Abstract
Whereas oncogenic retroviruses are common in animals, human T-lymphotropic virus 1 (HTLV-1) is the only transmissible retrovirus associated with cancer in humans and is etiologically linked to adult T-cell leukemia. The leukemogenesis process is still largely unknown, but relies on extended survival and clonal expansion of infected cells, which in turn accumulate genetic defects. A common feature of human tumor viruses is their ability to stimulate proliferation and survival of infected pretumoral cells and then hide by establishing latency in cells that have acquired a transformed phenotype. Whereas disruption of the DNA repair is one of the major processes responsible for the accumulation of genomic abnormalities and carcinogenesis, the absence of DNA repair also poses the threat of cell-cycle arrest or apoptosis of virus-infected cells. This study describes how the HTLV-1 p30 viral protein inhibits conservative homologous recombination (HR) DNA repair by targeting the MRE11/RAD50/NBS1 complex and favors the error-prone nonhomologous-end–joining (NHEJ) DNA-repair pathway instead. As a result, HTLV-1 p30 may facilitate the accumulation of mutations in the host genome and the cumulative risk of transformation. Our results provide new insights into how human tumor viruses may manipulate cellular DNA-damage responses to promote cancer.
Introduction
Homologous recombination (HR) is a major pathway of double-strand break (DSB) repair in mammalian cells.1 Faithful recombination is critical to avoid genetic and genomic aberrations, and involves a complex and orderly assembly of many checkpoints and repair factors. DSBs frequently occur as a result of exposure to irradiation and chemicals. In response to DSBs, activation of ataxia telangiectasia mutated (ATM) initiates a cascade of events, including phosphorylation of histone H2AX (referred to as γ-H2AX) and downstream effectors such as structural chromosome maintenance 1 (SCM1) and checkpoint kinase 2 (Chk2).2,3 Chk2 phosphorylates p53, disrupting its interaction with Mdm2 and stabilizing the p53 protein,4 which pauses the cell cycle so that the cell can attempt to repair its damaged DNA. H2AX phosphorylation plays an important role in both DNA-damage–checkpoint activation and deactivation of the checkpoint signal to allow the cell cycle to resume. HR is very important during DNA replication in the S phase, when DSBs are generated during lagging strand synthesis or when unrepaired lesions cause replication-fork stalling.5,6 Initiation of HR involves the recruitment of the MRE11/RAD50/NBS1 (MRN) complex to DNA-damaged sites that can be visualized by accumulation of γ-H2AX as foci.7 In contrast, DSBs created during the G1 or M phase are preferentially repaired by a nonconservative, nonhomologous-end–joining (NHEJ) pathway. The NHEJ pathway has been shown to be Ku80 and DNA-dependent protein kinase (DNA-PK) dependent.8 The switch from HR to NHEJ has not been fully elucidated, but can in part be explained by the fact that MRE11-resection activity generates single-stranded DNA for which Ku80 has a poor affinity, allowing for the assembly of the MRN complex and HR repair. Therefore, regulation of DSB access to MRE11 or Ku80 is likely decisive in the fate and type of DNA repair.
HTLV-1 is a human retrovirus associated with adult T-cell leukemia/lymphoma, an aggressive disease with a dismal prognosis.9 Whereas the majority of HTLV-1–infected individuals remain asymptomatic, upwards of 5% of patients ultimately develop adult T-cell leukemia/lymphoma. The molecular mechanisms of HTLV-1 oncogenesis are poorly understood. HTLV-1 disrupts cell-cycle checkpoints, tumor suppressors, and Notch signaling and reactivates telomerase.10-13 Unlike animal oncoretroviruses, HTLV-1 does not transduce a protooncogene and does not integrate at specific sites in the human genome, thereby excluding insertional mutagenesis. The end of the HTLV-1 proviral genome encodes for the regulatory proteins p12, p30, and the HTLV-1 bZIP factor (HBZ), which are involved in virus infectivity, immune escape, and establishment of latency.14-17 HTLV-1 leukemic cells often present numerous genomic alterations, but the genesis and contribution of these chromosomal defects are presently unclear. The viral oncoprotein Tax plays an important role in the initiation of cellular transformation. In addition, several studies have shown that Tax inhibits the nucleotide excision–repair pathway, DNA β-polymerase, and DNA topoisomerase.18-20 Recently, Tax has been proposed to induce constitutive signaling of the DNA-PK pathway21 and to attenuate the ATM-mediated cellular DNA-damage response,22 and ATM has been shown to be hyperphosphorylated in HTLV-1–transformed cells.23 However, a role for other HTLV-1 viral proteins in genomic stability has been overlooked. In the present study, we report for the first time that HTLV-1 p30 is specifically redistributed from the nucleolus to the nucleoplasm upon DNA damage. We found that p30 inhibits HR repair of either chemically induced or naturally occurring DNA DSBs during replication of the DNA in the S phase. We also demonstrate that p30 binds to Nbs1 and Rad50 and interferes with the formation of the MRN complex and the recruitment of Nbs1 and Rad50 to p-H2AX DSB foci. Finally, we demonstrate that in p30-expressing cells, the error-prone NHEJ DNA repair is used instead of HR during the S phase. These findings suggest that p30 may promote the accumulation of genetic mutations/lesions, and provide a novel possible mechanism to explain why HTLV-1 human retrovirus transformation occurs at a very low frequency and after several decades of incubation.
Methods
Cells, plasmids, and lentiviral particles
HeLa, 293T, and Jurkat T cells were obtained from the American Type Culture Collection). PolyFect (QIAGEN) and calcium phosphate (Invitrogen) were used to transfect HeLa and 293T cells, respectively. Amaxa (Lonza) was used for the transfection of Jurkat T cells using Nucleofector Kit V and program X005. HTLV-1 p30 and HTLV-2 p28 were cloned in-frame with the hemagglutinin (HA) tag of pMH vectors. GFP-p30, GFP-p30 C-terminal deletion mutants, p30-HA, and p30-HA mutants have been described previously.24,25 pMH-p30T232A or pEGFPC1-p30T232A and HTLV-2 p28 Pro 40 to Thr (p28 P40T) were cloned by PCR and sequenced. B23-GFP and nucleolin-GFP were a gift from Dr M. Dundr.26 DR-GFP, pCAGS, and pSceI for the HR assay were provided by Dr M. Jasin.1 The NHEJ reporter system EJ5-GFP was provided by Dr J. M. Stark.27 MRE11-myc and Nbs1-myc expression vectors were provided by Dr Xiaohua Wu.28 The Rad50-Flag expression vector was provided by Dr Tanya Paull.29 Lentiviral particles expressing p30-myc were prepared by transfection of 293T cells with HR-CMV-p30-myc, along with pDLN and VSV-G, as described previously.30,31 The virus was collected, concentrated, and titered to reach 100% of cells infected. The HTLV-1 infectious molecular clones ACH and ACH-Δp30 were described previously.32
In vivo HR and NHEJ DNA-repair assay
The DR-GFP reporter vector uses a modified gene for green fluorescent protein (GFP) as a recombination reporter and the I-SceI endonuclease for the introduction of DSBs. The DR-GFP was transfected into HeLa cells either with the negative-control vector or with the Sce I–expressing vector, along with the vectors expressing the proteins indicated in Figure 3A. Forty-eight hours after transfection, cells were collected and GFP+ cells were counted using an LSR II flow cytometer (BD Biosciences). EJ5-GFP has the puromycin sequence inserted into the coding sequence of GFP between 2 generated I-SceI restriction sites. In the presence of I-SceI endonuclease, the puromycin sequence is excised and the expression of GFP is restored only after NHEJ repair of both I-SceI ends. HeLa cells were transfected with EJ5-GFP and pSceI expression vector with or without the p30 expression vector. The percentage of GFP+ cells was counted using an LSR II flow cytometer.
Irradiation, drug treatment, and cell-cycle analysis
Irradiation experiments were carried out at 2, 5, or 10 Gy, as indicated, and cells were immediately incubated at 37°C. Cells were collected for Western blot or immunofluorescence analysis as indicated in the figure legends. Synchronization experiments were performed using hydroxyurea (2mM for 24 hours), etoposide (100mM for 24 hours), or nocodazole (40 ng/mL). PD98059 (5μM) was used to inhibit the MAPK pathway. Bleomycin (10 μg/mL for 24 hours) was used to induce irreversible DSBs. Nu7026 (20μM) was used as a specific inhibitor for DNA-PK. Cell-cycle analyses were performed by standard propidium iodide staining using an LSR II flow cytometer.
Cell-imaging microscopy
HeLa cells were transfected with plasmids as indicated in the figure legends (Figures 1, 2, 4, and 6). GFP-expressing cells were fixed in 3.7% paraformaldehyde for 15 minutes at room temperature, washed with PBS, and mounted using mounting medium (2.5% DABCO [Sigma-Aldrich], 200mM Tris-HCl, pH 8.6, and 90% glycerol). For immunocytochemistry staining, after being fixed, cells were permeabilized on ice for 5 minutes with 0.5% Triton X-100 and blocked for 1 hour in PBS with 0.5% gelatin and 0.25% bovine serum albumin. Cells were then incubated overnight with primary antibody, washed 3 times with PBS plus gelatin (0.2%), and then incubated with Alexa Fluor–conjugated secondary antibody (Invitrogen), washed, and mounted. Images were captured using a 100× objective with the TE2000E and Ti-S epifluorescence Nikon microscopes. For foci counting, 10 z-stacks (1 μm each) for each cell were collected and stitched together.
Immunoprecipitation, Western blots, and antibodies
Cell extracts were prepared using lysis buffer (1% NP-40, 50mM Tris-HCl, pH 7.5, 150mM NaCl, phosphatases, and protease inhibitors). Immunoprecipitation was performed overnight at 4°C. Primary antibodies used were: goat polyclonal anti–actin (sc-1615; Santa Cruz Biotechnology), rabbit polyclonal anti–cyclin B1 (H-433; Santa Cruz Biotechnology), mouse monoclonal anti–phospho(ser139)-H2AX (clone 2F3; BioLegend), rabbit polyclonal anti–phospho(ser139)-H2AX (2775S; Cell Signaling Technology), mouse monoclonal anti–MRE11 (clone 12D7; Gentex), mouse monoclonal anti–Rad50 (clone 13B3; Gentex), anti–c-Myc tag (clone 9E10; Roche), mouse monoclonal anti–HA (clone 3F10; Roche), mouse monoclonal anti–Flag (Roche), mouse monoclonal anti–DNA-PK (Neomarkers), and phospho-threonine-proline–specific antibody (9391; Cell Signaling Technology). Secondary antibodies used were: goat anti–rabbit Alexa Fluor 488–conjugated and goat anti–mouse Texas Red–conjugated (Molecular Probes, Invitrogen). Rabbit polyclonal anti–Nbs1 (3002; Cell Signaling Technology) and rabbit polyclonal anti–Nbs1 (poly6138; BioLegend) antibodies did not work in any of our immunofluorescence assays.
Results
Nucleoplasmic redistribution of HTLV-1 p30 in response to DSB DNA damage
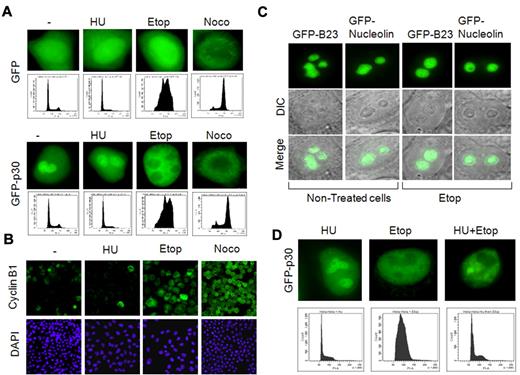
We have previously shown that the HTLV-1 p30 RNA-binding protein is a nucleolar resident protein that negatively regulates viral gene expression to favor viral latency. Because the nucleolus is a dynamic structure reorganized during the cell cycle, we investigated how p30 localization was affected. HeLa cells were transfected with GFP or GFP-p30 and treated with hydroxyurea, etoposide, or nocodazole to synchronize cells in the G1, S-G2, or M phase, respectively, as shown by cell-cycle analysis (Figure 1A) and immunofluorescence of cyclin B1, a cell cycle–regulated protein used as a marker of the cell-cycle phases (Figure 1B). Our results demonstrated that p30 was redistributed to the nucleus when cells were synchronized in the S phase with etoposide. Consistent with the fact that nucleolar structures are disassembled at the end of the G2 phase, p30 was relocalized to the nucleoplasm when cells were arrested in G2/M (Figure 1A). The nucleolar delocalization of p30 was independent of GFP fusion and was also observed with p30-HA–tagged protein (data not shown). To demonstrate that the etoposide effect on p30 cellular distribution was specific, we investigated the response of 2 nucleolar resident proteins, nucleoplasmin (B23) and nucleolin, which have been shown to colocalize with p30.25 In contrast to p30, the nucleolar localization of neither B23 nor nucleolin was affected by treatment with etoposide (Figure 1C), whereas, as expected, nocodazole treatment relocalized both proteins to the nucleoplasm (data not shown). These data suggest that etoposide treatment induces a specific redistribution of the HTLV-1 p30 protein to the nucleoplasm. We then investigated whether p30 nucleolar relocalization was the result of cellular entry into S phase or of effects inherent to etoposide treatment but independent of cell-cycle synchronization. To differentiate between these 2 hypotheses, we synchronized and blocked cells in the G1 phase and then treated them with etoposide.
Nucleolar delocalization of the HTLV-1 p30 protein. (A) Nucleolar delocalization of p30 in cells treated with etoposide. HeLa cells were transfected with GFP or GFP-p30 expression vectors, and 10 hours later were either left untreated or treated for 24 hours with hydroxyurea, etoposide, or nocodazole to synchronize them, respectively, in G1, G2, and mitosis. Cells were then fixed, mounted, and the localization of p30 was observed by epifluorescence microscopy (upper panels). Distribution of treated cells into cell-cycle phases was also accessed by flow cytometry (lower panels) or by immunofluorescence staining of cyclin B1, a cell-cycle marker exclusively expressed in G2/M (B). (C) Specificity of nucleolar delocalization of p30. HeLa cells were transfected with GFP-B23 or GFP-nucleolin expression vectors (2 structural nucleolar proteins) for 10 hours, and then either left untreated or treated with etoposide for 24 hours. The nucleolar localization of GFP-B23 and GFP-nucleolin was observed in the same conditions described in panel A. (D) Nucleolar delocalization of p30 was independent of the cell cycle and caused by etoposide treatment. HeLa cells were transfected with GFP-p30 and treated with hydroxyurea, etoposide, or hydroxyurea, followed by etoposide (top row) and cell-cycle analysis showing the distribution of cells treated under these conditions (bottom row).
Nucleolar delocalization of the HTLV-1 p30 protein. (A) Nucleolar delocalization of p30 in cells treated with etoposide. HeLa cells were transfected with GFP or GFP-p30 expression vectors, and 10 hours later were either left untreated or treated for 24 hours with hydroxyurea, etoposide, or nocodazole to synchronize them, respectively, in G1, G2, and mitosis. Cells were then fixed, mounted, and the localization of p30 was observed by epifluorescence microscopy (upper panels). Distribution of treated cells into cell-cycle phases was also accessed by flow cytometry (lower panels) or by immunofluorescence staining of cyclin B1, a cell-cycle marker exclusively expressed in G2/M (B). (C) Specificity of nucleolar delocalization of p30. HeLa cells were transfected with GFP-B23 or GFP-nucleolin expression vectors (2 structural nucleolar proteins) for 10 hours, and then either left untreated or treated with etoposide for 24 hours. The nucleolar localization of GFP-B23 and GFP-nucleolin was observed in the same conditions described in panel A. (D) Nucleolar delocalization of p30 was independent of the cell cycle and caused by etoposide treatment. HeLa cells were transfected with GFP-p30 and treated with hydroxyurea, etoposide, or hydroxyurea, followed by etoposide (top row) and cell-cycle analysis showing the distribution of cells treated under these conditions (bottom row).
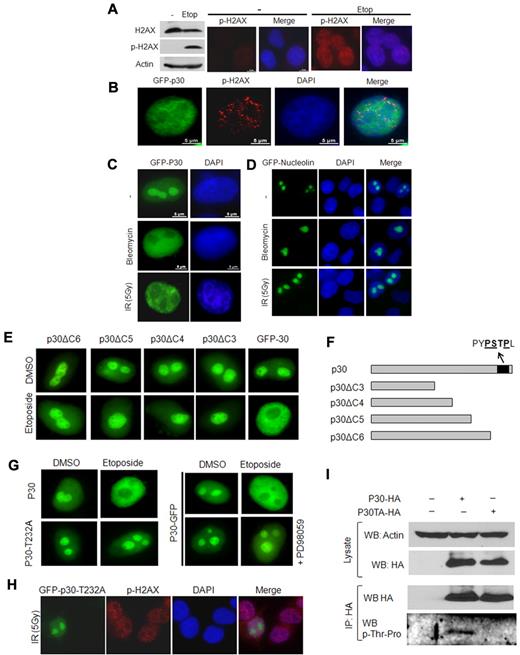
These experiments showed that p30 was still relocalized to the nucleoplasm (Figure 1D). Cell-cycle analysis confirmed that treated cells were in fact arrested in G1 (Figure 1D), suggesting that a cell-cycle–independent effect of etoposide was involved. Etoposide is a known inducer of DNA DSBs, and, as expected in the experimental conditions described in “Irradiation, drug treatment, and cell-cycle analysis”, it induced the phosphorylation of H2AX and the formation of DSB foci (Figure 2A). To further demonstrate that DSBs breaks may be responsible for p30 delocalization, cells were exposed to γ-irradiation or to bleomycin, an agent that induces nonrepairable DSBs. In both conditions, p30 was redistributed to the nucleoplasm (Figure 2B-C). In contrast, nucleolar localization of nucleolin was not affected by formation of DSBs under the same experimental conditions (Figure 2D), again confirming a specific effect for HTLV-1 p30.
p30 protein relocalizes on DNA-damage–induced DSBs. (A) Etoposide induces DNA DSBs. HeLa cells were left untreated or treated with etoposide for 24 hours, followed by SDS-PAGE and Western blot analyses of the phosphorylated form (S139) of H2AX (p-H2AX marker of DSB), H2AX, and actin proteins. Control or etoposide-treated cells were also immunostained with anti–phospho-H2AX antibody. (B) p30 nucleolar delocalization is associated with DSB triggering. HeLa cells transfected with GFP-p30 (green) were treated with etoposide as described in panel A and immunostained with anti–phospho-H2AX antibody showing DSBs (red). (C-D) p30 nucleolar delocalization is specifically induced by DNA-damage agents. HeLa cells transfected with GFP-p30 or GFP-nucleolin (control) were either treated with bleomycin (which induces irreversible DNA breaks) or γ-irradiated (5 Gy). (E) The C-terminal end of the p30 protein includes the response motif for p30 nucleolar delocalization. HeLa cells were transfected with a series of C-terminal–deleted mutants of p30 all known to localize in the nucleolus (GFP-p30delC6, GFP-p30delC5, GFP-p30delC4, and GFP-p30delC3), and treated with etoposide as described in panel A. Only cells expressing wild-type p30 responded to the etoposide treatment. (F) Representation of p30 C-terminal–deleted mutants. (G) Thr232 is required for p30 delocalization in response to DSB. HeLa cells were transfected with GFP-p30 or GFP-p30T232A and treated with etoposide in the presence or the absence of PD98059, a specific MAPK inhibitor. (H) HeLa cells were transfected with GFP-p30 or the GFP-p30T232A mutant, γ-irradiated, and stained with p-H2AX antibody. (I) The threonine at position 232 is phosphorylated. 293T cells were transfected with pMH-, p30-HA-, or p30-T232A-HA–expressing vectors. After transfection (48 hours), the cells were irradiated (5 Gy), and 2 hours later they were lysed and cell extracts were immunoprecipitated with anti-HA antibody. Western blot analysis was then performed with an anti–phospho-threonine-proline–specific antibody.
p30 protein relocalizes on DNA-damage–induced DSBs. (A) Etoposide induces DNA DSBs. HeLa cells were left untreated or treated with etoposide for 24 hours, followed by SDS-PAGE and Western blot analyses of the phosphorylated form (S139) of H2AX (p-H2AX marker of DSB), H2AX, and actin proteins. Control or etoposide-treated cells were also immunostained with anti–phospho-H2AX antibody. (B) p30 nucleolar delocalization is associated with DSB triggering. HeLa cells transfected with GFP-p30 (green) were treated with etoposide as described in panel A and immunostained with anti–phospho-H2AX antibody showing DSBs (red). (C-D) p30 nucleolar delocalization is specifically induced by DNA-damage agents. HeLa cells transfected with GFP-p30 or GFP-nucleolin (control) were either treated with bleomycin (which induces irreversible DNA breaks) or γ-irradiated (5 Gy). (E) The C-terminal end of the p30 protein includes the response motif for p30 nucleolar delocalization. HeLa cells were transfected with a series of C-terminal–deleted mutants of p30 all known to localize in the nucleolus (GFP-p30delC6, GFP-p30delC5, GFP-p30delC4, and GFP-p30delC3), and treated with etoposide as described in panel A. Only cells expressing wild-type p30 responded to the etoposide treatment. (F) Representation of p30 C-terminal–deleted mutants. (G) Thr232 is required for p30 delocalization in response to DSB. HeLa cells were transfected with GFP-p30 or GFP-p30T232A and treated with etoposide in the presence or the absence of PD98059, a specific MAPK inhibitor. (H) HeLa cells were transfected with GFP-p30 or the GFP-p30T232A mutant, γ-irradiated, and stained with p-H2AX antibody. (I) The threonine at position 232 is phosphorylated. 293T cells were transfected with pMH-, p30-HA-, or p30-T232A-HA–expressing vectors. After transfection (48 hours), the cells were irradiated (5 Gy), and 2 hours later they were lysed and cell extracts were immunoprecipitated with anti-HA antibody. Western blot analysis was then performed with an anti–phospho-threonine-proline–specific antibody.
A p30 C-terminal motif is involved in DSB-mediated delocalization
Using a series of p30 C-terminal–truncated mutants fused to GFP (Figure 2E), we delineated that the p30 domain responsive to DSBs was located within the last 50–amino acid residues (Figure 2F).
Additional mutants further delineated C-terminal motif “PSTPLLPHPENL” of the p30 protein as critical for its response to DSBs. This sequence contained a proline-rich PxxP (PSTP), a motif that is found in proteins that interact with the classic SH3 domain present in other partner proteins and that mediates the assembly of specific protein complexes. In addition, the PSTP motif in that region also constitutes a potential MAPK consensus sequence with a putative threonine phosphorylation site (Figure 2F). We used a broad MAPK inhibitor to determine whether MAPK signaling may play a role in the p30 response to DSBs. As shown in Figure 2G, treatment with PD98059 abrogated p30 relocalization on the DNA-damage signal. To further demonstrate that the putative MAPK site was critical, we performed site-directed mutagenesis to generate a specific mutant, p30T232A. Interestingly, our data clearly demonstrated that nucleolar localization of this mutant was no longer affected by ionizing radiation, etoposide, or bleomycin (Figure 2G-H). To formally demonstrate that this Thr was phosphorylated in response to DSBs, 293T cells were transfected with p30, and p30T232A cells were subjected to γ-irradiation. Cell lysates were immunoprecipitated using anti-HA antibody and probed with a specific p-Thr-Pro antibody. The only site recognized by this antibody and present in p30 is located at position Thr232. As demonstrated by our results in Figure 2I, p30, but not p30T232A, was phosphorylated on Thr232.
p30 inhibits DSB HR DNA repair in vivo
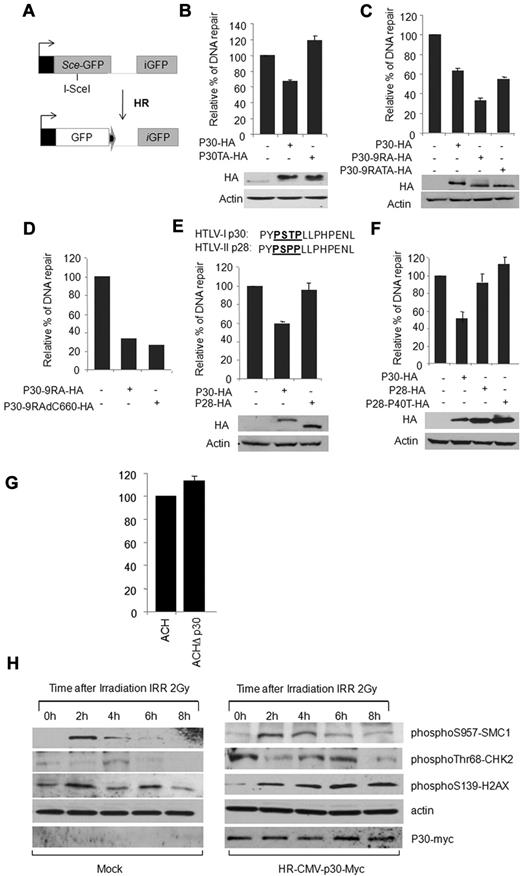
We then tested the effect of p30 on DSB DNA repair. We used an established in vivo HR assay by cotransfection of p30, along with a DR-GFP HR reporter vector and FACS analysis. The system uses a modified gene for GFP as a recombination reporter and expression of Sce I endonuclease for the creation of DSBs (Figure 3A).1 In this assay, p30 reproducibly led to a 35% decrease in efficiency of HR DNA repair, whereas the p30T232A had no significant effect on HR DNA repair (Figure 3B). The effect of p30 was statistically significant and comparable to that of MDM2 (data not shown), which was previously reported to inhibit HR through the binding of Nbs1. An important limitation to the in vivo HR assay is that it does not activate the ATM DNA-damage signal, and our data clearly show that in the absence of a DNA-damage signal, p30 expression remains nucleolar (Figure 2). In contrast, activation of a DNA-damage signal results in rapid (within minutes after γ-irradiation) and almost complete delocalization of p30 to the nucleoplasm (Figure 2). To address this, we used a p30 mutant in which the nucleolar retention signal had been mutated, resulting in a nucleoplasmic-localized p30 mutant, p309RA. We previously showed that this mutant is functional and suppresses viral replication as efficiently as the wild-type protein.24 In support of our hypothesis, p309RA was more potent than the wild-type p30, and resulted in approximately 60% inhibition of DSB DNA repair in the in vivo HR assay (Figure 3C).
p30 inhibits DSB HR DNA repair. (A) Schematic representation of the in vivo HR assay with DR-GFP HR reporter vector (see “In vivo HR and NHEJ DNA-repair assay”). (B-F) HeLa cells were cotransfected with DR-GFP vector either with the control vector or with the pI-SceI and with the vector expressing p30 and p30T232A (B); p30, p30-9RA, and p30-9RA-TA (C); p30-9RA and p30-9RA-delC660 (D); p30 and HTLV-2 p28 (E); or with p30, HTLV-2 p28, and HTLV-2 p28-P40T (F). Forty-eight hours later, the expression of GFP was assessed by FACS analysis. The relative percentage of GFP-expressing cells was represented by histograms corresponding to the average of 5 independent experiments. Western blots show p30, p30T232A, p30-9RA, p30-9RA-TA, p28, and p28-P40T expression in the assay. (G) HTLV-I molecular clones ACH and ACHΔp30 were transfected into Jurkat T cells to evaluate the effect of p30 on HR DNA repair when expressed in the context of the whole virus. Results demonstrated an increase of 13.3% of HR when p30 was ablated from an HTLV-I molecular clone. Data are derived from 2 independent transfection experiments and the standard deviation value for ACHΔp30 was 3. (H) p30 delays the response to DNA damage. HeLa cells infected with mock or p30-expressing lentivirus were γ-irradiated (2 Gy) and collected at 0, 2, 4, 6, and 8 hours after irradiation. Shown are Western blot analyses of pS957-SMC1, pThr68-CHK2, and p-H2AX checkpoints involved in the response to DNA damage.
p30 inhibits DSB HR DNA repair. (A) Schematic representation of the in vivo HR assay with DR-GFP HR reporter vector (see “In vivo HR and NHEJ DNA-repair assay”). (B-F) HeLa cells were cotransfected with DR-GFP vector either with the control vector or with the pI-SceI and with the vector expressing p30 and p30T232A (B); p30, p30-9RA, and p30-9RA-TA (C); p30-9RA and p30-9RA-delC660 (D); p30 and HTLV-2 p28 (E); or with p30, HTLV-2 p28, and HTLV-2 p28-P40T (F). Forty-eight hours later, the expression of GFP was assessed by FACS analysis. The relative percentage of GFP-expressing cells was represented by histograms corresponding to the average of 5 independent experiments. Western blots show p30, p30T232A, p30-9RA, p30-9RA-TA, p28, and p28-P40T expression in the assay. (G) HTLV-I molecular clones ACH and ACHΔp30 were transfected into Jurkat T cells to evaluate the effect of p30 on HR DNA repair when expressed in the context of the whole virus. Results demonstrated an increase of 13.3% of HR when p30 was ablated from an HTLV-I molecular clone. Data are derived from 2 independent transfection experiments and the standard deviation value for ACHΔp30 was 3. (H) p30 delays the response to DNA damage. HeLa cells infected with mock or p30-expressing lentivirus were γ-irradiated (2 Gy) and collected at 0, 2, 4, 6, and 8 hours after irradiation. Shown are Western blot analyses of pS957-SMC1, pThr68-CHK2, and p-H2AX checkpoints involved in the response to DNA damage.
We next investigated whether the lack of the p30 T232A effect on DSB repair was due to its inability to relocate to the nuclear compartment or to the presence of the T → A mutation. Introduction of a T232A mutation in p309RA did not prevent inhibition of HR, suggesting that Thr232 is mostly involved in relocalization of p30 and not HR inhibition (Figure 3C). To further confirm these results, we constructed a C-terminal deletion (from 660 to 723 bp) of p30-9RA to generate a p30-9RA-del660 mutant that no longer included the Thr232. In vivo HR assays (Figure 3D) indicated that both mutants had similar inhibitory effects, confirming that the C-terminus of p30 is not involved in HR inhibition.
HTLV-2 is nonpathogenic in humans and is not associated with leukemia. The reasons for this are not clear, and few differences between the 2 viruses have been reported.34 In contrast to p30, its HTLV-2 homolog, p28, was expressed exclusively in the nuclear compartment (like our mutant, p309RA). Unexpectedly, the putative MAPK domain present in p30 had a natural polymorphism and a conserved T232P mutation in p28 (Figure 3E). When p28 was tested in an in vivo HR assay, its expression had no significant effect on DSB DNA repair (Figure 3E). These data demonstrate that HTLV-1, but not HTLV-2, has evolved a regulatory protein affecting the HR DNA-repair pathway. As expected from the results in Figure 3E, replacement of the Pro by a Thr in the PSPP motif of HTLV-2 p28 had no significant effect on HR activity (Figure 3F). Finally, the effect of p30 on the HR DNA-repair pathway and downstream effectors was analyzed. We next investigated the effects of p30 when expressed at physiologic levels in the context of an infectious HTLV-1 molecular clone. ACH and ACHΔp30 were transfected into Jurkat T cells, along with reporter plasmids for HR assays (Figure 3G). Our results demonstrated a significant increase of 13.3% over ACH controls when ACHΔp30 was tested, which is consistent with a p30-inhibitory function on the HR DNA-repair pathway. In these experimental settings, reduced effects of p30 on HR were expected because the levels of p30 expression from molecular clones were also lower. However, the difference observed (13.3% ± 3%) was statistically significant.
Consistent with results presented in Figure 3B, Western blotting analyses of some activated checkpoints (pS957-SMC1, pThr68-CHK2, and p-H2AX) detected in response to DNA damage indicated a lasting response in the presence of p30 compared with the control extracts (Figure 3H). These results further demonstrate that the DNA-damage response is impaired by p30. The levels of p-SMC1 and p-CHK2 were higher in p30-expressing cells even in the absence of γ-irradiation (time T0). We believe that this observation is related to the ability of p30 to inhibit HR on naturally occurring breaks in the S phase during DNA replication (see Figure 6A). It is also possible that p30 affects these kinases directly or other upstream effectors such as ATM/ATR, which warrants further study.
p30 disrupts the formation of the MRN complex on DSB DNA foci in induced and physiologically occurring DNA breaks in the S phase
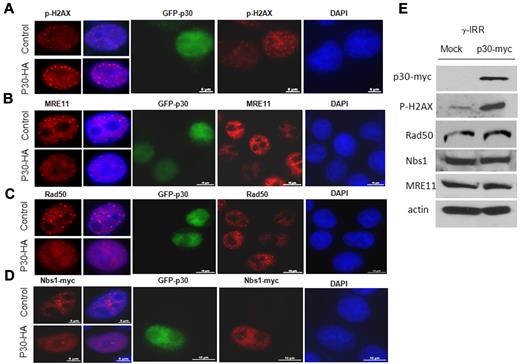
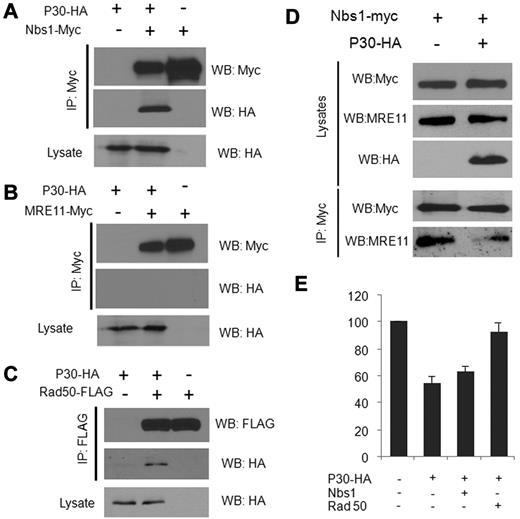
Because a direct correlation has been reported between the number of p-H2AX foci and the number of DSBs, it appears that the inhibition of HR by p30 is global rather than occurring at specific DNA sites, as suggested by the accumulation of numerous larger p-H2AX foci throughout the nucleus in the presence of p30 (Figure 4A). The MRN protein complex is involved in the sensing of DSBs and is recruited onto the DNA lesions, where it tethers together and stabilizes broken chromosomes. Long-lasting (8 hours or more), large foci mark the location of lesions that are particularly difficult to repair. Our results show that p30 expression itself does not induce DNA DSBs (data not shown). Shortly after γ-irradiation (15-30 minutes), p30 did not affect quantitatively or qualitatively the foci formation, suggesting that p30 does not make cells prone to accumulating DSBs (data not shown). In contrast, 6-7 hours after irradiation, p-H2AX foci were significantly increased in number and significantly bigger in size in the presence of p30, suggesting a defect or delay in DNA repair (Figure 4A and 4E). When we examined in vivo MRN foci complex formation several hours after γ-irradiation, the accumulation of MRE11, Nbs1, and Rad50 in foci was greatly impaired in p30-expressing cells (Figure 4B-D). In p30-expressing cells, the lack of foci formation and diffuse expression gave a false impression of lower expression; however, relative expression of MRE11, Nbs1, and Rad50 was not affected, as shown by Western blot analyses (Figure 4E). To determine how the formation of MRN complex foci was impaired, we investigated whether p30 interacts individually in vivo with the MRN complex members. As shown in Figure 5, coimmunoprecipitation experiments indicated a specific interaction between p30 with Nbs1 and Rad50, but not with MRE11 (Figure 5A-C). These results suggest that p30 does not interact with the MRN complex, but rather with some of its components, thereby preventing the assembly of a functional MRN complex onto DSBs. Because p30 was not found to interact with MRE11, and because the latter interacts with Nbs1 and Rad50, this suggested that p30 competes with MRE11 for the same binding site on Nbs1 and Rad50. Consistent with this model, the interaction of Nbs1 with MRE11 was significantly reduced when p30 was present (Figure 5D). These data indicate that p30 interferes with the formation of a functional MRN complex and its recruitment to p-H2AX DSB foci. Because p30 interacts with Nbs1 and Rad50 and competes with MRE11 for Rad50 interactions, we investigated whether overexpression of Rad50 and/or Nbs1 can rescue HR activity. As shown in Figure 5E, Rad50 had a significant effect and restored HR activity.
p30 disrupts the formation of the MRN complex on DSB DNA foci. HeLa cells nontransfected or transfected with GFP-p30 (green) were γ-irradiated (10 Gy) and incubated at 37°C. Seven hours later, cells were fixed and stained with antibodies (red) directed against p-H2AX (A), MRE11 (B), Rad50 endogenous proteins (C), or expressed Nbs1-myc tagged protein (D). Cells expressing GFP-p30 have diffused MRN complex proteins compared with nontransfected cells. (E) Relative expression of MRE11, Nbs1, and Rad50 was not affected by the expression of p30. HeLa cells were infected with mock or p30-myc–expressing lentivirus. The titer of the virus was calculated to have 100% of cells infected, and Western blot analysis was performed on the cell extracts using antibodies directed against p-H2AX, MRE11, and Rad50 endogenous proteins or expressed Nbs1-myc.
p30 disrupts the formation of the MRN complex on DSB DNA foci. HeLa cells nontransfected or transfected with GFP-p30 (green) were γ-irradiated (10 Gy) and incubated at 37°C. Seven hours later, cells were fixed and stained with antibodies (red) directed against p-H2AX (A), MRE11 (B), Rad50 endogenous proteins (C), or expressed Nbs1-myc tagged protein (D). Cells expressing GFP-p30 have diffused MRN complex proteins compared with nontransfected cells. (E) Relative expression of MRE11, Nbs1, and Rad50 was not affected by the expression of p30. HeLa cells were infected with mock or p30-myc–expressing lentivirus. The titer of the virus was calculated to have 100% of cells infected, and Western blot analysis was performed on the cell extracts using antibodies directed against p-H2AX, MRE11, and Rad50 endogenous proteins or expressed Nbs1-myc.
p30 interacts in vivo with Nbs1 and Rad50 but not with MRE11 proteins. 293T cells were transfected for 48 hours with p30-HA and the Nbs1-myc (A), MRE11-myc (B), or Rad50-flag (C) expression vectors. Immunoprecipitation of cell extracts was performed with the indicated antibodies, followed by Western blot analysis of the corresponding proteins. (D) p30 affects the interaction of the MRN complex components in vivo. 293T cells were transfected with either Nbs1-myc or Nbs-1-myc and p30-HA–expressing vectors and irradiated 48 hours after transfection. Nbs1-myc was immunoprecipitated with an anti-myc antibody, followed by Western blot analysis with an anti-MRE11 antibody. (E) Rescue of the HR DNA repair by ectopic expression of MRN complex components. HeLa cells were transfected with the HR assay vectors (DR-GFP and pSceI) and either with p30 alone or with p30 and Nbs1 or p30 and Rad50 expression vectors together. After 48 hours of transfection, the percentage of GFP+ cells was measured by FACS and is represented by the histograms of 3 independent experiments.
p30 interacts in vivo with Nbs1 and Rad50 but not with MRE11 proteins. 293T cells were transfected for 48 hours with p30-HA and the Nbs1-myc (A), MRE11-myc (B), or Rad50-flag (C) expression vectors. Immunoprecipitation of cell extracts was performed with the indicated antibodies, followed by Western blot analysis of the corresponding proteins. (D) p30 affects the interaction of the MRN complex components in vivo. 293T cells were transfected with either Nbs1-myc or Nbs-1-myc and p30-HA–expressing vectors and irradiated 48 hours after transfection. Nbs1-myc was immunoprecipitated with an anti-myc antibody, followed by Western blot analysis with an anti-MRE11 antibody. (E) Rescue of the HR DNA repair by ectopic expression of MRN complex components. HeLa cells were transfected with the HR assay vectors (DR-GFP and pSceI) and either with p30 alone or with p30 and Nbs1 or p30 and Rad50 expression vectors together. After 48 hours of transfection, the percentage of GFP+ cells was measured by FACS and is represented by the histograms of 3 independent experiments.
p30 induces a switch from the HR to the NHEJ DNA-repair pathway during the S phase
HR DNA repair is exclusively used during the S phase, and MRE11 foci colocalize with p-H2AX on natural breaks. These DSBs are very transient and are rapidly repaired and removed by the MRN complex. The requirement for the MRN complex during DNA replication in the absence of induced DNA damage has already been demonstrated. Therefore, we investigated whether p30 affects the DSB-repair response in nonirradiated cells when breaks are naturally occurring during DNA replication in the S phase. As shown in Figure 6A, disruption of the MRN complex formation by p30 was evident by the lack of colocalization between MRE11 and p-H2AX on physiologically occurring DNA breaks during the S phase in the absence of any treatment.
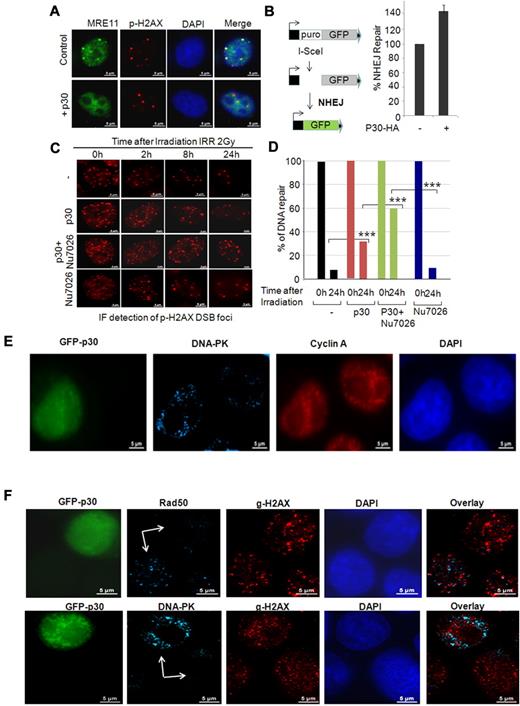
p30 induces a switch from the HR to the NHEJ DNA-repair pathway during the S phase. (A) p30 hampers the recruitment of the MRN complex on naturally occurring breaks. HeLa control or p30-expressing cells were stained with anti-MRE11 and anti p-H2AX antibodies to reveal natural breaks. (B) p30 favors NHEJ DNA repair. Representation of the in vivo NHEJ assay based on the EJ5-GFP reporter vector. HeLa cells were transfected with EJ5-GFP and pSceI expression vector with or without p30 expression vector. The percentage of GFP+ cells was estimated by FACS and is represented by the histograms of 3 independent experiments with standard deviations. (C) p30 favors unfaithful DNA repair. HeLa cells infected with mock lentivirus or p30-expressing lentivirus particles were treated or not with the DNA-PK inhibitor Nu7026 (20μM), a specific inhibitor of the NHEJ-repair pathway. Cells were γ-irradiated and the time course of the DNA-repair rate at 0, 2, 8, and 24 hours was performed by immunofluorescence staining with anti–p-H2AX antibody showing the number of breaks in each time condition. (D) DSB foci were counted in at least 30 cells collected by z-stack acquisition at 0 and 24 hours of each condition. Results represent average values. ***P < .001. (E) NHEJ DNA repair is increased in p30-expressing cells. HeLa cells transfected with GFP-p30 were synchronized with hydroxyurea and released for 7 hours to have the majority of cells in the S phase. The cells were then irradiated (10 Gy) and 2 hours later stained with anti-cyclin A (a marker of the S phase) or anti–DNA-PK (a marker of NHEJ) antibodies, followed by Alexa Fluor 568 (red) or Alexa Fluor 647 (blue)–conjugated secondary antibodies, respectively. (F) p30 favors a switch from the HR to the NHEJ DNA-repair. HeLa cells transfected with GFP-p30 were synchronized with hydroxyurea and released for 7 hours to have the majority of cells in the S phase. Cells were irradiated (10 Gy) and 2 hours later stained with anti–p-H2AX (DSB) and either Rad50 (HR-specific) or DNA-PK (NHEJ-specific) antibodies.
p30 induces a switch from the HR to the NHEJ DNA-repair pathway during the S phase. (A) p30 hampers the recruitment of the MRN complex on naturally occurring breaks. HeLa control or p30-expressing cells were stained with anti-MRE11 and anti p-H2AX antibodies to reveal natural breaks. (B) p30 favors NHEJ DNA repair. Representation of the in vivo NHEJ assay based on the EJ5-GFP reporter vector. HeLa cells were transfected with EJ5-GFP and pSceI expression vector with or without p30 expression vector. The percentage of GFP+ cells was estimated by FACS and is represented by the histograms of 3 independent experiments with standard deviations. (C) p30 favors unfaithful DNA repair. HeLa cells infected with mock lentivirus or p30-expressing lentivirus particles were treated or not with the DNA-PK inhibitor Nu7026 (20μM), a specific inhibitor of the NHEJ-repair pathway. Cells were γ-irradiated and the time course of the DNA-repair rate at 0, 2, 8, and 24 hours was performed by immunofluorescence staining with anti–p-H2AX antibody showing the number of breaks in each time condition. (D) DSB foci were counted in at least 30 cells collected by z-stack acquisition at 0 and 24 hours of each condition. Results represent average values. ***P < .001. (E) NHEJ DNA repair is increased in p30-expressing cells. HeLa cells transfected with GFP-p30 were synchronized with hydroxyurea and released for 7 hours to have the majority of cells in the S phase. The cells were then irradiated (10 Gy) and 2 hours later stained with anti-cyclin A (a marker of the S phase) or anti–DNA-PK (a marker of NHEJ) antibodies, followed by Alexa Fluor 568 (red) or Alexa Fluor 647 (blue)–conjugated secondary antibodies, respectively. (F) p30 favors a switch from the HR to the NHEJ DNA-repair. HeLa cells transfected with GFP-p30 were synchronized with hydroxyurea and released for 7 hours to have the majority of cells in the S phase. Cells were irradiated (10 Gy) and 2 hours later stained with anti–p-H2AX (DSB) and either Rad50 (HR-specific) or DNA-PK (NHEJ-specific) antibodies.
Because p30 can disrupt DSB foci formation during the S phase, and because p30-expressing cells do not arrest in the S phase, we hypothesized that p30-mediated inhibition of HR was compensated for by activation of the error-prone NHEJ DNA-repair pathway during the S phase. We used a previously established in vivo NHEJ-repair assay based on GFP expression (Figure 6B), and showed that p30 expression increased NHEJ repair by more than 40% (Figure 6B). To further confirm the effect of p30 on NHEJ, we then irradiated p30-expressing cells in the presence or absence of an NHEJ-specific DNA-PK inhibitor. As expected, the number of DNA break foci revealed a noticeable delay in DNA repair in p30-expressing cells in the S phase compared with normal cells (Figure 6C-D). Our data also revealed that p30-expressing cells in the S phase that were treated with an NHEJ inhibitor were significantly impaired in their ability to repair DNA breaks compared with p30 alone (40% compared with 65%, respectively; Figure 6C-D). These findings suggest that in p30-expressing cells, DNA breaks that occur during DNA replication–associated breaks are repaired by the error-prone NHEJ pathway. To confirm these results, GFP-p30–expressing cells were synchronized and released to increase the number of cells in the S phase. Two hours after irradiation, cells were stained with cyclin A (a specific marker of the S phase) and DNA-PK (a specific marker for NHEJ). Our results demonstrated that in the S phase (ie, cyclin A–positive), p30-expressing cells were also positive for DNA-PK (NHEJ), whereas p30-negative cells did not have DNA-PK foci (Figure 6E). Because Rad50 is specific for HR and DNA-PK is specific for NHEJ, we analyzed the presence of these factors on DSB γ-H2AX foci in the presence or absence of p30 (Figure 6F). GFP-p30–expressing cells were synchronized and released to increase the number of cells in the S phase, and 2 hours after irradiation were fixed and analyzed. As expected for HR repair in the S phase, γ-H2AX foci and Rad50 foci colocalized in the absence of p30. In contrast, in the presence of p30, γ-H2AX foci colocalized with DNA-PK foci and Rad50 foci were inhibited, suggesting a switch to NHEJ repair in the S phase when p30 is present.
Discussion
We have demonstrated previously that HTLV-1 p30 is a negative regulator of HTLV-1 replication and may favor the establishment of viral latency. Although p30 is mainly a nucleolar resident protein, to date, no function has been assigned to this localization, because mutation of the nucleolar retention signals led to a nuclear protein with similar functions as the wild-type.
It is possible that the virus has evolved p30 retention in the nucleolus to serve as a reservoir that is rapidly available as needed. In fact, our results indicate that p30 was specifically delocalized from the nucleolus within minutes when cells were subjected to a DNA-damage signal induced by ionizing radiation. Consistent with the fact that p30 may be needed during the S phase, nucleolar storage of p30 would allow the virus to respond to the swift assembly of DNA repair complexes and prevent HR repair of DNA breaks. In the present study, we demonstrate that p30 interferes with the assembly of a functional MRN complex and prevents HR repair of naturally occurring and induced breaks during DNA replication. A fundamental role for the MRN complex in the DSB response is well established. MRN plays an essential role during normal DNA replication and prevents the accumulation of broken replication forks.35-37 Our current results suggest that p30 prevents the accumulation of Nbs1 and Rad50 into DSB foci upon irradiation, suggesting that p30 blocks the sensing of DNA damage. As a DSB-specific “guardian of the genome,” the MRN complex may in part function in the suppression of transformation. It is therefore exciting that a transforming retrovirus targets this complex. MDM2 has been shown to inhibit early DNA-damage–signaling events, resulting in a delay in DSB DNA repair. MDM2 interference with DSB DNA repair is independent of p53 and p14ARF; instead, MDM2 is recruited to the DNA DSB-repair complex (MRE11, Rad50, and Nbs1) through interaction with Nsb1.37 Our data show that HTLV-1 p30 forms specific complexes with Nbs1 and Rad50 but not MRE11. We also found that p30 competes with Nbs1 for binding to MRE11, suggesting that p30 prevents the assembly of a functional MRN complex. In the presence of p30, Rad50 was unable to assemble in foci with the MRN complex. It is possible that the Nbs1 complexes lacking Rad50 are involved in the DNA-damage checkpoint, whereas complexes that contain all proteins function in DSB repair. In such a model, depletion of Rad50 would leave Nbs1 and the checkpoint intact, but block repair of DSBs. In this scenario, p30 would affect DSB repair but tolerate cell proliferation. Our results suggest that p30 does not arrest cells in the S phase after DNA damage, but rather activates a shift from the HR to the error-prone NHEJ DNA-repair pathway. We also found that p30 prevents the formation of the MRN complex in physiologic conditions on naturally occurring DNA-replication–associated breaks in the absence of external treatment. The effects of p30 on HR DNA repair were not only seen in transient transfection assays, but also when p30 was expressed at more physiologic levels in the context of an infectious molecular clone. These results suggest that p30 may play an important role during leukemogenesis in vivo and warrant further studies. We also found that the HTLV-2 homolog p28 protein does not affect the HR pathway; this finding, coupled with the fact that HTLV-2 is not associated with malignancies in humans, is intriguing and warrants further studies. Consistent with our model, we demonstrated that cells in the S phase exposed to DSBs have DNA-PK foci only in p30-expressing cells. We also demonstrated that S-phase cells subjected to γ-irradiation display γ-H2AX foci that are colocalized with DNA-PK (NHEJ) in p30-expressing cells, whereas γ-H2AX foci are colocalized with Rad50 (HR) when p30 is absent.
Our results reveal a novel strategy that may be used by the oncogenic retrovirus HTLV-1 to increase the accumulation of mutations in the host genome by preventing HR to repair breaks occurring during DNA replication and switching to NHEJ. Our findings are consistent with the long latency preceding the development of the disease, and highlight once again that retroviral proteins have multifunctional purposes in the virus life cycle. The model proposed here also offers a rational explanation for the very low incidence of HTLV-1–associated leukemia compared with other known animal retroviruses.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Elizabeth Jenkins for editorial assistance.
This work was supported by the National Cancer Institute (NCI) and the National Institute of Allergy and Infectious Diseases (NIAID; grants CA106258 and AI058944 to C.N.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCI, NIAID, or the National Institutes of Health.
National Institutes of Health
Authorship
Contribution: H.H.B. performed the research described in Figures 1,Figure 2,Figure 3,Figure 4,Figure 5–6; J.P. performed the research described in Figure 5; and C.N. conceived of the experiments, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christophe Nicot, University of Kansas Medical Center, Department of Pathology and Laboratory Medicine, 2001 Lied, 3901 Rainbow Blvd, Kansas City, KS 66160; e-mail: cnicot@kumc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal