Abstract
Hepatitis C virus (HCV) is associated with the B-cell lymphoproliferative disorders mixed cryoglobulinemia (MC) and non-Hodgkin lymphoma. We have previously reported that HCV+MC+ patients have clonal expansions of hypermutated, rheumatoid factor–bearing marginal zone-like IgM+CD27+ peripheral B cells using the VH1-69 gene. Here we coupled transcriptional profiling with immunophenotypic and functional studies to ascertain these cells' role in MC pathogenesis. Despite their fundamental role in MC disease, these B cells have overall transcriptional features of anergy and apoptosis instead of neoplastic transformation. Highly up-regulated genes include SOX5, CD11C, galectin-1, and FGR, similar to a previously described FCRL4+ memory B-cell subset and to an “exhausted,” anergic CD21low memory B-cell subset in HIV+ patients. Moreover, HCV+MC+ patients' clonal peripheral B cells are enriched with CD21low, CD11c+, FCRL4high, IL-4Rlow memory B cells. In contrast to the functional, rheumatoid factor–secreting CD27+CD21high subset, the CD27+CD21low subpopulation exhibits decreased calcium mobilization and does not efficiently differentiate into rheumatoid factor–secreting plasmablasts, suggesting that a large proportion of HCV+MC+ patients' clonally expanded peripheral B cells is prone to anergy and/or apoptosis. Down-regulation of multiple activation pathways may represent a homeostatic mechanism attenuating otherwise uncontrolled stimulation of circulating HCV-containing immune complexes. This study was registered at www.clinicaltrials.gov as #NCT00435201.
Introduction
Hepatitis C virus (HCV) chronically infects approximately 170 million people worldwide and is the leading indicator for liver transplantation in the United States. Although hepatocytes are the primary target for HCV infection, the B-cell lymphoproliferative disorder mixed cryoglobulinemia (MC) affects up to 50% of HCV patients.1 MC is characterized by the aberrant production of monoclonal rheumatoid factor (RF)–containing immune complexes that deposit on vascular endothelium of organs, such as skin, kidneys, and peripheral nerves, eliciting a complement C1q-mediated vasculitis.2 HCV has also been associated with B-cell non-Hodgkin lymphoma (NHL),3 most frequently of low-grade marginal zone or mucosa-associated lymphoid tissue subtypes, although associations with higher-grade NHL have been reported.
HCV-induced B-cell dysregulation probably represents a continuum from the relatively benign clonal B-cell expansion of MC to overt NHL. The continued presence of HCV is necessary for abnormal B-cell lymphoproliferation, as eradication of HCV typically results in resolution of both HCV-related MC and NHL.4 Clonal B-cell populations are present in the liver and peripheral blood of HCV+MC+ patients5 ; such B cells demonstrate biased usage of the RF-encoding VH1-69 and Vκ3-20 gene segments,6 as do B cells isolated from lymph nodes of HCV-NHL patients.7 It remains unclear why B cells undergo clonal proliferation during chronic HCV infection.
It is probable that HCV-induced B-cell lymphoproliferation is not the result of direct B-cell infection or transformation, but rather, an indirect process arising from chronic antigenic stimulation of a limited pool of preexisting autoreactive B cells. We have proposed that persistently high levels of HCV-containing immune complexes stimulate the proliferation of RF-bearing B cells,6 but the precise antigen(s) and stimulatory mechanisms have remained elusive. We have previously shown that HCV+MC+ patients' clonal B cells are predominantly IgM memory B cells expressing modestly hypermutated immunoglobulin genes; phylogenetic analysis supports a process of antigen-directed affinity maturation. However, many of these clonal cells have decreased expression of CD21, the CR2 complement receptor.6 Because CD21 augments B-cell receptor (BCR)-mediated signaling as part of the B-cell coreceptor complex, its down-regulation may confer a state of relative anergy to these cells, as has been demonstrated among CD21low naive B cells from patients with chronic variable immunodeficiency and rheumatoid arthritis.8
To better understand how HCV elicits the expansion of autoreactive B-cell clones, we have performed transcriptional, immunophenotypic, and functional analyses on HCV+MC+ patients' clonal B cells. Contrary to expectations, these cells have a global transcriptional profile suggestive of anergy and apoptosis, and a large proportion of them have immunophenotypic features of anergy. Taken together, our data suggest that, although HCV+MC+ patients clearly have expanded peripheral B cells capable of differentiating into RF-secreting plasmablasts, these cells do not have transcriptional features of neoplastic transformation, and a significant proportion of this clonal population may be refractory to ongoing antigenic stimulation.
Methods
Patients
The studies were approved by the Institutional Review Boards at the Rockefeller University and New York Presbyterian Hospitals. Donors gave written informed consent according to the Declaration of Helsinki before enrollment. We enrolled HCV Ab−, HCV Ab+/HCV RNA+, and HCV Ab+/HCV RNA− volunteers. No subjects received interferon or immunosuppressive therapy within 6 months of enrollment. Blood was obtained by peripheral blood draw and leukapheresis. Peripheral blood mononuclear cells (PBMCs) were prepared as previously described.6
Clinical tests
HCV RNA was quantified clinically by the Roche Amplicor assay (Version 2.0; Roche Diagnostics); results are standardized to international units. Liver biopsies were evaluated by pathologists according to the Scheuer system. These tests, in addition to serum alanine aminotransferase measurements, were performed as part routine medical care. Testing for MC was performed as previously described.6
IgM+κ+CD27+ B-cell isolation
IgM+κ+ B cells were isolated from PBMCs by negative selection to minimize transcriptional changes effected by BCR signaling. All steps were performed at 4°C. B cells were immunomagnetically isolated using a B Cell Isolation Kit (Miltenyi Biotec). These were incubated with phycoerythrin-conjugated anti-IgG, anti-IgA, and anti-λ, then with anti–phycoerythrin-conjugated microbeads, and the negative fraction was magnetically purified. The CD27+ fraction was immunomagnetically isolated using anti–CD27-conjugated microbeads.
RNA extraction, cDNA synthesis, amplification, and labeling
RNA was extracted from 5000 to 10 000 cells using the RNeasy Plus Micro Kit (QIAGEN) with on-column DNase digestion. RNA integrity and concentration were determined using Lab on a Chip Pico. Samples with RNA integrity numbers > 9.0 were used for downstream processing. A total of 2 ng RNA was reverse-transcribed with random hexamers as primers and amplified using the WT-Ovation Pico Kit (Nugen), and 5 μg cDNA labeled using uracyl-N-glycosylase (Epicentre Biotechnologies) and biotinylated aldehyde-reactive probe.
Microarray procedures
Human V3 BeadChips (Illumina) were hybridized with 1.5 μg cDNA. Chips were scanned on an Illumina Beadstation and analyzed with Illumina BeadStudio software (Version 3.2). Datasets were analyzed using GeneSpring GX Version 11.1 (Agilent Technologies). Raw signal values were log-transformed, chips were normalized to the 50th percentile, and genes normalized to the median signal. This dataset was filtered to include genes with signals above background. Welch t test (P = .05, Benjamini-Hochberg false discovery rate = 0.05) was used to test for differences in genes between groups. The resulting set was filtered to include genes that were 2-fold up- or down-regulated. Hierarchical clustering was performed using the weighted pairwise group method with centroid average, using the Pearson correlation as the distance metric. Statistics were calculated using GeneSpring GX and Prism (GraphPad Software).
Quantitative RT-PCR
RNA was prepared from isolated B cells, as described under “RNA extraction, cDNA synthesis, amplification, and labeling.” Random-primed cDNA was synthesized using Superscript III (Invitrogen). Primers (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) were constructed using the PrimerBank Database (www.pga.mgh.harvard.edu/primerbank) and were designed to span exon-intron borders to reduce the possibility of genomic DNA amplification. SYBR Green PCR Master Mix (Applied Biosystems) was used for quantitative reverse-transcribed polymerase chain reaction (RT-PCR). We normalized all samples to RPS11 and to the target gene in Universal Human Reference RNA (Stratagene). Fold change expression was calculated using the 2−ΔΔCt method. Groups were compared using the Kruskal-Wallis test; when Kruskal-Wallis P < .05, the HCV+MC+ and HCV+MC− groups were compared using the Dunn post-test.
Flow cytometry
Cells were stained with monoclonal antibodies (mAbs) in phosphate-buffered saline supplemented with 2% (weight/volume) bovine serum albumin (Fraction V; Fisher Biotech) and 0.02% NaN3. All antibodies and reagents were from BD Biosciences, except for G6 (provided by R. Jefferis), F(ab′)2 anti-FCRL4 biotin (provided by G. Erhardt and M. Cooper), and Ki-67 fluorescein isothiocyanate (Invitrogen). Conjugation of G6 to biotin and AlexaFluor-594 was performed using commercial kits (Pierce Chemical, Invitrogen). Analysis was performed within 1 hour on a BD LSRII flow cytometer (BD Biosciences).
G6+ B-cell subset isolation
PBMCs were stained with anti-CD20, anti-CD27, anti-CD21, G6 mAbs, and 4,6-diamidino-2-phenylindole (to exclude dead cells). Live CD27+/−CD21high/low G6+, CD20+ B cells were bulk-sorted on a BD FACSAria II (BD Biosciences). For assessment of postsort viability, sorted samples were restained with 4,6-diamidino-2-phenylindole and were reanalyzed by flow cytometry. Postsort analysis confirmed more than 85% viability and more than 99% purity of sorted populations.
Cell cycle analysis
B cells were negatively isolated from PBMCs using EasySep Human B Cell Enrichment Kit (StemCell Technologies) and incubated with biotinylated G6 mAb. G6+ and G6− B-cell subsets were purified using streptavidin-conjugated immunomagnetic beads. Cells were fixed in 80% ethanol. After incubation with FITC-labeled Ki-67, cells were resuspended in PBS containing 10 mg/mL RNase A and incubated at 37°C. Propidium iodide 20 μg/mL was added before flow cytometry.
Electron microscopy
Cells were fixed in 2% glutaraldehyde, incubated in 1% osmium, dehydrated in a graded alcohol series, embedded in spur resin, and then treated with 2% uranyl acetate and Reynold lead citrate. Transmission electron microscopy was performed at ×2000 and ×10 000 magnification.
Calcium mobilization assay
A total of 2 × 106 B cells were incubated with 1μM Indo-1 for 30 minutes. Cells were then labeled with anti-CD19, IgG, CD27, and CD21 mAbs and suspended in HBSS with Ca2+ and 1% bovine serum albumin. Emission at 405 and 495 nm was measured to obtain a baseline, and then for 5 minutes. After addition of 10 μg/mL goat F(ab′)2 anti-IgM, 405/495 nm emission ratios of IgG− B-cell subsets were analyzed with FlowJo software Version 9.2 (TreeStar).
Annexin V apoptosis assay
A total of 2 × 106 PBMCs were incubated with 1 μg/mL anti-CD95 or mouse IgG1 and 2 μg/mL Protein G (Invitrogen) in RPMI/10% fetal calf serum at 37°C for 0 and 6 hours. Cells were washed with phosphate-buffered saline and resuspended in 0.01M N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (pH 7.4), 0.14M NaCl, 2.5mM CaCl2, and 2% fetal calf serum. After incubation with annexin V–phycoerythrin and 7-amino-actinomycin at room temperature, cells were stained with anti-CD20 FITC anti-CD21–allophycocyanin, and G6-biotin at room temperature. After staining cells with streptavidin-Cy7-allophycocyanin, flow cytometry was performed.
Immunoglobulin secretion assays
Cells (50 000/well in 96-well round-bottom plates) were cultured for 6 days in RPMI supplemented with 10% fetal calf serum, 2mM l-glutamine, 100 U/mL penicillin/streptomycin, and 0.25 μg/mL amphotericin B, with the addition of 6 U/mL IL-2 (R&D Systems), 200 ng/mL IL-10 (R&D Systems), and 1 μg/mL flag-tagged CD40L with 2 μg/mL mouse IgG1 anti-flag Ab (Alexis Biochemicals).
For ELISPOT, cells were washed with RPMI, placed on MultiScreen filter plates (Millipore), coated with goat F(ab′)2 anti–human IgM (Jackson ImmunoResearch Laboratories), and incubated at 37°C for 6 hours. Plates were then incubated with horseradish peroxidase-labeled anti–human IgM, and the assays were developed with 3-amino-9-ethylcarbazole (Sigma-Aldrich). Spots were counted using an ImmunoSpot Analysis Instrument (Cellular Technology).
For ELISA, cell culture supernatants were added to MaxiSorb plates (Nunc) coated with anti-IgM (Bethyl Laboratories), or for RF assay, human IgG1λ (Sigma-Aldrich), and incubated at room temperature for 1 hour. Plates were then incubated with HRP-labeled goat-anti–human IgM, and the assays were developed with TMB (BioFX Laboratories). After stopping reactions with 1N H2SO4, A450 was measured on a FLUOstar Omega microplate reader (BMG Laboratories).
Accession numbers
Microarray data are accessible through NCBI Gene Expression Omnibus accession number GSE18084 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE18084).
Results
Characteristics of study subjects
Five HCV Ab− volunteers, 7 sustained virologic responders (SVRs), and 27 persons with chronic HCV infection were enrolled for gene transcriptional analyses. Three additional HCV+MC− and 2 additional HCV+MC+ subjects were enrolled only for immunophenotypic and functional analyses (Table 1). Sixteen HCV+ subjects were MC+, and all 16 had evidence of clonal B-cell populations, as demonstrated by complementarity-determining region 3 PCR.6 Four of these subjects had lymphadenopathy; lymph node biopsies performed by their physicians confirmed low-grade B-NHL (subjects 1116, 1308, 1716, and ECH 529). Fifteen of 16 patients had evidence of clonal IgM gammopathy by serum immunofixation electrophoresis and were classified as being IgM MC+. Subject ECH 529 had evidence of clonal IgA gammopathy by immunofixation electrophoresis. In addition, cervical lymph node biopsy in this patient revealed abnormal numbers of IgA+κ+ B cells. Plasma from 10 of 12 HCV RNA− and 25 of 27 HCV RNA+ patients had detectable anti-Epstein-Barr virus nuclear antigen 1 IgG, indicating previous exposure to Epstein-Barr virus. HCV+ IgM MC+ subjects had significantly expanded populations of IgM+κ+ peripheral B cells, although overall B-cell numbers were not increased, consistent with our earlier report6 (supplemental Figure 1).
Characteristics of the study patients
| Condition/subject no. . | Age, y . | Sex . | Ethnicity . | HCV RNA . | GT . | Stage (0-4) . | Treatment history . | Clonal CDR3 . |
|---|---|---|---|---|---|---|---|---|
| HCV Ab- | ||||||||
| LDU 099 | 35 | Male | White | NA | NA | NA | NA | No |
| LDU 128 | 40 | Male | Black | NA | NA | NA | NA | No |
| ECH 503 | 35 | Male | Asian American | NA | NA | NA | NA | No |
| ECH 527 | 50 | Female | White/Hispanic | NA | NA | NA | NA | No |
| ECH 528 | 51 | Female | White | NA | NA | NA | NA | No |
| SVR | ||||||||
| 543 | 48 | Female | White | < 50 | 1 | 4 | pIFN/RBV 2001 | No |
| 731 | 52 | Female | White | < 50 | 2b | 2 | pIFN/RBV 2003 | No |
| 856 | 61 | Female | White | < 50 | 1a | 2 | pIFN/RBV 2003 | No |
| 1154 | 56 | Female | Asian American | < 50 | 1a | 2 | pIFN/RBV 2003 | No |
| 1197 | 38 | Female | White | < 50 | 2a | 1 | pIFN/RBV 2003 | No |
| ECH 521 | 43 | Male | White | < 50 | 1 | ND | pIFN/RBV 2005 | No |
| ECH 542 | 38 | Female | Black | < 50 | 1 | ND | pIFN/RBV 2007 | No |
| HCV+, IgM MC− | ||||||||
| 1235 | 44 | Female | White | 3.85 × 106 | 2b | ND | Naive | No |
| 1330 | 54 | Female | White/Hispanic | > 7 × 105 | 1a | 2 | Naive | No |
| 1419 | 48 | Female | White | 0.39 × 106 | 1 | 3 | Naive | No |
| 1864 | 58 | Male | White | 0.02 × 106 | 4 | 3 | Naive | No |
| LDU 107 | 50 | Male | White/Hispanic | > 7 × 105 | 2 | 2 | Naive | No |
| ECH 507 | 54 | Male | White/Hispanic | > 7 × 105 | 1 | ND | Naive | No |
| ECH 512* | 62 | Male | White | > 7 × 105 | 1 | ND | Naive | No |
| ECH 516 | 57 | Male | White/Hispanic | 2.4 × 106 | 1b | 2 | Naive | No |
| ECH 519 | 52 | Female | White/Hispanic | > 7 × 105 | 1 | ND | Naive | No |
| ECH 520 | 57 | Male | Black | > 5 × 106 | 1b | 2 | Naive | No |
| ECH 522 | 50 | Female | White/Hispanic | > 7 × 105 | 1 | ND | Naive | No |
| ECH 529†‡ | 52 | Female | White | > 7 × 105 | 3 | 4 | Naive | Yes |
| ECH 530 | 49 | Male | White/Hispanic | > 7 × 105 | 1 | ND | Naive | No |
| ECH 536* | 50 | Male | White/Hispanic | 0.2 × 106 | 1 | 1 | Naive | No |
| ECH 537* | 54 | Male | Black | 0.9 × 106 | 1a | ND | Naive | No |
| HCV+, IgM MC+ | ||||||||
| 110 | 53 | Male | White/Hispanic | 1.70 × 106 | 1 | 3 | Naive | Yes |
| 880 | 54 | Female | White | 3 × 106 | 1b | 2 | Naive | Yes |
| 1116‡ | 37 | Male | White | 3 × 104 | 2b | 2 | Naive | Yes |
| 1308‡ | 55 | Female | White | 0.72 × 106 | 1 | 1 | Naive | Yes |
| 1403 | 44 | Female | White/Hispanic | 0.50 × 106 | 1 | 2 | Naive | Yes |
| 1432 | 69 | Female | White | 0.03 × 106 | 1 | 4 | Naive | Yes |
| 1540 | 37 | Male | White/Hispanic | 0.28 × 106 | 1 | ND | Naive | Yes |
| 1716‡ | 30 | Female | White | 1.45 × 106 | 1a | ND | Naive | Yes |
| 1931 | 56 | Female | White | 2.2 × 106 | 1b | 2 | Naive | Yes |
| 92200 | 57 | Female | White | 2 × 106 | 2 | 2 | Naive | Yes |
| LDU 125 | 60 | Male | White | 0.2 × 106 | 3 | ND | Naive | Yes |
| ECH 531 | 56 | Male | White | > 7 × 105 | 1 | ND | Naive | Yes |
| ECH 532 | 51 | Female | Black | > 7 × 105 | 1 | ND | Naive | Yes |
| ECH 533 | 63 | Male | White | 3.8 × 106 | 1 | 0 | pIFN/RBV 2007 | Yes |
| ECH 535 | 57 | Male | White | 0.74 × 106 | 1 | ND | Naive | Yes |
| ECH 546* | 52 | Male | White | > 7 × 105 | 1 | 3 | pIFN/RBV 2008 | ND |
| ECH 559* | 58 | Female | White/Hispanic | 1.27 × 106 | 1 | 4 | pIFN/RBV 2001 | ND |
| Condition/subject no. . | Age, y . | Sex . | Ethnicity . | HCV RNA . | GT . | Stage (0-4) . | Treatment history . | Clonal CDR3 . |
|---|---|---|---|---|---|---|---|---|
| HCV Ab- | ||||||||
| LDU 099 | 35 | Male | White | NA | NA | NA | NA | No |
| LDU 128 | 40 | Male | Black | NA | NA | NA | NA | No |
| ECH 503 | 35 | Male | Asian American | NA | NA | NA | NA | No |
| ECH 527 | 50 | Female | White/Hispanic | NA | NA | NA | NA | No |
| ECH 528 | 51 | Female | White | NA | NA | NA | NA | No |
| SVR | ||||||||
| 543 | 48 | Female | White | < 50 | 1 | 4 | pIFN/RBV 2001 | No |
| 731 | 52 | Female | White | < 50 | 2b | 2 | pIFN/RBV 2003 | No |
| 856 | 61 | Female | White | < 50 | 1a | 2 | pIFN/RBV 2003 | No |
| 1154 | 56 | Female | Asian American | < 50 | 1a | 2 | pIFN/RBV 2003 | No |
| 1197 | 38 | Female | White | < 50 | 2a | 1 | pIFN/RBV 2003 | No |
| ECH 521 | 43 | Male | White | < 50 | 1 | ND | pIFN/RBV 2005 | No |
| ECH 542 | 38 | Female | Black | < 50 | 1 | ND | pIFN/RBV 2007 | No |
| HCV+, IgM MC− | ||||||||
| 1235 | 44 | Female | White | 3.85 × 106 | 2b | ND | Naive | No |
| 1330 | 54 | Female | White/Hispanic | > 7 × 105 | 1a | 2 | Naive | No |
| 1419 | 48 | Female | White | 0.39 × 106 | 1 | 3 | Naive | No |
| 1864 | 58 | Male | White | 0.02 × 106 | 4 | 3 | Naive | No |
| LDU 107 | 50 | Male | White/Hispanic | > 7 × 105 | 2 | 2 | Naive | No |
| ECH 507 | 54 | Male | White/Hispanic | > 7 × 105 | 1 | ND | Naive | No |
| ECH 512* | 62 | Male | White | > 7 × 105 | 1 | ND | Naive | No |
| ECH 516 | 57 | Male | White/Hispanic | 2.4 × 106 | 1b | 2 | Naive | No |
| ECH 519 | 52 | Female | White/Hispanic | > 7 × 105 | 1 | ND | Naive | No |
| ECH 520 | 57 | Male | Black | > 5 × 106 | 1b | 2 | Naive | No |
| ECH 522 | 50 | Female | White/Hispanic | > 7 × 105 | 1 | ND | Naive | No |
| ECH 529†‡ | 52 | Female | White | > 7 × 105 | 3 | 4 | Naive | Yes |
| ECH 530 | 49 | Male | White/Hispanic | > 7 × 105 | 1 | ND | Naive | No |
| ECH 536* | 50 | Male | White/Hispanic | 0.2 × 106 | 1 | 1 | Naive | No |
| ECH 537* | 54 | Male | Black | 0.9 × 106 | 1a | ND | Naive | No |
| HCV+, IgM MC+ | ||||||||
| 110 | 53 | Male | White/Hispanic | 1.70 × 106 | 1 | 3 | Naive | Yes |
| 880 | 54 | Female | White | 3 × 106 | 1b | 2 | Naive | Yes |
| 1116‡ | 37 | Male | White | 3 × 104 | 2b | 2 | Naive | Yes |
| 1308‡ | 55 | Female | White | 0.72 × 106 | 1 | 1 | Naive | Yes |
| 1403 | 44 | Female | White/Hispanic | 0.50 × 106 | 1 | 2 | Naive | Yes |
| 1432 | 69 | Female | White | 0.03 × 106 | 1 | 4 | Naive | Yes |
| 1540 | 37 | Male | White/Hispanic | 0.28 × 106 | 1 | ND | Naive | Yes |
| 1716‡ | 30 | Female | White | 1.45 × 106 | 1a | ND | Naive | Yes |
| 1931 | 56 | Female | White | 2.2 × 106 | 1b | 2 | Naive | Yes |
| 92200 | 57 | Female | White | 2 × 106 | 2 | 2 | Naive | Yes |
| LDU 125 | 60 | Male | White | 0.2 × 106 | 3 | ND | Naive | Yes |
| ECH 531 | 56 | Male | White | > 7 × 105 | 1 | ND | Naive | Yes |
| ECH 532 | 51 | Female | Black | > 7 × 105 | 1 | ND | Naive | Yes |
| ECH 533 | 63 | Male | White | 3.8 × 106 | 1 | 0 | pIFN/RBV 2007 | Yes |
| ECH 535 | 57 | Male | White | 0.74 × 106 | 1 | ND | Naive | Yes |
| ECH 546* | 52 | Male | White | > 7 × 105 | 1 | 3 | pIFN/RBV 2008 | ND |
| ECH 559* | 58 | Female | White/Hispanic | 1.27 × 106 | 1 | 4 | pIFN/RBV 2001 | ND |
GT indicates genotype; CDR3, Ig complementarity determining region 3; pIFN/RBV, pegylated interferon/ribavirin; NA, not applicable; and ND, not done.
Subjects used for immunophenotypic and functional assays only.
Subject with κ+ IgA MC.
Subjects with marginal zone B-cell NHL (documented on lymph node biopsy).
IgM+κ+CD27+ B cells from HCV+MC+ patients have a distinct transcriptional profile
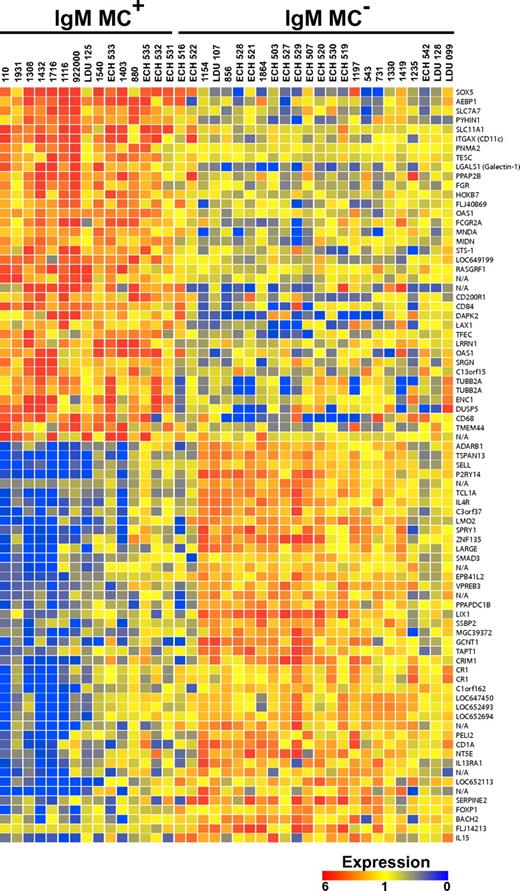
A total of 69 unique genes (33 up-regulated, 36 down-regulated) were found to be more than 2-fold differentially expressed in IgM+κ+CD27+ B cells from IgM MC+, compared with IgM MC−, subjects (Figure 1). Notably, the transcriptional profile of 2 HCV RNA+MC− patients (ECH 516 and ECH 522) shared several features with that of the MC+ population.
Transcriptome analysis of IgM+κ+CD27+ B cells. The transcripts differentially expressed by IgM+κ+CD27+ B cells from HCV+MC+ and MC− patients are displayed. Up- and down-regulated transcripts are indicated in red and blue, respectively. The magnitude of expression is indicated by the color bar.
Transcriptome analysis of IgM+κ+CD27+ B cells. The transcripts differentially expressed by IgM+κ+CD27+ B cells from HCV+MC+ and MC− patients are displayed. Up- and down-regulated transcripts are indicated in red and blue, respectively. The magnitude of expression is indicated by the color bar.
Several of the differentially expressed genes were grouped according to broad function (Table 2). The overall transcriptional pattern was suggestive not of oncogenesis, but of dampened activation and augmented proapoptotic pathways. Unsurprisingly, several IFN-induced genes were up-regulated: growth interferon-inducible protein X (PYHIN1), myeloid nuclear differentiation antigen (MNDA), and 2′, 5′-oligoadenylate synthetase 1 (OAS1). Several genes associated with B-cell anergy were up-regulated: galectin-1 (LGALS1), lymphocyte transmembrane adapter 1 (LAX1), and CD200 receptor 1 (CD200R1), an inhibitory receptor highly expressed on memory B cells and plasmablasts. Significantly up-regulated proapoptotic genes included: galectin-1, the interferon-response gene, PYHIN1, death-associated protein kinase 2 (DAPK2), and MNDA. The prosurvival gene, T-cell lymphoma 1A (TCL1A), was markedly down-regulated.
Functional grouping of differentially expressed genes
| Functional group/gene . | Fold difference . | P . | Significance . |
|---|---|---|---|
| Interferon response | |||
| PYHIN1 (IFIX), (growth interferon-inducible protein X) | 2.8 | .000052 | Interferon-inducible HIN-200 gene family member17 |
| MNDA (myeloid nuclear differentiation antigen) | 2.2 | .0092 | HIN-200 gene family member17,19 |
| OAS1 (2′, 5′ oligoadenylate cyclase) | 2.1 | .000052 | IFN-inducible gene which activates RNaseL |
| B-cell anergy | |||
| LGALS1 (galectin-1) | 3.6 | .00038 | Negatively regulates B-cell proliferation upon BCR ligation14 ; up-regulated in anergic murine B cells12 |
| CD200R1 (CD200 receptor 1) | 2.4 | .0034 | Inhibitory receptor highly expressed on memory B cells and plasmablasts; definitive role in B cell activation not established11 |
| LAX1 (lymphocyte transmembrane adapter 1) | 2.1 | .032 | Dampens B-cell response to BCR engagement16 |
| B-cell apoptosis | |||
| LGALS1 | 3.6 | .00038 | Expressed in IgM+ memory cells, reported to enhance apoptosis via inhibition of Akt phosphorylation and up-regulation of Bim13 |
| PYHIN1 | 2.8 | .000052 | As an HIN-200 gene family member, contains an N-terminal PAAD/DAPIN/Pyrin domain that mediates binding with proteins involved in apoptotic NF-κB and caspase signaling17 |
| DAPK2 (death-associated protein kinase 2) | 2.4 | .043 | Calcium/calmodulin-dependent protein kinase, which induces apoptosis15 |
| MNDA | 2.2 | .0092 | As an HIN-200 gene family member, contains an N-terminal PAAD/DAPIN/Pyrin domain that mediates binding with proteins involved in apoptotic NF-κB and caspase signaling17 |
| B-cell survival | |||
| TCL1A (T-cell lymphoma antigen 1A) | 0.34 | .000075 | Reported to enhance B-cell survival by induction of Mcl-113 |
| B-cell lymphomagenesis | |||
| SOX5 (SRY-box 5) | 3.6 | .00062 | Reported to be up-regulated in splenic follicular lymphoma20 |
| ITGAX, (CD11C), (integrin, α X) | 2.9 | .00066 | High expression associated with splenic marginal zone lymphoma21 |
| MNDA | 2.2 | .0092 | Expressed in marginal zone B cells, up-regulated in marginal zone lymphoma22 |
| BACH2 | 0.46 | .022 | Down-regulated in Waldenström macroglobulinemia10 |
| FOXP1 | 0.44 | .00072 | Up-regulation of FOXP1 in mucosa-associated lymphoid tissue lymphoma may be associated with neoplastic transformation23 |
| TCL1A (T-cell lymphoma 1A) | 0.34 | .00075 | Up-regulated in splenic marginal zone lymphoma18 |
| SELL (L-selectin) | 0.34 | .00031 | Up-regulated in splenic marginal zone lymphoma18 |
| IL-4R (IL-4 receptor) | 0.31 | .00017 | Down-regulated in mantle cell lymphoma9 and Waldenström macroglobulinemia10 |
| LMO2 (LIM domain only 2) | 0.31 | .0010 | Up-regulation corresponds to increased survival in diffuse large B-cell lymphoma24 |
| Functional group/gene . | Fold difference . | P . | Significance . |
|---|---|---|---|
| Interferon response | |||
| PYHIN1 (IFIX), (growth interferon-inducible protein X) | 2.8 | .000052 | Interferon-inducible HIN-200 gene family member17 |
| MNDA (myeloid nuclear differentiation antigen) | 2.2 | .0092 | HIN-200 gene family member17,19 |
| OAS1 (2′, 5′ oligoadenylate cyclase) | 2.1 | .000052 | IFN-inducible gene which activates RNaseL |
| B-cell anergy | |||
| LGALS1 (galectin-1) | 3.6 | .00038 | Negatively regulates B-cell proliferation upon BCR ligation14 ; up-regulated in anergic murine B cells12 |
| CD200R1 (CD200 receptor 1) | 2.4 | .0034 | Inhibitory receptor highly expressed on memory B cells and plasmablasts; definitive role in B cell activation not established11 |
| LAX1 (lymphocyte transmembrane adapter 1) | 2.1 | .032 | Dampens B-cell response to BCR engagement16 |
| B-cell apoptosis | |||
| LGALS1 | 3.6 | .00038 | Expressed in IgM+ memory cells, reported to enhance apoptosis via inhibition of Akt phosphorylation and up-regulation of Bim13 |
| PYHIN1 | 2.8 | .000052 | As an HIN-200 gene family member, contains an N-terminal PAAD/DAPIN/Pyrin domain that mediates binding with proteins involved in apoptotic NF-κB and caspase signaling17 |
| DAPK2 (death-associated protein kinase 2) | 2.4 | .043 | Calcium/calmodulin-dependent protein kinase, which induces apoptosis15 |
| MNDA | 2.2 | .0092 | As an HIN-200 gene family member, contains an N-terminal PAAD/DAPIN/Pyrin domain that mediates binding with proteins involved in apoptotic NF-κB and caspase signaling17 |
| B-cell survival | |||
| TCL1A (T-cell lymphoma antigen 1A) | 0.34 | .000075 | Reported to enhance B-cell survival by induction of Mcl-113 |
| B-cell lymphomagenesis | |||
| SOX5 (SRY-box 5) | 3.6 | .00062 | Reported to be up-regulated in splenic follicular lymphoma20 |
| ITGAX, (CD11C), (integrin, α X) | 2.9 | .00066 | High expression associated with splenic marginal zone lymphoma21 |
| MNDA | 2.2 | .0092 | Expressed in marginal zone B cells, up-regulated in marginal zone lymphoma22 |
| BACH2 | 0.46 | .022 | Down-regulated in Waldenström macroglobulinemia10 |
| FOXP1 | 0.44 | .00072 | Up-regulation of FOXP1 in mucosa-associated lymphoid tissue lymphoma may be associated with neoplastic transformation23 |
| TCL1A (T-cell lymphoma 1A) | 0.34 | .00075 | Up-regulated in splenic marginal zone lymphoma18 |
| SELL (L-selectin) | 0.34 | .00031 | Up-regulated in splenic marginal zone lymphoma18 |
| IL-4R (IL-4 receptor) | 0.31 | .00017 | Down-regulated in mantle cell lymphoma9 and Waldenström macroglobulinemia10 |
| LMO2 (LIM domain only 2) | 0.31 | .0010 | Up-regulation corresponds to increased survival in diffuse large B-cell lymphoma24 |
IFN indicates interferon; PAAD/DAPIN, Pyrin, AIM (Absent in Melanoma), ASC (Apoptosis-associated-speck-like protein containing a caspase recruitment domain [CARD]) and death-domain (DD)-like Domain in Apoptosis and Interferon response; and NFκB, nuclear factor-κB.
Bonferroni corrected.
In addition to having an overall transcriptional program suggestive of B-cell anergy and apoptosis, HCV+MC+ patients' IgM+κ+CD27+ B cells demonstrated differential regulation of several genes previously reported increased in patients with NHL. Up-regulated genes included: SRY-box 5 (SOX5), α-X integrin (ITGAX, CD11C), and MNDA. Down-regulated genes included: L-selectin (SELL), LIM only 2 (LMO2), forkhead box 1 (FOXP1), and TCL1A. Also down-regulated was IL-4 receptor (IL-4R), polymorphisms of which have been associated with diffuse large B-cell lymphoma25 and which may be down-regulated in mantle cell lymphoma.9 In addition, BTB and CNC homology 1, basic leucine zipper transcription factor 2 (BACH2) was down-regulated; both BACH2 and IL-4R are reported to be down-regulated in B cells from patients with Waldenström macroglobulinemia.10
Because many of the up-regulated genes (eg, CD11C, CD84, CD200R1, and bone morphogenetic protein receptor 1A [BMPR1A]) are known to be expressed in activated and/or memory B cells,11,26-28 we hypothesized that their up-regulation reflected a particular stage of differentiation of HCV+MC+ patients' clonally expanded B cells. Several of the most significantly up-regulated genes (SOX5, Gardner-Rasheed feline sarcoma viral oncogene homolog [FGR], and CD11C) have previously been found to be highly up-regulated in FCRL4-expressing tonsillar B cells, a recently described memory B-cell subset thought to play an important role in mucosal defense.29 Despite this transcriptional similarity, our microarray data did not reveal differences in FCRL4 transcript between MC+ and MC− patients' IgM+κ+CD27+ peripheral B cells. However, quantitative RT-PCR of unamplified cDNA confirmed the up-regulated expression of SOX5, FGR, and CD11C in HCV+MC+ patients' expanded IgM+κ+CD27+ B cells (Figure 2; supplemental Table 2). In addition, quantitative RT-PCR confirmed the up-regulation of galectin-1 and OAS1 and the down-regulation of IL-4R and TCL1A, and it detected no significant difference in FCRL4 expression. We did not detect Epstein-Barr virus nuclear antigen 2 or latent membrane protein 1 transcripts by quantitative RT-PCR (data not shown).
Relative expression of selected genes in IgM+κ+CD27+ B cells determined by quantitative RT-PCR. Values are normalized to RPS11 for cDNA content and to a universal standard RNA. Comparisons between groups were made using the Kruskal-Wallis test, and P values for HCV+MC− compared with HCV+MC+ subjects were computed using Dunn post test. N.S. indicates not significant (P > .05).
Relative expression of selected genes in IgM+κ+CD27+ B cells determined by quantitative RT-PCR. Values are normalized to RPS11 for cDNA content and to a universal standard RNA. Comparisons between groups were made using the Kruskal-Wallis test, and P values for HCV+MC− compared with HCV+MC+ subjects were computed using Dunn post test. N.S. indicates not significant (P > .05).
A significant proportion of clonal cells from IgM MC+ subjects are CD21low, CD11c+, FCRL4high, IL-4Rlow memory B cells
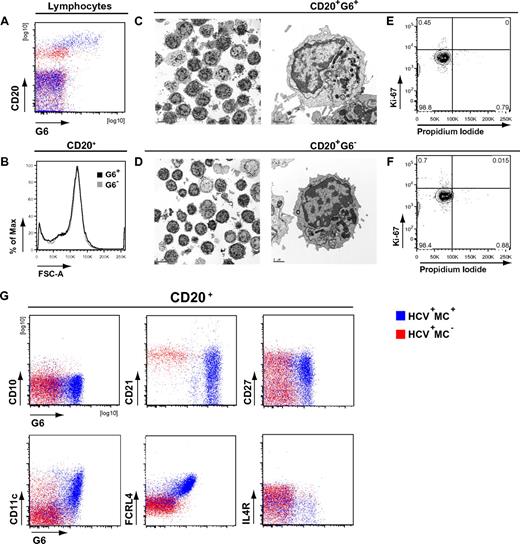
We and others have previously shown that clonally expanded IgM+κ+CD27+ B cells from IgM MC+ patients preferentially use the VH1–69 gene segment.6,30 We used the G6 mAb, which recognizes the complementarity-determining region 2 of Ig VH1-69,31 to more precisely immunophenotype HCV+MC+ patients' clonally expanded B cells. We have previously confirmed the specificity of G6 for VH1-69 by RT-PCR.32 These G6+ B cells from HCV+MC+ patients are frequently CD21low and IgM+κ+ (supplemental Figure 2). They are also morphologically normal, nonproliferating, and predominantly CD20high, CD10−, CD21low, CD27+, CD11c+, FCRL4high, and IL-4Rlow (Figure 3). We immunophenotyped B cells from 11 MC− (SVR = 2, HCV Ab− = 3, HCV RNA+ = 6) and 9 HCV RNA+MC+ subjects (Figure 4). As expected, we found that MC+ patients had a significant (P < .005) expansion of G6+ B cells (median, 25.9% of B cells) compared with HCV RNA+MC− patients (median, 5.1%), SVR (median, 3.0%), and HCV Ab− (median, 3.8%) patients. In 3 MC+ patients (LDU 125, 110, and 1432), more than 50% of total peripheral B cells were G6+. When we examined G6+ and G6− B cells from each person, we confirmed that G6+, compared with G6−, B cells from HCV+MC+ patients were predominantly CD21low, CD11c+, FCRL4high, and IL-4Rlow. Interestingly, G6+, compared with G6−, B cells from MC− persons were also disproportionately CD21low. However, they did not have significantly increased expression of CD11c or FCRL4, nor did they have decreased expression of IL-4R.
Phenotypic analysis of HCV+MC+ patients' clonal B cells. PBMCs from one HCV−MC− (ECH 542, red) and one HCV+MC+ patient (LDU 125, blue) representative of 6 samples each are shown. (A) Staining of PBMCs with anti-CD20 and G6 mAbs. (B) Forward light scatter analysis of G6+ and G6− B cells from the HCV+MC+ person. Scanning electron micrographs of immunomagnetically sorted G6+ and G6− B cells from the HCV+MC+ person. (C-D) Scale bars on the low and high power magnifications represent 5 μm and 1 μm, respectively. Propidium iodide and Ki-67 staining of immunomagnetically sorted G6+ and G6− B cells from the HCV+MC+ person for Ki-67 and propidium iodide (E-F). Staining for cell surface CD10, CD21, CD27, CD11c, FCRL4, and IL-4R (G). Electron micrographs (original magnification ×10 000) were taken with a FEI Tecnai G2 Spirit BioTWIN Transmission Electron Microscope equipped with a Gatan ES500W Erlangshen CCD camera and DigitalMicrograph Software.
Phenotypic analysis of HCV+MC+ patients' clonal B cells. PBMCs from one HCV−MC− (ECH 542, red) and one HCV+MC+ patient (LDU 125, blue) representative of 6 samples each are shown. (A) Staining of PBMCs with anti-CD20 and G6 mAbs. (B) Forward light scatter analysis of G6+ and G6− B cells from the HCV+MC+ person. Scanning electron micrographs of immunomagnetically sorted G6+ and G6− B cells from the HCV+MC+ person. (C-D) Scale bars on the low and high power magnifications represent 5 μm and 1 μm, respectively. Propidium iodide and Ki-67 staining of immunomagnetically sorted G6+ and G6− B cells from the HCV+MC+ person for Ki-67 and propidium iodide (E-F). Staining for cell surface CD10, CD21, CD27, CD11c, FCRL4, and IL-4R (G). Electron micrographs (original magnification ×10 000) were taken with a FEI Tecnai G2 Spirit BioTWIN Transmission Electron Microscope equipped with a Gatan ES500W Erlangshen CCD camera and DigitalMicrograph Software.
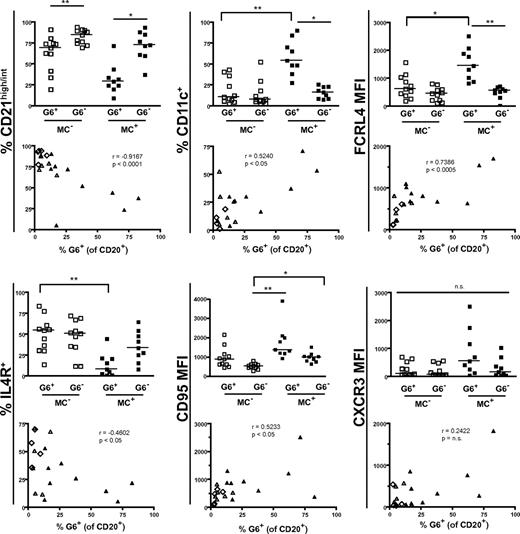
Immunophenotypic profiles of MC+ and MC− patients' G6+, G6−, and total B cells. Data are flow cytometric analyses of PBMCs from HCV+MC+ (n = 9) and MC− (n = 11; SVR = 2, HCV Ab− = 2, HCV RNA+ = 6) subjects. Cell surface marker expression in each subject's G6+ and G6− B cells is shown in the column graphs. The scatterplots represent expression of cell surface markers versus the proportion of total B cells that are G6+. In the scatterplots: ▴ represents MC+ patients; ▵, HCV RNA+ patients; and ◊, HCV RNA− patients. MFI indicates geometric mean fluorescence intensity. The P values for the column graphs were determined using the Kruskal-Wallis and Dunn multiple comparison tests. *P < .05. **P < .01. n.s. indicates not significant (P > .05). Black bars represent medians. For the scatterplots, R was calculated using the Spearman correlation coefficient.
Immunophenotypic profiles of MC+ and MC− patients' G6+, G6−, and total B cells. Data are flow cytometric analyses of PBMCs from HCV+MC+ (n = 9) and MC− (n = 11; SVR = 2, HCV Ab− = 2, HCV RNA+ = 6) subjects. Cell surface marker expression in each subject's G6+ and G6− B cells is shown in the column graphs. The scatterplots represent expression of cell surface markers versus the proportion of total B cells that are G6+. In the scatterplots: ▴ represents MC+ patients; ▵, HCV RNA+ patients; and ◊, HCV RNA− patients. MFI indicates geometric mean fluorescence intensity. The P values for the column graphs were determined using the Kruskal-Wallis and Dunn multiple comparison tests. *P < .05. **P < .01. n.s. indicates not significant (P > .05). Black bars represent medians. For the scatterplots, R was calculated using the Spearman correlation coefficient.
Reflective of having G6+ B-cell expansions, HCV+MC+ patients had increased percentages of total B cells that were CD21low, CD11c+, FCRL4high, and IL-4Rlow. These markers were significantly correlated with the percentage of peripheral B cells that were G6+. We did not detect any significant differences in any of these cell surface markers among the total B cells of SVR, HCV Ab−, and HCV RNA+ patients, probably reflective of the low proportion of G6+ B cells in these MC− persons.
FCRL4+ memory B cells from normal tonsils and HIV-viremic blood are reported to be CD20highCD11c+CD95+CD21low.29,33 In healthy tonsils, these FCRL4+ B cells have been variously reported to be CD27− or CD27+ memory B cells.34,35 Although we detected increased FCRL4 expression on G6+, compared with G6−, B cells, we did not detect significant differences in FCRL4 expression among CD27+, CD27−, CD21high, or CD21low B-cell subsets (supplemental Figure 3). In an attempt to reconcile the findings of unchanged FCRL4 transcript levels and increased FCRL4 protein expression in MC+ compared with MC− B cells, we performed quantitative RT-PCR on RNA isolated from the bulk-sorted FCRL4+ and FCRL4− G6+ B cells from subject LDU 125. However, we failed to detect any difference in FCRL4 mRNA between these 2 groups (data not shown).
CD21low B cells from healthy, HCV+MC−, and HCV+MC+ persons are anergic to BCR-mediated stimulation
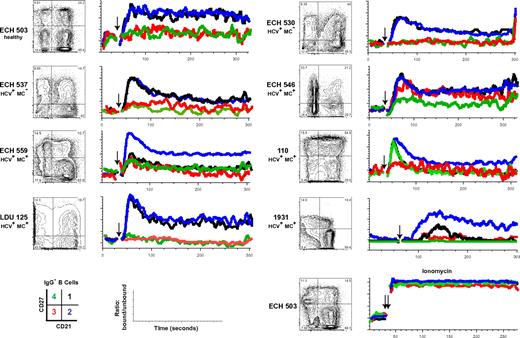
CD21low memory B cells have been described in HIV-viremic patients as being relatively anergic, “exhausted” B cells.33 We sought to determine whether IgG−, CD21low B cells had attenuated signaling on BCR ligation with anti–human IgM. Data for one healthy subject with an elevated (22%) CD21low B-cell frequency (ECH 503), 2 HCV+MC− (ECH 530, ECH 537) volunteers and 5 HCV+MC+ (ECH 546, ECH 559, 110, LDU 125, and 1931) volunteers are shown in Figure 5. In all but one (110), the CD27+CD21low subset had decreased Ca2+ mobilization; the reason for this outlier is unclear. Furthermore, in all subjects, the CD27−CD21low B-cell subset had diminished amplitude of Ca2+ mobilization; ECH 546, for unknown reasons, had an intermediate response to BCR triggering. Finally, in ECH 559 and 1931, CD27+CD21high cells had low responses to BCR stimulation; IgA may possibly be a prominent non-IgG BCR in their CD27+CD21high subsets. Overall, these data suggest that CD21low B cells, whether from healthy, HCV+MC−, or HCV+MC+ persons, are anergic to BCR-mediated stimulation.
CD27+CD21low, compared with CD27+CD21high, B cells have attenuated Ca2+ responses after BCR cross-linking. Analyses of B cells from one healthy volunteer (ECH 503), 2 HCV+MC− patients (ECH 530 and ECH 537), and 5 HCV+MC+ patients (ECH 546, ECH 559, 110, LDU 125, and 1931) are shown. PBMCs from 1931 and LDU 125 were collected 4 and 10 months, respectively, after the cells collected for the microarray and primary immunophenotyping experiments. (Left panels) CD27 and CD21 staining of IgG− B cells. Indo-1-AM-loaded cells were stained with anti-CD19, anti-IgG, anti-CD27, and anti-CD21, warmed to 37°C, and, after establishing a baseline for 30 seconds, stimulated with 10 μg/mL goat F(ab′)2 anti–human IgM. Kinetic graphs represent ratios of bound/unbound Indo-1 over time for CD27+CD21high, CD27+CD21low, CD27−CD21high, and CD27−CD21low B-cell populations. Single arrows indicate injection of F(ab′)2 anti–human IgM; and double arrow, injection of 10 μg/mL ionomycin.
CD27+CD21low, compared with CD27+CD21high, B cells have attenuated Ca2+ responses after BCR cross-linking. Analyses of B cells from one healthy volunteer (ECH 503), 2 HCV+MC− patients (ECH 530 and ECH 537), and 5 HCV+MC+ patients (ECH 546, ECH 559, 110, LDU 125, and 1931) are shown. PBMCs from 1931 and LDU 125 were collected 4 and 10 months, respectively, after the cells collected for the microarray and primary immunophenotyping experiments. (Left panels) CD27 and CD21 staining of IgG− B cells. Indo-1-AM-loaded cells were stained with anti-CD19, anti-IgG, anti-CD27, and anti-CD21, warmed to 37°C, and, after establishing a baseline for 30 seconds, stimulated with 10 μg/mL goat F(ab′)2 anti–human IgM. Kinetic graphs represent ratios of bound/unbound Indo-1 over time for CD27+CD21high, CD27+CD21low, CD27−CD21high, and CD27−CD21low B-cell populations. Single arrows indicate injection of F(ab′)2 anti–human IgM; and double arrow, injection of 10 μg/mL ionomycin.
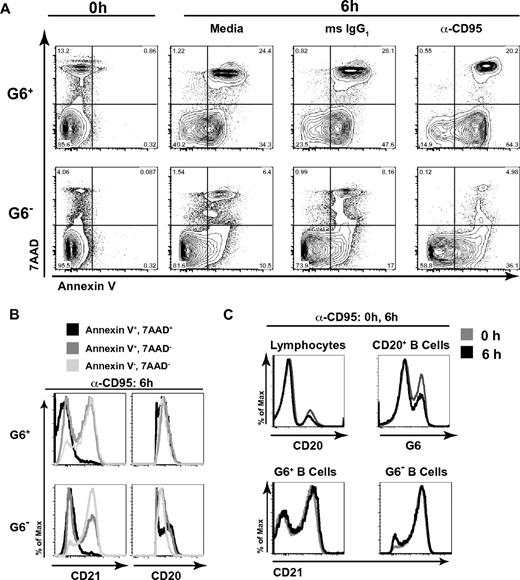
G6+ B cells are preferentially prone to death and apoptosis
Our microarray data suggested that HCV+MC+ patients' IgM+κ+CD27+ B cells up-regulate several genes associated with apoptosis. We tested whether these cells were prone to apoptosis in vitro by incubating them with either media, anti-CD95 mAb, or isotype control IgG (Figure 6). After 6 hours of incubation, PBMCs were stained with annexin V (to measure apoptosis), 7-amino-actinomycin D (7-AAD; to measure cell death), anti-CD20, and-CD21, and G6 Abs. After incubation with media alone, isotype control, or anti-CD95, G6+, compared with G6−, B cells had significantly higher binding of annexin V, either alone or in combination with 7-AAD. The percentage of annexin V+/7-AAD− cells was highest in the anti-CD95-stimulated G6+ B-cell subset. However, whereas G6− cells demonstrated a 2.1-fold increase in annexin V binding on stimulation with anti-CD95, compared with isotype control, G6+ cells had a 1.4-fold increase (Figure 6A). This indicates that G6+ B cells' propensity for apoptosis is not fully dependent on CD95, although CD95 was upregulated on several HCV+MC+ patients' G6+ cells. A larger percentage of apoptotic annexin V+7-AAD− cells had lower CD21 expression compared with annexin V−7-AAD− cells (Figure 6B). This was true for both G6+ and G6− populations. Surface CD20 expression was similar among all subsets, except for G6− annexin V+7-AAD+ cells. Importantly, 6 hours of incubation with anti-CD95 did not significantly alter the percentages of CD20+ B cells that were G6+ (37 vs 29% at 0 and 6 hours, respectively), nor did it cause down-regulation of CD21 on G6+ or G6− B cells (Figure 6C), indicating that cell surface CD20, CD21, and G6 expression was stable and not significantly affected by apoptosis or cell death. Taken together, these data suggest that, in the absence of survival signals, G6+ B cells, and in particular, the CD21low G6+ subset, are prone to spontaneous cell death and apoptosis.
G6+ B cells are prone to apoptosis and cell death, and the apoptotic cells are disproportionately CD21low. PBMCs were incubated for 6 hours in the presence of media alone, mouse IgG1 (isotype control), or 1 μg/mL anti-CD95 with 2 μg/mL protein G. Cells were then stained with annexin V, 7-AAD, anti-CD20, anti-CD21, and G6 mAbs and were analyzed by flow cytometry. Analyses of G6+ and G6− CD20+ B cells are shown. (A) Annexin V and 7-AAD staining of cells at baseline and after 6 hours of stimulation. (B) Analysis of surface CD21 and CD20 expression on cells incubated for 6 hours with α-CD95/protein G; annexin V+7-AAD+, annexin V+7-AAD−, and annexin V−7-AAD− subsets. (C) Cell surface anti-CD20 (of total lymphocytes), G6 (of total B cells), and anti-CD21 (of G6+ and G6− B cells) staining at baseline, compared with 6 hours, of stimulation. PBMCs from LDU 125 were collected 10 months after the cells collected for the microarray and primary immunophenotyping experiments. Data are representative of 3 independent experiments.
G6+ B cells are prone to apoptosis and cell death, and the apoptotic cells are disproportionately CD21low. PBMCs were incubated for 6 hours in the presence of media alone, mouse IgG1 (isotype control), or 1 μg/mL anti-CD95 with 2 μg/mL protein G. Cells were then stained with annexin V, 7-AAD, anti-CD20, anti-CD21, and G6 mAbs and were analyzed by flow cytometry. Analyses of G6+ and G6− CD20+ B cells are shown. (A) Annexin V and 7-AAD staining of cells at baseline and after 6 hours of stimulation. (B) Analysis of surface CD21 and CD20 expression on cells incubated for 6 hours with α-CD95/protein G; annexin V+7-AAD+, annexin V+7-AAD−, and annexin V−7-AAD− subsets. (C) Cell surface anti-CD20 (of total lymphocytes), G6 (of total B cells), and anti-CD21 (of G6+ and G6− B cells) staining at baseline, compared with 6 hours, of stimulation. PBMCs from LDU 125 were collected 10 months after the cells collected for the microarray and primary immunophenotyping experiments. Data are representative of 3 independent experiments.
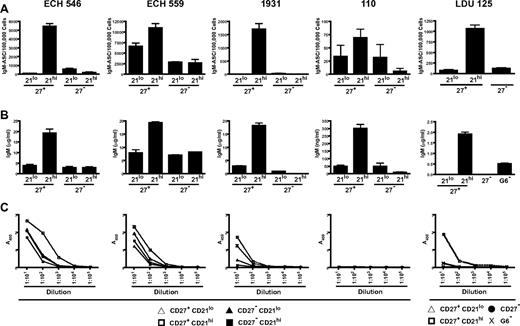
CD27+CD21high, but not CD27+CD21low, G6+ B cells from HCV+MC+ patients efficiently differentiate into antibody-secreting plasmablasts
To determine whether CD27+CD21low B cells could proliferate and differentiate in response to stimulatory signals, we stimulated sorted CD27+CD21high, CD27+CD21low, CD27−CD21high, and CD27−CD21low G6+ B cells from 4 HCV+MC+ subjects. For a fifth patient, CD27+CD21low, CD27+CD21high, CD27− G6+ B cells and total G6− B cells were sorted. We used 2 independent samples to test whether our cell sorting procedure caused early cell death; in all subsets, more than 85% of cells were viable, as measured by 4,6-diamidino-2-phenylindole staining (supplemental Figure 4). Cells were cultured for 6 days with CD40L, IL-2, and IL-10 and analyzed for differentiation toward antibody-secreting cells. Using IgM ELISPOT, we found that the CD27+CD21high, compared with CD27+CD21low and CD27− G6+ B-cell subsets, were more efficient at differentiating to IgM-secreting plasmablasts on CD40L/IL-2/IL-10 stimulation (Figure 7A). To confirm these findings, we quantitated IgM in the cell culture supernatants using ELISA. The CD27+CD21high cell culture supernatants all had higher levels of IgM compared with their CD27+CD21low and CD27− counterparts (Figure 7B). Class switch did not account for the decreased IgM in these subsets, as we consistently detected < 150 ng/mL IgG in cell culture supernatants.
CD27+CD21low, compared with CD27− and CD27+CD21high, G6+ B cells from HCV+MC+ patients demonstrate decreased differentiation to IgM RF-secreting plasmablasts on CD40L/IL-2/IL-10 stimulation. Analyses of 5 HCV+MC+ patients' B cells are shown. B cells from LDU 125 were collected 10 months after the cells collected for the microarray and primary immunophenotyping experiments. CD27+CD21low, CD27+CD21high, CD27−CD21low, and CD27+CD21high G6+ B cells were bulk-sorted and plated in a 96-well dish. For patient LDU 125, CD27+CD21low, CD27+CD21high, CD27− G6+ B cells and total G6− B cells were sorted. After 6 days of incubation in media supplemented with CD40L, IL-2, and IL-10, cells and supernatants were collected. (A) IgM enzyme-linked immunospot of stimulated B cells. (B) IgM ELISA of cell culture supernatants. (C) IgM RF ELISA of cell culture supernatants.
CD27+CD21low, compared with CD27− and CD27+CD21high, G6+ B cells from HCV+MC+ patients demonstrate decreased differentiation to IgM RF-secreting plasmablasts on CD40L/IL-2/IL-10 stimulation. Analyses of 5 HCV+MC+ patients' B cells are shown. B cells from LDU 125 were collected 10 months after the cells collected for the microarray and primary immunophenotyping experiments. CD27+CD21low, CD27+CD21high, CD27−CD21low, and CD27+CD21high G6+ B cells were bulk-sorted and plated in a 96-well dish. For patient LDU 125, CD27+CD21low, CD27+CD21high, CD27− G6+ B cells and total G6− B cells were sorted. After 6 days of incubation in media supplemented with CD40L, IL-2, and IL-10, cells and supernatants were collected. (A) IgM enzyme-linked immunospot of stimulated B cells. (B) IgM ELISA of cell culture supernatants. (C) IgM RF ELISA of cell culture supernatants.
We confirmed that the IgM produced by the stimulated G6+ B-cell subsets had RF activity by demonstrating that they could bind human IgG1 (Figure 7C). As a control, we tested patient LDU 125's G6− B-cell supernatant. Although IgM was present at a total concentration of approximately 0.5 μg/mL, we detected no significant RF activity. Taken together, these results suggest that differentiation to pathogenic RF-secreting plasmablasts on CD40L/IL-2/IL-10 stimulation is less efficient in the CD27+CD21low, compared with the CD27+CD21high, G6+ B-cell subset.
Discussion
We previously reported that HCV+MC+ patients have clonal expansions of IgM+κ+CD27+ peripheral B cells.6 Here we have transcriptionally profiled IgM+κ+CD27+ B cells isolated from HCV+MC+ patients, and we have identified a set of 69 genes that are differentially expressed. This set is notable for interferon response genes, B-cell activation markers, transcription factors, and glycoprotein synthesis genes. Although several genes previously implicated in B-cell neoplasia (eg, SOX5, CD11C, and MNDA) are up-regulated among these cells, others (LMO2, IL-4R, SELL, TCL1A, FOXP1, and BACH2) are down-regulated. Thus, although IgM+κ+CD27+ B cells are responsible for MC pathogenesis, their overall gene expression pattern suggests that there exists a subpopulation that is biased toward anergy and/or apoptosis.
The most highly up-regulated gene was the β-galactoside–binding lectin galectin-1, which is intriguing given its known pleiotropic roles in innate and adaptive immune responses. Galectin-1 (and CD11C) transcripts have been identified as being up-regulated in anergic murine B cells.12 Moreover, galectin-1 has been shown to be overexpressed in IgM+CD27+ B cells, is furtherinduced on BCR ligation, has been associated with Bim-mediated apoptosis,13 and may negatively regulate B-cell proliferation and BCR-mediated tyrosine phosphorylation.14 Also up-regulated were CD200R1, a potentially inhibitory immune receptor that is up-regulated in human tonsillar memory B cells and plasmablasts,11 and DAPK2, a calcium/calmodulin-dependent serine/threonine kinase responsible for IFN-γ, TNF-α, and Fas ligand–induced apoptosis.15 Similarly up-regulated was LAX1, a negative regulator of BCR-mediated calcium flux, Akt activation, and cell survival and proliferation.16 PYHIN1 (IFIX) and MNDA, 2 members of the IFN-inducible HIN-200 gene family implicated in apoptosis,17 were also up-regulated. One of the most down-regulated genes was TCL1A, a proto-oncogene up-regulated in splenic marginal zone lymphoma cells.18 TCL1A is induced in naive B cells on BCR ligation, and this may protect B cells from apoptosis.13 The down-regulation of IL-4R in HCV+MC+ patients' clonally expanded B cells is additionally supportive of a proapoptotic state, given that IL-4 is an effective antiapoptotic cytokine for B cells and that IL-4R engagement can abrogate Fas-mediated B cell apoptosis.36
We found that HCV+MC+ patients' IgM+κ+CD27+ B cells had up-regulation of CD11C, SOX5, and FGR, similar to what was recently reported in an FCRL4+ CD27− memory tonsillar B-cell subset. FCRL4 is an orphan receptor whose intracellular domain contains 3 immunoreceptor tyrosine-based inhibitory motifs that can inhibit BCR-mediated signaling.37 The primary location of FCRL4+ cells in the marginal zone equivalents of tonsillar and Peyer patch epithelia and in mucosa-associated lymphoid tissue lymphomas34 suggests that these cells may play a role in mucosal defense against pathogens. FCRL4+ B cells are reported to be large CD20high, CD11c+, CXCR3+, CD95+, CD21low proliferating B cells.29,33 Although we did not detect increased FCRL4 transcript in clonally expanded G6+ peripheral B cells, they shared several features with FCRL4+ cells, as they expressed FCRL4 protein and were predominantly CD20high, CD21low, CD11c+ B cells. However, in contrast to the concordance between FCRL4 mRNA transcript and FCRL4 protein expression that has been reported in healthy persons' tonsillar B cells,35 FCRL4 protein, but not mRNA, was elevated in HCV+MC+ patients' peripheral B cells. We speculate that an increased level of FCRL4 mRNA is not necessary to maintain the relatively increased surface FCRL4 protein expression on these cells. Further research is necessary to investigate whether HCV+MC+ patients' mucosal-associated and peripheral B cells share common transcriptional, immunophenotypic, and functional characteristics.
Decreased CD21 expression is observed on apoptotic38 B cells. Increased CD21low B cells are seen in systemic lupus erythematosis,39 chronic variable immunodeficiency,8,40 and rheumatoid arthritis,8 and increases of CD21low, CD27−, FCRL4+ B cells have been described in HIV-viremic33 and Plasmodium falciparum–exposed persons.41 Interestingly, the expanded B cells in chronic variable immunodeficiency patients are activated, have up-regulated SOX5 expression, and diminished calcium signaling and proliferative responses to BCR or CD40L stimulation.8,40 In HIV-viremic patients, they have up-regulated CD95 expression and are prone to apoptosis.42 Moreover, these cells may be functionally “exhausted,” as they demonstrate less ability to proliferate on BCR ligation with or without T-cell help and they have decreased ability to differentiate into antibody-secreting cells on stimulation with Staphylococcus aureus Cowan and CpG.33
Given these facts, and because CD21, as part of the B-cell coreceptor complex, augments BCR-mediated signaling, we suspected that HCV+MC+ patients' expanded CD21low G6+ B-cell subset represented a relatively anergic B-cell population. In addition, because transcriptional profiling revealed the up-regulation of several proapoptotic genes, we hypothesized that these B cells were prone to apoptosis. Indeed, we found that G6+, compared with G6−, B cells were more prone to apoptosis and cell death and that these cells were skewed toward the CD21low subset. Our data are consistent with earlier reports suggesting that HCV+ patients' memory B cells are prone to apoptosis and that this may serve as a feedback inhibition mechanism to prevent exaggerated autoreactive B-cell responses.43,44 In addition, CD21low B-cell subpopulations from healthy, HCV+MC−, or HCV+MC+ persons were relatively hyporesponsive to BCR stimulation, as measured by Ca2+ mobilization. Thus, the frequent down-regulation of CD21 among G6+ B cells suggests that these cells are relatively anergic. Moreover, the G6+CD21low subpopulation was only weakly induced to differentiate to IgM-secreting plasmablasts on stimulation with CD40L, IL-2, and IL-10, suggesting that these cells in vivo may be refractory to stimulation by T cell–mediated signals in the context of BCR engagement. Whether other stimuli (eg, TLR agonists or IFN-α) can induce these cells to differentiate remains an open question. We surmise that the down-regulation of CD21 is a homeostatic control mechanism that attenuates autoreactive RF+ B-cell responses to chronic HCV-containing immune complex stimulation.
To summarize, we have found that HCV+MC+ patients' clonally expanded peripheral B cells have global transcriptional features not of proto-oncogenesis, but rather of stimulatory hyporesponsiveness and anergy. Immunophenotypically, they resemble activated memory B cells and share several features with previously described CD21low and FCRL4+ B-cell populations. Although we have shown that the overall clonal population is capable of differentiating into RF-secreting plasmablasts, the expanded CD21low fraction is prone to anergy. Together, our results suggest overall attenuation mechanisms present in these autoreactive-prone B cells that limit their pathogenic expansion and differentiation in vivo. Several clinical lines of evidence suggest that these B-cell attenuation mechanisms may serve to limit disease activity in vivo. First, the incidence of clinically apparent MC during chronic HCV infection is rather low (1 per 1000 person-years in US veterans3 ), even in the setting of decades of infection. Second, clinical signs and symptoms of MC frequently wax and wane, with no apparent correlation with fluctuations in HCV RNA or liver inflammation.45 Third, even among patients with MC, progression to overt B-NHL is a relatively rare phenomenon (6.6 per 1000 person-years in an Italian cohort46 ).
Most importantly, these attenuation mechanisms may, in part, explain the relatively ineffective anti-HCV humoral response. It has been shown that VH169 partially encoded Abs are enriched in anti-influenza47,48 and anti-HIV49 responses. One of the most potent cross-neutralizing anti-HCV mAbs, CBH5, is partially encoded by VH169.50 VH169 encodes a distinct hydrophobic complementarity-determining region-H2 loop, and it has been speculated that this confers antiviral activity by binding to hydrophobic viral targets.47 As autoreactive RF is partially encoded by VH169, it is tempting to speculate that homeostatic mechanisms that are up-regulated to attenuate self-reactivity have the unintended consequence of abrogating effective VH169-mediated anti-HCV responses.
It remains unclear why only some HCV-infected persons develop MC and why these attenuation mechanisms fail to prevent the development of NHL in some HCV+MC+ patients. It must be emphasized that we have studied patients' peripheral B cells; it remains to be seen whether B cells at other anatomic sites, such as the liver or perihepatic lymph nodes, display similar features of attenuation. Further patient-oriented research will be necessary to answer these central questions, as well as to determine whether the autoregulatory mechanisms described here depend on anatomic context, are specific for HCV+MC+ patients or are common to other autoimmune diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patient volunteers for their generosity; Roy Jefferis for G6 monoclonal antibody; Götz Ehrhardt for F(ab′)2 anti-FCRL4 antibody; Donna Brassil, Veronica Whalen, and Rhonda Kost of the Rockefeller University Center for Clinical and Translational Science for assistance with subject enrollment and study management; Natasha Levenkova for statistical advice; and Santa Maria Di Vittorio for administrative assistance.
This study was supported in part by the National Institutes of Health/National Institute of Allergy and Infectious Diseases (R01AI60561, L.B.D.; and K08AI075031, E.D.C.), the Irma T. Hirschl/Monique Weill-Caulier Trust (L.B.D.), Center for Translational Science Award (Pilot Grant CCL3001018) (E.D.C.), and Center for Translational Science Award (grant UL1 RR024143, to Rockefeller University), from the National Center for Research Resources, a component of National Institutes of Health. Sorting on the FACSAria II was made possible by support from the Empire State Stem Cell Fund (New York State Department of Health contract C023046).
Opinions expressed here are solely those of the authors and do not necessarily reflect those of the Empire State Stem Cell Fund, the New York State Department of Health, or the state of New York.
National Institutes of Health
Authorship
Contribution: E.D.C., C.B., and L.B.D. devised and conducted the experiments; E.D.C., K.M., A.H.T., and I.M.J. provided patient referrals; E.D.C., L.B.D., C.M.R., S.M., K.D.R., K.M., A.H.T., and I.M.J. interpreted results; and E.D.C. and L.B.D. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lynn B. Dustin, Center for the Study of Hepatitis C, Laboratory of Virology and Infectious Disease, Rockefeller University, Box 64, 1230 York Ave, New York, NY 10065; e-mail: dustinl@rockefeller.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal