expression of BLIMP1α (gray) and LMP-1 (red) were mutually exclusive in B95.8 cells. Magnification ×800. Images acquired with 60×/1.40 NA oil immersion objective lens (Nikon Ltd).
Differential regulation of BLIMP1α target genes by LMP-1 in GC B cells
Having shown that LMP-1 could down-regulate BLIMP1α in GC B cells, we next investigated whether LMP-1 could also modulate the expression of BLIMP1α target genes. We focused on 3 B cell-associated transcription factors, MYC, BCL6, and PAX5, which are repressed, and one, IRF4 which is up-regulated by BLIMP1α. The silencing of MYC, BCL6, and PAX5 and the up-regulation of IRF4 are necessary for plasma cell differentiation. After their separate transfection into primary GC B cells, both BLIMP1α and LMP-1 were shown to down-regulate BCL6 and PAX5 and to up-regulate IRF4. However, whereas MYC was down-regulated by BLIMP1α, it was up-regulated by LMP-1 in GC B cells (Figure 3).
Differential regulation of BLIMP1α target genes by LMP-1 in GC B cells. Quantitative RT-PCR was used to measure the relative quantity of BCL6, PAX5, IRF4, and MYC mRNA in LMP-1-expressing GC B cells or BLIMP1α-expressing GC B cells presented as 2−ΔΔ CT values compared with vector only-transfected GC B cells. The expression of BCL6 and PAX5 was down-regulated, and that of IRF4 up-regulated by both LMP-1 and BLIMP1α. In contrast, whereas MYC was down-regulated by BLIMP1α, it was up-regulated by LMP-1.
Differential regulation of BLIMP1α target genes by LMP-1 in GC B cells. Quantitative RT-PCR was used to measure the relative quantity of BCL6, PAX5, IRF4, and MYC mRNA in LMP-1-expressing GC B cells or BLIMP1α-expressing GC B cells presented as 2−ΔΔ CT values compared with vector only-transfected GC B cells. The expression of BCL6 and PAX5 was down-regulated, and that of IRF4 up-regulated by both LMP-1 and BLIMP1α. In contrast, whereas MYC was down-regulated by BLIMP1α, it was up-regulated by LMP-1.
LMP-1 partially disrupts the BLIMP1α transcriptional program in GC B cells
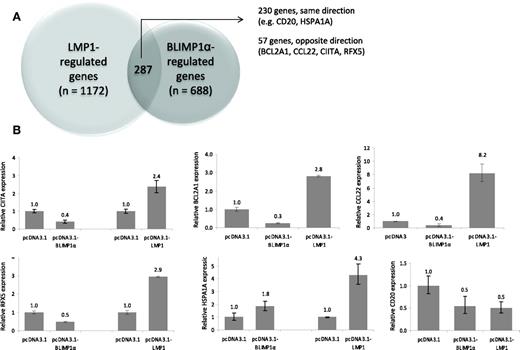
Having shown that the down-regulation of BLIMP1α by LMP-1 in GC B cells was accompanied by the aberrant induction of MYC, we next investigated the influence of LMP-1 on the BLIMP1α transcriptional program using genome-wide expression profiling. Transfection of GC B cells with BLIMP1α was followed by the up-regulation of 321 genes and the down-regulation of 654 genes. As anticipated, the BLIMP1α transcriptional program in GC B cells was found to overlap substantially with that associated with plasma cell differentiation (supplemental Figure 1). Having defined BLIMP1α target genes, we next transfected GC B cells from the same donors with LMP-1; this was followed by the up-regulation of 365 genes and the down-regulation of 1094 genes. When these datasets were compared, 230 genes were found to be concordantly regulated by LMP-1 and BLIMP1α (Figure 4A). However, 57 genes were found to be down-regulated by BLIMP1α and up-regulated by LMP-1, or vice versa (these data are available in GEO database number GSE27670). Quantitative RT-PCR was used to confirm the transcriptional changes in selected genes. For example, we confirmed that HSPA1A and CD20 were concordantly regulated by LMP-1 and BLIMP1α, and that BCL2A1, CIITA, CCL22, and RFX5 were down-regulated by BLIMP1α but up-regulated by LMP-1 (Figure 4B). We conclude that LMP-1 can partially disrupt the BLIMP1α transcriptional program in GC B cells.
LMP-1 partially disrupts the BLIMP1α transcriptional program in GC B cells. (A) Genome-wide expression profiling revealed that BLIMP1α expression in GC B cells was followed by the up-regulation of 321 genes and the down-regulation of 654 genes. LMP-1 expression was followed by the up-regulation of 365 genes and the down-regulation of 1094 genes. A total of 230 genes were found to be concordantly regulated by LMP-1 and BLIMP1α. However, 57 genes were found to be down-regulated by BLIMP1α and up-regulated by LMP-1, or vice versa (B) Validation of genes from the microarray analysis. LMP-1 expression in GC B cells resulted in the up-regulation of a number of genes repressed by BLIMP1α, including CIITA, BCL2A1, RFX5, and CCL22. Both LMP-1 and BLIMP1α down-regulated CD20 and up-regulated HSPA1A.
LMP-1 partially disrupts the BLIMP1α transcriptional program in GC B cells. (A) Genome-wide expression profiling revealed that BLIMP1α expression in GC B cells was followed by the up-regulation of 321 genes and the down-regulation of 654 genes. LMP-1 expression was followed by the up-regulation of 365 genes and the down-regulation of 1094 genes. A total of 230 genes were found to be concordantly regulated by LMP-1 and BLIMP1α. However, 57 genes were found to be down-regulated by BLIMP1α and up-regulated by LMP-1, or vice versa (B) Validation of genes from the microarray analysis. LMP-1 expression in GC B cells resulted in the up-regulation of a number of genes repressed by BLIMP1α, including CIITA, BCL2A1, RFX5, and CCL22. Both LMP-1 and BLIMP1α down-regulated CD20 and up-regulated HSPA1A.
MYC represses BLIMP1α in transformed and untransformed GC B cells
We next addressed the mechanism by which LMP-1 down-regulates BLIMP1α expression in GC B cells. We considered the possibility that the up-regulation of MYC by LMP-1 might be responsible for the down-regulation of BLIMP1α. This seemed a reasonable proposition given that BLIMP1 is known to form reciprocal regulatory loops with other transcription factors involved in B-cell differentiation, including BCL6, PAX5, and IRF4, and also that MYC can suppress plasma cell differentiation by an undefined mechanism. To investigate whether MYC regulates BLIMP1α, we transfected MYC into primary GC B cells and measured BLIMP1α expression. In GC B cells isolated from 3 separate donors, ectopic MYC expression led to the robust down-regulation of BLIMP1α expression. Figure 5A-B shows a representative experiment. To further substantiate these findings, we used siRNA to knock down the expression of MYC in 2 BL cell lines, BL2 and DG75 (Figure 5C). Figure 5D shows that in both cell lines the knock-down of MYC led to the up-regulation of BLIMP1α expression. We conclude that MYC can repress BLIMP1α expression in GC B cells. Given that we had shown that MYC is up-regulated by LMP-1 in GC B cells, our observations provide a plausible explanation for the down-regulation of BLIMP1α by LMP-1 as well as potential new mechanism to explain, at least in part, the impaired differentiation that is characteristic of B-cell lymphomas harboring MYC abnormalities.
MYC represses BLIMP1α in transformed and untransformed GC B cells. (A) Representative Western blot for MYC and β-actin after the transfection of primary human GC B cells with MYC expression vector or control. (B) Quantitative RT-PCR for BLIMP1α in GC B cells after ectopic MYC expression (one representative experiment of 3 is shown). (C) RNAi-mediated knockdown of MYC in BL2 and DG75 BL lines using smartpool siRNA targeting MYC or scrambled control siRNA. (D) MYC knockdown led to the up-regulation of BLIMP1α in both BL lines.
MYC represses BLIMP1α in transformed and untransformed GC B cells. (A) Representative Western blot for MYC and β-actin after the transfection of primary human GC B cells with MYC expression vector or control. (B) Quantitative RT-PCR for BLIMP1α in GC B cells after ectopic MYC expression (one representative experiment of 3 is shown). (C) RNAi-mediated knockdown of MYC in BL2 and DG75 BL lines using smartpool siRNA targeting MYC or scrambled control siRNA. (D) MYC knockdown led to the up-regulation of BLIMP1α in both BL lines.
BLIMP1α induces the EBV lytic cycle in LCL
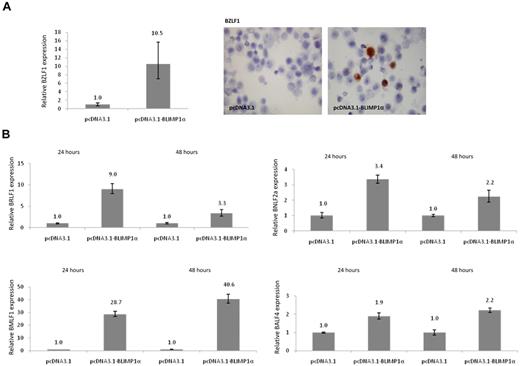
Given that EBV replication in B cells appears to be intimately linked to plasma cell differentiation, we next investigated whether BLIMP1α expression could induce the lytic cycle in EBV-transformed cells. To do this, 5 LCLs, including the GC-derived LCLs, were transfected with a BLIMP1α expression vector or vector only as a control. We observed that, in all cell lines, the expression of BLIMP1α was followed by the induction of BZLF1 mRNA, elevated BZLF1 promoter activity (data not shown), and increased numbers of BZLF1-expressing cells (Figure 6A), which correlated with the number of transfected cells as measured by immunohistochemistry for the HA tag (Table 1). BLIMP1α also induced the expression of other lytic genes, including the immediate-early viral gene BRLF1, the early lytic genes BNLF2a and BMLF1, and the late lytic gene BALF4 (Figure 6B). Consistent with previous reports suggesting that viral replication is abortive in most plasma cells, we observed that the late lytic proteins, gp350/220 and VCA were expressed in only a fraction of transfected cells.34 For example, in OKU-LCL, VCA expression could be detected in only 0.16% of BLIMP1α-expressing cells (data not shown). Additional evidence for the induction of the lytic cycle by BLIMP1α was provided by our immunohistochemical analysis of the B95.8 cell line, which has already been described; and which unusually expresses both BLIMP1α and the late viral genes.35 We found that, whereas the late lytic proteins VCA and gp350 are rarely expressed in BLIMP1α− B95.8 cells (0.4% and 0.6%, respectively), they are commonly expressed in BLIMP1α+ B95.8 cells (36.9% and 55.5%, respectively; supplemental Figure 2).
BLIMP1α up-regulates BZLF1 expression and induces lytic cycle in LCLs. (A) Quantitative RT-PCR analysis of BZLF1 mRNA levels in OKU-LCL after transfection with BLIMP1α or empty vector. In all transfected LCLs (n = 5, Table 1), the expression of BLIMP1α was followed by the up-regulation of BZLF1 mRNA. (Left panel) Immunostaining revealed that ectopic BLIMP1α expression increased the frequency of BZLF1-expressing cells. (B) Ectopic expression of BLIMP1α led to the increased expression at 24 and 48 hours of BRLF1, and of the lytic cycle genes, BMLF1, BNLF2a, and BALF4. Shown here are results of OKU-LCL; similar results were obtained after transfection of SAL-LCL and B95.8 cells (data not shown). Image is at magnification of ×450, and was acquired with 60×/1.40 NA oil immersion objective lens (Nikon Ltd).
BLIMP1α up-regulates BZLF1 expression and induces lytic cycle in LCLs. (A) Quantitative RT-PCR analysis of BZLF1 mRNA levels in OKU-LCL after transfection with BLIMP1α or empty vector. In all transfected LCLs (n = 5, Table 1), the expression of BLIMP1α was followed by the up-regulation of BZLF1 mRNA. (Left panel) Immunostaining revealed that ectopic BLIMP1α expression increased the frequency of BZLF1-expressing cells. (B) Ectopic expression of BLIMP1α led to the increased expression at 24 and 48 hours of BRLF1, and of the lytic cycle genes, BMLF1, BNLF2a, and BALF4. Shown here are results of OKU-LCL; similar results were obtained after transfection of SAL-LCL and B95.8 cells (data not shown). Image is at magnification of ×450, and was acquired with 60×/1.40 NA oil immersion objective lens (Nikon Ltd).
Increased frequency of BZLF1-expressing cells following expression of BLIMP1α in LCL
| Cell line . | BZLF1+ cells in vector only-transfected cells, % . | BZLF1+ cells in BLIMP1α-transfected cells, % . | HA-tag+ cells in BLIMP1α-transfected cells, % . |
|---|---|---|---|
| HK-LCL | 3.56 | 8.12 | 6.96 |
| OKU-LCL | 1.02 | 7.14 | 11.36 |
| SL1-LCL | 0.0 | 6.32 | 9.9 |
| PER213 | 3.02 | 5.68 | 2.25 |
| SAL-LCL | 1.22 | 2.55 | 4.13 |
| Akata-BL | 0.12 | 4.31 | 5.12 |
| Cell line . | BZLF1+ cells in vector only-transfected cells, % . | BZLF1+ cells in BLIMP1α-transfected cells, % . | HA-tag+ cells in BLIMP1α-transfected cells, % . |
|---|---|---|---|
| HK-LCL | 3.56 | 8.12 | 6.96 |
| OKU-LCL | 1.02 | 7.14 | 11.36 |
| SL1-LCL | 0.0 | 6.32 | 9.9 |
| PER213 | 3.02 | 5.68 | 2.25 |
| SAL-LCL | 1.22 | 2.55 | 4.13 |
| Akata-BL | 0.12 | 4.31 | 5.12 |
Immunohistochemistry was used to measure the frequency of BZLF1-expressing cells in BLIMP1α-transfected cell lines. At least 1000 cells were counted in each sample. Staining for the HA tag provided an estimate of the transfection efficiency.
BLIMP1α induces the EBV lytic cycle in Burkitt lymphoma cells
Finally, we investigated whether BLIMP1α could also induce the viral lytic cycle in EBV+ Akata BL cells. We observed that the ectopic expression of BLIMP1α induced transcription of the immediate early genes BZLF1 and BRLF1 as well as that of BNLF2a, BMLF1, and BALF4 (Figure 7A). However, in contrast to the LCL, immunohistochemistry revealed that VCA was expressed in 70.6% of BLIMP1α-expressing Akata cells and gp350 in 52.1% (Figure 7B). We repeated this experiment, but this time we measured virus genome replication after the ectopic expression of BLIMP1α expression and after BCR cross-linking. We found that the EBV genome copy number in supernatants of Akata cells (as measured by quantitative PCR after DNase I treatment) was increased in both BLIMP1α-transfected cells (6.03 × 104/mL; SD, 6.18 × 103) and after BCR cross-linking (6.31 × 105/mL; SD, 1.09 × 104), compared with vector-only transfected cells (3.00 × 104/mL; SD, 4.53 × 103). The use of DNase I discriminates encapsidated (ie, virion DNA), which is resistant to this treatment, from naked EBV DNA. The lower viral load in the BLIMP1α-transfected cells compared with that observed after BCR cross-link could represent either differences in the kinetics of BLIMP1α-induced viral replication or simply the consequence of the low number of BLIMP1α-expressing cells observed after transfection. We conclude that BLIMP1α can induce the viral lytic cycle in EBV+ BL cells and is sufficient for virus genome replication and virion release.
BLIMP1α induces EBV lytic cycle in Akata BL cells. (A) Quantitative RT-PCR demonstrates induction of BZLF1, BRLF1, BNLF2a, BMLF1, and BALF4 mRNA expression after BLIMP1α transfection into Akata BL cells. (B) Immunohistochemistry shows costaining for BLIMP1α HA tag (gray) and VCA or gp350 (red) in transfected Akata cells. Representative examples of double-positive cells taken from the same experiment are shown. Magnification ×800. Images acquired with 60×/1.40 NA oil immersion objective lens (Nikon Ltd).
BLIMP1α induces EBV lytic cycle in Akata BL cells. (A) Quantitative RT-PCR demonstrates induction of BZLF1, BRLF1, BNLF2a, BMLF1, and BALF4 mRNA expression after BLIMP1α transfection into Akata BL cells. (B) Immunohistochemistry shows costaining for BLIMP1α HA tag (gray) and VCA or gp350 (red) in transfected Akata cells. Representative examples of double-positive cells taken from the same experiment are shown. Magnification ×800. Images acquired with 60×/1.40 NA oil immersion objective lens (Nikon Ltd).
Discussion
In this study, we have shown that BLIMP1α, a key regulator of plasma cell differentiation, is down-regulated in primary GC B cells by the EBV oncogene LMP-1. Consistent with a role for LMP-1 in hijacking the B-cell transcriptional program, we found a striking overlap between the LMP-1 and BLIMP1α transcriptional programs in GC B cells. Although we have not investigated this further, the commonality between the transcriptional programs of BLIMP1α and LMP-1 is probably best explained by their concordant regulation of transcription factors, including BCL6, IRF4, and PAX5, all of which are important in regulating post-GC B-cell differentiation.15-20,36,37 However, we observed that the down-regulation of PAX5 and BCL6 that followed the ectopic expression of both LMP-1 and BLIMP1α in GC B cells, although reproducible, was relatively modest compared with the effects on MYC. These observations suggest that BLIMP1α and LMP-1 are alone insufficient to mediate the complete repression of PAX5 and BCL6 in GC B cells. This is consistent with previous studies showing that the repression of PAX5 that occurs during plasma cell differentiation is dependent not only on BLIMP1α but also on the presence of other signals, which suppress PAX5 independently of BLIMP1α.32 In particular, it has been shown that the repression of PAX5 and BCL6 that occurs early after the initiation of plasma cell differentiation in preplasmablasts does not require BLIMP1α.32 Our analysis also revealed other genes that were repressed by BLIMP1α but induced by LMP-1, including several genes that are known to be down-regulated during plasma cell differentiation (eg, BCL2A1, CIITA38,39 ) as well as MYC, the suppression of which has been shown to be essential for plasma cell differentiation.17 Our findings are consistent with the possibility that, although LMP-1 can drive the B-cell differentiation program, its down-regulation of BLIMP1α prevents plasma cell differentiation.
When interpreting the results of the LMP-1 transfection experiments, it is important to remember that in unsorted GC B cells the proportion of BLIMP1α+ cells is only approximately 15% to 20% (our own immunohistochemistry data).32,33 Therefore, one possible explanation of our data is that LMP-1 preferentially induces the death of BLIMP1-expressing GC B cells. Were this the case, then we might have expected LMP-1 to reduce the viability of GC B cells accordingly. However, we consistently observed no change in the viability of LMP-1-expressing GC B cells compared with empty vector-transfected cells (supplemental Figure 3). Therefore, we think that this explanation is unlikely.
Although we observed that BLIMP1α was also down-regulated after the infection of GC B cells with EBV, an alternative interpretation of these data is that infection of BLIMP1α+ cells results in viral replication and cell death, leading to the selective immortalization of BLIMP1α− GC B cells. It has been shown previously that the infection of BLIMP1+ multiple myeloma cells does not lead to cell death but to the down-regulation of BLIMP1 and to a partial reprogramming of these cells to a mature B-cell phenotype.40 However, it remains to be established whether EBV infection of BLIMP1-expressing GC B cells leads to a similar outcome.
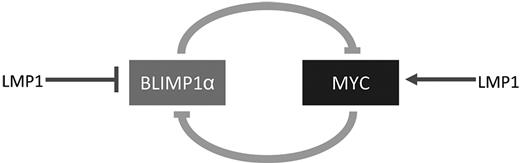
Inactivation of the PRDM1 gene encoding BLIMP1 has been detected in diffuse large B-cell lymphoma of the activated B-cell type.21,22 Translocations deregulating the BCL6 gene have not been found in BLIMP1-mutated diffuse large B-cell lymphoma but are restricted to unmutated cases, suggesting that BCL6 deregulation and BLIMP1 inactivation might represent alternative pathogenic mechanisms, both leading to a block in post-GC differentiation and, ultimately, to lymphomagenesis.21 Our observations suggest that LMP-1 expression in progenitor GC B cells might provide an alternative mechanism to block terminal B differentiation in EBV+ lymphomas. Because the repression of MYC is required for terminal B-cell differentiation,17 one possibility is that LMP-1 antagonizes plasma cell differentiation by up-regulating MYC, which then represses plasma cell differentiation by suppressing BLIMP1α. This not only provides a plausible explanation for the down-regulation of BLIMP1α by LMP-1 but also a potentially novel mechanism, which might explain the impaired differentiation characteristic of B-cell lymphomas harboring MYC abnormalities. Alternatively, it is possible that LMP-1 initially drives the down-regulation of BLIMP1α, which then leads to the up-regulation of MYC. In either case, LMP-1 would drive a reciprocal regulatory loop involving BLIMP1α and MYC, which would ultimately lead to the activation of MYC and the repression of BLIMP1α. Figure 8 shows a schematic illustration of the proposed regulation of this loop by LMP-1.
Schematic illustration of the potential regulation by LMP-1 of a reciprocal regulatory loop involving BLIMP1α and MYC. LMP-1 might act initially to down-regulate BLIMP1α, which would release the BLIMP1α-mediated suppression of MYC. The up-regulation of MYC would lead to further suppression of BLIMP1α. Alternatively, LMP-1 might initially activate MYC expression.
Schematic illustration of the potential regulation by LMP-1 of a reciprocal regulatory loop involving BLIMP1α and MYC. LMP-1 might act initially to down-regulate BLIMP1α, which would release the BLIMP1α-mediated suppression of MYC. The up-regulation of MYC would lead to further suppression of BLIMP1α. Alternatively, LMP-1 might initially activate MYC expression.
The failure to complete plasma cell differentiation in EBV+ B cells also has implications for the viral life cycle. We observed that the reexpression of BLIMP1α in EBV-transformed B cells and in EBV+ BL cells induced the viral lytic cycle. For herpesviruses, viral replication leading to the production of infectious virions ultimately results in cell death. Therefore, both terminal B-cell differentiation and lytic replication are probably incompatible with the transformed state. Thus, the ability of BLIMP1α to activate the EBV lytic cycle would seem to represent a hitherto undescribed tumor suppressor function for BLIMP1α in the context of EBV-associated lymphomas. However, we have yet to establish how BLIMP1α induces the EBV lytic cycle in B cells. One mechanism might involve an acute reactivation in which B cells respond to cellular stress by initiating virus replication, thereby allowing the virus to escape quickly before the cell dies.12,41 However, more likely is that BLIMP1α induces B cells into lytic cycle because it drives their differentiation toward plasma cells. This is supported by our observation that in GC B cells BLIMP1α expression induced changes characteristic of plasma cell differentiation. This latter mechanism might be mediated by XBP-1, a transcription factor known to be induced by BLIMP1α and has been shown to bind to the BZLF1 promoter and to activate the EBV lytic cycle.41-44
Our data are also consistent with previous reports showing that LMP-1 can block entry into lytic cycle in B cells and that it does so primarily by suppressing BZLF1 expression at the transcriptional level.45,46 In this context, it is interesting to compare the effects of LMP-1 with those of its cellular homolog, CD40. It has previously been shown that CD40 ligation can also suppress induction of the EBV lytic cycle in B cells.45 Furthermore, CD40 engagement can direct the differentiation of GC B cells toward memory B cells while at the same time suppressing plasma cell differentiation, an effect associated with the down-regulation of BLIMP1.47-49 Although in these respects LMP-1 would appear to mimic closely the effects of CD40, it should be noted that the LMP-1 signal is constitutive, whereas that induced by CD40 is regulated by the availability of ligand. It remains to be established how the constitutive nature of the signal influences the eventual outcome of LMP-1-driven post-GC B-cell differentiation.
In conclusion, our results suggest that the down-regulation of the tumor suppressor, BLIMP1α by LMP-1 could be an essential step in the pathogenesis of EBV-associated GC-derived lymphomas, preventing not only the terminal differentiation of GC-derived progenitors but also virus replication.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof Georg Bornkamm for kindly providing us with the c-Myc expression vectors, Naheema Gordon and Kaisheng Wen for technical assistance, Eszter Nagy for help with immunoblotting, John Arrand and Sim Sihota for microarray studies, and Michael Kuo for help in recruiting patients.
This work was supported by the Leukemia and Lymphoma Research Fund, in part by the Czech Ministry of Education (grant MSM 6198959216) and European Union (infrastructure support CZ.1.05/2.1.00/01.0030 and CZ.1.07/2.3.00/20.0019), the German Cancer Aid organization, the Stiftung der Georg-August-Universität aus Mitteln der Kubeschka/Stricker/Wirth-Stiftung (Union for International Cancer Control International Cancer Technology Transfer Fellowships; A.S.), and the Cancer Research United Kingdom, London (A.B., M.R.).
Authorship
Contribution: K.V., M.V., S.L., and A.S. performed the experiments; C.B.W., K.V., A.B., D.K., M.V., and W.W. analyzed the data; K.L.W., M.V., M.R., C.B.W., and P.G.M. designed the study; and K.V., P.G.M., and C.B.W. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paul G. Murray, School of Cancer Sciences, University of Birmingham, Vincent Dr, Birmingham, B15 2TT, United Kingdom; e-mail: p.g.murray@bham.ac.uk.

![Figure 2. LMP-1 down-regulates BLIMP1α expression in primary GC B cells and transformed B cells. (A) Ectopic expression of LMP-1 in GC B cells was confirmed by Western blotting or was published before.31 (B) Quantitative RT-PCR analysis of BLIMP1α mRNA in CD10+ GC B cells from 5 separate donors after transfection with LMP-1 (pSG5-LMP-1 or pcDNA3.1-LMP-1) or vector control only (pSG5 or pcDNA3.1); LMP-1 down-regulated BLIMP1α mRNA in all samples. (C) LMP-1 also down-regulated BLIMP1α mRNA in both CD77− (centrocytes [CCs]) and CD77+ (centroblasts [CBs]) subsets of CD10+ cells. (D) Immunohistochemistry revealed that the expression of BLIMP1α (gray) and LMP-1 (red) were mutually exclusive in B95.8 cells. Magnification ×800. Images acquired with 60×/1.40 NA oil immersion objective lens (Nikon Ltd).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/22/10.1182_blood-2010-09-307710/4/m_zh89991170760002.jpeg?Expires=1765892032&Signature=HyIgnpOLwmKR5CgJKLU9D8vJmCpQb10Xg5ciG2ryNGwDDN2U4ZDQpa8TRuQQNtvBtVWXmcBdRkK7TN4nP0YEHLT6QH2xpppbS2R5iTSY6OHn1cVKcLDUgYI5GT1UFl8IxpL131NW8h8hC1hQKzCORTvbPYgHpBUDhkzJuFcVIvLEOzRtyi14Pza5ZErScUHblRLGhvlXcsF~R8frSE8cS5LlJOVCcoGDQixNEiHAcTI08UMQy2M~wE-evTioDcl4ceoAj15oCa3dQM-Oy-Z7RspNfHL4xfrnU8lwqdqa5EUvllpc05hbNRONRzcSz~fpUDmG2G5f3MXeczbjAaJB9A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal