Abstract
Immunomodulatory derivatives of thalidomide (IMiD compounds), such as pomalidomide and lenalidomide, are highly active in multiple myeloma (MM) treatment. However, the precise mechanisms of action and resistance in MM are unresolved. Here we show that IMiD compounds down-regulate CCAAT/enhancer-binding protein-β (C/EBPβ) resulting in abrogation of cell proliferation. Overexpression of C/EBPβ rescued MM cells from IMiD-induced inhibition of proliferation, indicating that C/EBPβ is critical in mediating antiproliferative effects. IMiD-induced decrease of C/EBPβ protein led to impaired transcription of interferon regulatory factor 4 (IRF4). Down-regulation of IRF4 by lenalidomide was confirmed by longitudinal studies of bone marrow samples from 23 patients obtained before and during lenalidomide treatment using CD138+/IRF4+ double labeling. In contrast to down-regulation of C/EBPβ protein, IMiD compounds did not alter C/EBPβ mRNA levels or protein stability, suggesting translational regulation of C/EBPβ. We could demonstrate that C/EBPβ protein expression is under eIF4E-translational control in MM. Furthermore, inhibition of the eIF4E-C/EBPβ axis by IMiD compounds was not observed in IMiD-resistant MM cells. However, targeting translation at a different level by inhibiting eukaryotic translation initiation factor 4E-binding protein 1 phosphorylation overcame resistance, suggesting that this pathway is critical and might be a target to overcome drug resistance.
Introduction
Immunomodulatory derivatives of thalidomide (IMiD compounds), such as lenalidomide and pomalidomide, represent a novel class of agents with potent activity against multiple myeloma (MM).1,2 IMiD compounds were shown to induce antitumor, antiangiogenic, and anti-inflammatory effects. However, the precise mechanisms by which IMiD compounds induce the direct anti-MM effect have not been determined.3 Escoubet-Lozach et al demonstrated that IMiD compounds induced G0-G1 growth arrest in MM cells because of increased p21WAF-1 mRNA and protein expression through a histone demethylase LSD1-mediated epigenetic mechanism.4 Other tumor suppressor genes, such as the early growth response genes (Egr 1-3) and cyclin-dependent kinase inhibitors (p15, p27), are also induced by lenalidomide alone and synergistically when used in combination with dexamethasone in MM cells.5 The recent work by Xu et al showed that modulation of guanosine triphosphatase activity resulting in alteration of the cytoskeleton is another mechanism by which IMiD compounds regulate the interplay between tumor cells, their environment, and host immune cells.6 In this study, we focused on the direct effects of IMiD compounds on MM cells and on genes critical for the pathogenesis and drug resistance of MM.
CCAAT/enhancer-binding proteins (C/EBPs) belong to a large family of leucine zipper transcription factors (TFs) termed bZip proteins, which have a basic DNA-binding domain linked to a leucine zipper dimerization motif.7 C/EBPβ is widely expressed and exists in 3 distinct isoforms: termed LAP* (full-length protein), LAP (contains a 21-amino acid truncation at the N terminus), and LIP (contains a large N-terminal truncation and can act as a dominant-negative).8 The C/EBPβ isoforms are generated by alternative initiation of protein translation from a single mRNA transcript. C/EBPβ, also called NF-IL6, regulates a variety of genes involved in diverse functions, such as cellular differentiation processes, including adipogenesis,9 cell survival,10 tumor invasiveness,11 and hematopoiesis.12 Deletion of the C/EBPβ gene in mice results in impaired generation of B lymphocytes,13 and it has been shown that C/EBPβ contributes to the induction of the antiapoptotic protein, BCL2, in t(14;18) lymphoma cells.10 Earlier studies identified C/EBPβ as a regulator of IL-6 and demonstrated that C/EBPβ itself is also induced by IL-6,14 the most important survival factor for MM cells. Recently, we have shown that C/EBPβ regulates IRF4 gene expression in MM.15 Our studies showed that down-regulation of IRF4 by C/EBPβ further decreased expression of the TFs XBP1 and B-lymphocyte–induced maturation protein (BLIMP1). These TFs have been shown to be critical for the pathogenesis of MM.16 Further, IRF4 participates in the immune response resulting in lymphocyte activation and generation of immunoglobulin-secreting plasma cells, and has been reported to be required for myeloma cell survival.16-19
Previous studies reported that translational regulation of the ratio of C/EBPβ isoforms determines differentiation and proliferation in a number of cell types.12 For example, C/EBPβ isoform expression is differentially regulated at the translational level in prostate cancer cells with an increased usage of the translation start site, which generates the active full-length C/EBPβ in androgen-independent prostate cancer compared with androgen-dependent prostate cancer cells.20 The mammalian target of rapamycin (mTOR) signaling pathway controls the ratio of C/EBPβ isoform expression through mRNA cap-binding eIF4E,12 which is rate limiting for cap-dependent translation.21 It has been proposed that the translational efficiency of mRNA with highly complex 5′-untranslated regions is especially dependent on eIF4E levels.22 An increase in eIF4E level or activity does not lead to increased rates of global translation but instead results in increased translation of mRNAs with highly complex 5′-untranslated regions.23 Several genes, including Myc, fibroblast growth factor, and VEGF involved in tumorigenesis, are regulated at the translational level by eIF4E,24-26 suggesting that eIF4E might be an attractive target in antitumor treatment.
In this paper we show, for the first time, that IMiD compounds block C/EBPβ translation through the down-regulation of eIF4E, indicating the critical role of activated translation in MM cells. The decreased C/EBPβ protein levels further led to impaired transcription of downstream IRF4, a critical TF for MM cells. Overexpression of C/EBPβ induced resistance to IMiD compounds, further supporting the role of C/EBPβ in mediating drug resistance.
Methods
Chemicals and antibodies
Cell culture media, sera, and penicillin-streptomycin were purchased from Invitrogen. Lenalidomide and pomalidomide were obtained from Celgene. Antibodies were purchased from the following vendors: anti-C/EBPβ (C-19) and anti-XBP1 (C-20) from Santa Cruz Biotechnology; anti-IRF4 antibody, anti–4E-binding protein 1 (4EBP1) antibody, anti–phospho-4EBP1 antibody, and anti-eIF4E antibody from Cell Signaling; anti-BLIMP1 from R&D Systems; and anti–β-actin monoclonal antibody, dimethyl sulfoxide (DMSO), and cycloheximide from Sigma-Aldrich.
Cell culture and cell selection
MM cell lines RPMI 8226 and H929 were purchased from ATCC. OPM2 was provided by Dr Klaus Podar (Dana-Farber Cancer Institute, Boston, MA) and MM.1S from Dr Steven Rosen (Northwestern University, Chicago, IL). The MM cell lines MM.1S, RPMI 8226, H929, and OPM2 were cultured at 37°C in a humidified atmosphere in the presence of 5% CO2. Cells were cultured in RPMI 1640 medium with l-glutamine, 1 × penicillin/streptomycin, and 10% fetal bovine serum. Selection of primary MM cells was performed using Ficoll (Invitrogen) for isolation of mononuclear cells according to the manufacturer's instructions. Primary myeloma cells were further selected using CD138+ antibody-specific microbeads followed by magnetic separation according to the manufacturer's protocol (Miltenyi Biotec) as described before.27,28 The negative population was considered as CD138−.
All patient samples were obtained after informed consent was given in accordance with the Declaration of Helsinki, and all studies were approved by the University of Pittsburgh School of Medicine Institutional Review Board.
Cell proliferation assays
MM.1S, OPM2, and RPMI 8226 were incubated in 96-well plates and treated with DMSO or with various concentrations of lenalidomide or pomalidomide in RPMI 1640 medium containing 10% fetal bovine serum at 37°C and 5% CO2 for 2 days. DNA synthesis was measured by 3H-thymidine incorporation (NEN Products, PerkinElmer Life and Analytical Sciences). Cells were pulsed with 3H-thymidine (1 μCi/well or 0.037 MBq) for the last 8 hours of culture, harvested onto glass-fiber filter mats (Wallac) using an automatic cell harvester (Tomtec Harvester 96, Mach III), and counted using a Wallac TriLux Beta plate scintillation counter (PerkinElmer Life and Analytical Sciences).28,29
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis
Protein was extracted from cells using 1× RIPA containing a Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). Cell lysate proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinyl difluoride membranes (Bio-Rad). The blots were incubated with the appropriate antibodies to detect the protein level of interest, and the immune complexes were visualized using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) as described before.30
IRF4 and CD138+ double antigen labeling IHC
Double antigen labeling immunohistochemistry (IHC) studies to assess IRF4 expression in CD138+ plasma cells were performed on bone marrow biopsy sections from 23 patients before and during treatment with lenalidomide. Patients with newly diagnosed MM were treated with 25 mg/day lenalidomide on days 1 to 21 in combination with dexamethasone 40 mg on days 1, 8, 15, and 22. In brief, IHC was performed using B-Plus fixed, paraffin-embedded, decalcified bone marrow tissue sections that were deparaffinized, rehydrated, and treated with hydrogen peroxidase to inactivate endogenous peroxidase activity. Sections were incubated with a primary antibody against IRF4 (Dako North America), and positive reactions were visualized using the ImPRESS polymerized reporter enzyme staining system (Vector Laboratories). Incubation with nickel enhanced diaminobenzidine (Vector Laboratories) produced a nuclear black reaction product when positive for IRF4. Sections were incubated overnight with the second primary antibody against CD138 (Dako North America). Positive reactions were developed using the ImPRESS reagent and Vector NovaRED substrate (Vector Laboratories) to yield a red reaction product when positive for CD138 that highlighted the membrane of plasma cells. The marrow biopsy sections were counterstained with Shandon hematoxylin (Thermo Scientific). A score for IRF4 reactivity was determined for the CD138+ plasma cells on deidentified slides. A total of 100 individual CD138+ cells were rated for IRF4 reactivity as follows: 3 indicates strong; 2, moderate; and 1, negative/weak. The score for IRF4 reactivity was the sum of 100 cells. A pathologist determined the score in triplicate, and the final score was the mean ± SD of the 3 determinations.
Real-time quantitative PCR analysis
For the determination of mRNA levels of C/EBPβ, IRF4, and eIF4E, total RNAs were isolated from cells using Trizol reagent (Invitrogen) following the manufacturer's instructions. Total RNA was converted into cDNA using Superscript III RT (Invitrogen). Quantitative reverse-transcribed PCR was performed using a MyiQ2 Two-Color Real-Time PCR Detection System (Bio-Rad). Reverse-transcribed PCR was carried out with the SYBR Green PCR master mix (Bio-Rad) using 1 μL cDNA in a 25-μL final reaction mixture (10 minutes at 95°C, 40 cycles of 15 seconds at 95°C, and 1 minute at 60°C). The average threshold cycle (Ct) for each gene was determined from triplicate reactions, and the target gene expression was normalized to β-actin using the difference between their Ct values to generate the ΔCt value. This was then compared with the control sample in each experiment to give the ΔΔCt value, where the control had a ΔΔCt value of 0. The fold change in target gene expression with treatment, compared with the control sample, is given by the formula 2−ΔΔCt. The following PCR primer sets were used: C/EBPβ, 5′-AACTCTCTGCTTCTCCCTCTG-3′; and 5′-AAGCCCGTAGGAACATCTTT-3′; IRF4, 5′-TTAATTCTCCAAGCGGATGC-3′; and 5′-AAGGAATGAGGAAGCCGTTC-3′; β-actin, 5′-GGACTTCGAGCAAGAGATGG-3′; and 5′-AGCACTGTGTTGGCGTACAG-3′ (Real Time Primers); and eIF4E, 5′-ACAAGTCAGTCTGAAACCATCGAAC-3′; and 5′-CTTCATCCTCTTCGGCCACTCCTCC-3′.31
Transfection of empty vector (pcDNA3), WT-C/EBPβ
MM.1S cells (2 × 106) were transfected by electroporation with 10 μg of the empty vector pcDNA3.1 or wild-type (WT)–C/EBPβ plasmids.15 Expression vectors for the full-length WT-C/EBPβ (pcNF-IL6) was generated by inserting the respective coding regions into pcDNA3.1 and provided by Dr Philip. E. Auron (Duquesne University, Pittsburgh, PA).8 Electroporation was accomplished using the Cell Line Nucleofector Kit V according to the manufacturer's instructions (Lonza Walkersville). Transfected cells were selected for resistance to G418 (1 mg/mL) treatment. Selected cells were treated with DMSO or IMiD compounds and analyzed by Western blotting or cell proliferation assay.
eIF4E shRNA knockdown assay
Lentiviral shRNA was used to knock down eIF4E expression in MM cells. HIV-based lentivirus particles were provided by Dr Robert W. Sobol (Hillman Cancer Center, Pittsburgh, PA). The sequences used were as follows: CCGGCCACTCTGTAATAGTTCAGTACTCGAGTACTGAACTATTACAGAGTGGTTTTTG and CCGGCGGCTGATCTCCAAGTTTGATCTCGAGATCAAACTTG GAGATCAGCCGTTTTTG. MM.1S cells were incubated with lentivirus particle and polybrene 8 μg/mL for 16 hours and then washed with media. A second infection was repeated 24 hours later. Cells were selected by culturing for 10 days in puromycin (500 ng/mL). Transfected cells were then lysed, and whole cell lysates were analyzed by Western blotting. β-Actin expression was probed for loading control.
Apoptosis assays
Apoptosis was analyzed by annexin V-fluorescein isothiocyanate staining using the AlexaFluor-488 annexin V kit (Invitrogen). Briefly, 0.5 × 106 cells/mL were harvested, washed once with cold phosphate-buffered saline, and then resuspended in 1× annexin-binding buffer. Cell survival was determined by annexin V-fluorescein isothiocyanate/propidium iodide double staining. Samples were analyzed on FACSCalibur (BD Bioscience) using the software program CellQuest 3 (BD Biosciences).
Cell cycle assays
MM.1S (1 × 106 cells/mL) were cultured for 3 days at 37°C in RPMI 1640 medium with DMSO or various concentrations of lenalidomide or pomalidomide. The cells were harvested, washed with ice-cold phosphate-buffered saline, fixed with 70% ethanol for 1 hour at 4°C, and pretreated with RNase (Worthington) for 30 minutes at 37°C. Cells were stained with propidium iodide (20 μg/mL; Sigma Aldrich). Analyses were performed on a BD FACSCalibur flow cytometer and analyzed using ModFit LT2.0 and Cellquest 3 software (BD Biosciences).
Image acquisition and manipulation
Slides were evaluated using an Olympus BX45 microscope equiped with a 100×/1.35 NA oil objective (Olympus). Images were captured as .tif files using SPOT Insight Digital Camera and SPOT Advanced software (Diagnostic Instruments). The images were opened and then sized and placed into figures using Adobe Photoshop Version 7.0 Professional (Adobe Systems).
Statistical analyses
Each experiment was repeated at least 3 times, and all quantitative data are presented as mean plus or minus SD. Statistical differences were determined by Student t test. The results were considered statistically significant with P values < .05. The LAP/LIP ratio was measured as follows: The images were opened and then sized and placed into figures using ImageJ 1.44 (available at www.rsb.info.nih.gov/ij) or Adobe Photoshop Version 7.0 Professional (Adobe Systems).
Results
IMiD compounds down-regulate C/EBPβ protein
In this study, we focused on the direct effects of IMiD compounds on MM cells. Proliferation studies using analysis of DNA synthesis by measuring thymidine uptake revealed that both pomalidomide and lenalidomide significantly inhibited proliferation of MM.1S and OPM2 cells at concentrations as low as 0.01μM (P < .05; Figure 1A). The proliferation change was a result of G0/G1 growth arrest (supplemental Figure 1) with only a slight increase in apoptosis (supplemental Figure 2). Because our previous work demonstrated that C/EBPβ is a critical TF in MM that controls growth and proliferation of myeloma cells,15 we analyzed the effects of IMiD compounds on C/EBPβ expression in MM cell lines and primary MM cells (n = 3). In Western blot studies, both pomalidomide and lenalidomide significantly decreased the protein level of C/EBPβ isoforms (LAP, LAP*, and LIP) in MM.1S, H929, and OPM2, as well as in primary CD138+ MM cells (Figure 1B), and in a time- and dose-dependent manner (Figure 1C-D). The LAP* isoform is visible as a double band beneath the LAP isoform. The ratio of LAP, LAP*, and LIP did not differ significantly between lenalidomide or pomalidomide treatment.
IMiD compounds down-regulate C/EBPβ in MM cells in a time- and dose-dependent manner. (A) MM.1S and OPM2 cells (6 × 104/well) were cultured with lenalidomide or pomalidomide at different concentrations for 2 days. DMSO (0.01%) was used as the control treatment. DNA synthesis was measured by 3H-thymidine incorporation, and the amount of DNA synthesis in treated cells relative to the amount in control cells (as a percentage) was plotted. Results are shown as triplicates of mean ± SD. (B) MM.1S, H929, OPM2, or primary myeloma cells were cultured with DMSO, pomalidomide, or lenalidomide at the indicated concentrations for 3 days. Representative results from 3 independent experiments are shown. Cells were then lysed, and cell lysates were analyzed for C/EBPβ expression by Western blotting. β-actin expression was probed for loading control. The ratio of LAP/LIP is measured using ImageJ 1.44 software. (C-D) MM.1S cells were incubated with lenalidomide (C) or pomalidomide (D) at different concentrations with fixed time period of 4 days, or for different time periods with fixed concentration of 10μM, or with DMSO 0.01% as control. Cells were lysed and cell lysates were analyzed for C/EBPβ expression by Western blotting. β-Actin expression was probed for loading control.
IMiD compounds down-regulate C/EBPβ in MM cells in a time- and dose-dependent manner. (A) MM.1S and OPM2 cells (6 × 104/well) were cultured with lenalidomide or pomalidomide at different concentrations for 2 days. DMSO (0.01%) was used as the control treatment. DNA synthesis was measured by 3H-thymidine incorporation, and the amount of DNA synthesis in treated cells relative to the amount in control cells (as a percentage) was plotted. Results are shown as triplicates of mean ± SD. (B) MM.1S, H929, OPM2, or primary myeloma cells were cultured with DMSO, pomalidomide, or lenalidomide at the indicated concentrations for 3 days. Representative results from 3 independent experiments are shown. Cells were then lysed, and cell lysates were analyzed for C/EBPβ expression by Western blotting. β-actin expression was probed for loading control. The ratio of LAP/LIP is measured using ImageJ 1.44 software. (C-D) MM.1S cells were incubated with lenalidomide (C) or pomalidomide (D) at different concentrations with fixed time period of 4 days, or for different time periods with fixed concentration of 10μM, or with DMSO 0.01% as control. Cells were lysed and cell lysates were analyzed for C/EBPβ expression by Western blotting. β-Actin expression was probed for loading control.
Down-regulation of IRF4 expression in MM cells in vitro and in lenalidomide- treated patients
We have previously shown that IRF4, which is critical for myeloma cell survival,17-19 is an important downstream transcription target of C/EBPβ. Chromatin immunoprecipitation analysis indicated that C/EBPβ directly binds to the endogenous IRF4 promoter in MM cells.15 Given the fact that C/EBPβ is down-regulated by lenalidomide and pomalidomide, we analyzed whether its downstream target IRF4 was also affected by IMiD compound treatment. Our data showed that both pomalidomide and lenalidomide down-regulate the IRF4 protein level in MM.1S, OPM2, H929, and primary CD138+ cells (Figure 2A) in a time- and dose-dependent manner (Figure 2B-C).
IMiD compounds down-regulate IRF4 in MM cells in vitro and in lenalidomide-treated patients. (A) MM.1S, H929, OPM2, or primary myeloma cells were cultured with DMSO, pomalidomide, or lenalidomide at the indicated concentrations for 3 days. (B-C) MM.1S cells were incubated with lenalidomide (B) or pomalidomide (C) at different concentrations with a fixed time period of 4 days, or for different time periods with fixed concentration of 10μM, or with DMSO 0.01% as control. Cells were lysed, and cell lysates were analyzed for IRF-4 expression by Western blotting. β-actin expression was probed for loading control. (D) Double staining immunohistochemistry on paraffin-embedded marrow biopsy sections for IRF4 (black nuclear staining) and CD138+ (red staining) was performed before and during treatment with lenalidomide. Cells were rated for IRF4 reactivity as follows: 3 indicates strong; 2, moderate; and 1, negative/weak. The score for a sample is the sum of 100 cells (original magnification ×1000). (E) IRF4 scores of bone marrow biopsy samples, before and during treatment with lenalidomide. Lines connect scores from the same patients. (F) MM.1S cells were incubated with lenalidomide or pomalidomide for 2 or 3 days at 100μM. BLIMP1 and XBP1 expression was analyzed by Western blotting. β-actin expression was probed for loading control.
IMiD compounds down-regulate IRF4 in MM cells in vitro and in lenalidomide-treated patients. (A) MM.1S, H929, OPM2, or primary myeloma cells were cultured with DMSO, pomalidomide, or lenalidomide at the indicated concentrations for 3 days. (B-C) MM.1S cells were incubated with lenalidomide (B) or pomalidomide (C) at different concentrations with a fixed time period of 4 days, or for different time periods with fixed concentration of 10μM, or with DMSO 0.01% as control. Cells were lysed, and cell lysates were analyzed for IRF-4 expression by Western blotting. β-actin expression was probed for loading control. (D) Double staining immunohistochemistry on paraffin-embedded marrow biopsy sections for IRF4 (black nuclear staining) and CD138+ (red staining) was performed before and during treatment with lenalidomide. Cells were rated for IRF4 reactivity as follows: 3 indicates strong; 2, moderate; and 1, negative/weak. The score for a sample is the sum of 100 cells (original magnification ×1000). (E) IRF4 scores of bone marrow biopsy samples, before and during treatment with lenalidomide. Lines connect scores from the same patients. (F) MM.1S cells were incubated with lenalidomide or pomalidomide for 2 or 3 days at 100μM. BLIMP1 and XBP1 expression was analyzed by Western blotting. β-actin expression was probed for loading control.
To confirm our in vitro findings, we analyzed IRF4 expression of myeloma cells in longitudinal bone marrow biopsy samples of 23 patients before and during lenalidomide therapy (“IRF4 and CD138+ double antigen labeling IHC”). IHC for IRF4 staining was performed on bone marrow samples of patients treated within the randomized trial “A randomized clinical trial of lenalidomide (CC-5013) and dexamethasone with and without autologous peripheral blood stem cell transplant in patients with newly diagnosed multiple myeloma.” Bone marrow biopsy sections of patients were stained by double labeling (CD138+/IRF4+) IHC to analyze intensity of IRF4 expression (black nuclear staining) in CD138+ MM cells (red cytoplasmic staining, Figure 2D left panel). The IHC showed that the nuclear IRF4 staining intensity of CD138+ MM cells significantly decreased in patients during treatment with lenalidomide (Figure 2D right panel). This was semiquantified using a scoring system for IRF4 staining intensity (“IRF4 and CD138+ double antigen labeling IHC”). The mean ± SD staining score significantly decreased from 176 ± 31 to 152 ± 27 (P < .001; Figure 2E). The overall response rate within this trial was 94.4% and was assessed after completion of treatment, which either included 8 cycles of lenalidomide/dexamethasone or 4 cycles of lenalidomide/dexamethasone followed by transplantation. All bone marrow samples were taken during cycle 4 of lenalidomide/dexamethasone treatment. This was at a relatively early time point of the treatment because most of the patients showed their best response later than cycle 4 or after transplantation. Because bone marrow samples still presented plasma cells at cycle 4 and the response rate of the patients was very high with 94.4% complete remission/very good partial remission/partial remission, we assume that down-regulation of IRF4 in MM cells precedes MM cell death.
Previously, we have shown that C/EBPβ directly binds to the promoter of BLIMP1 and indirectly regulates XBP1 via regulation of the IRF4 gene.15 Analysis of the effects of pomalidomide and lenalidomide on BLIMP1 and XBP1 protein expression showed that both IMiDs significantly down-regulate the protein level of BLIMP1 and XBP1 after 48 and 72 hours of treatment (Figure 2F). These data indicate that IMiDs down-regulate C/EBPβ protein levels and downstream TFs, including IRF4, BLIMP1, and XBP1.
Overexpression of C/EBPβ induces resistance to IMiD compounds
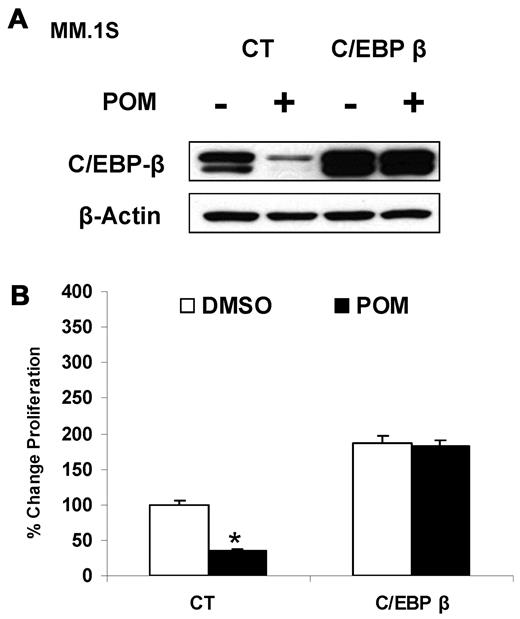
To further support our hypothesis that IMiD compounds affect proliferation of MM cells via targeting C/EBPβ, we overexpressed C/EBPβ in MM.1S cells. Pomalidomide down-regulated endogenous C/EBPβ protein in the control cells, whereas forced C/EBPβ expression was resistant to down-regulation (Figure 3A), suggesting that the down-regulation of C/EBPβ by IMiD compounds is mediated by altered regulation of C/EBPβ mRNA or protein through an IMiD compound response region that may not be present in the transfected C/EBPβ plasmid. Overexpression of C/EBPβ rescued MM cells from pomalidomide-induced inhibition of proliferation (Figure 3B), indicating that C/EBPβ is critical for control of proliferation. Alternatively, excess exogenous C/EBPβ mRNA and protein may out-compete the negative regulatory signal from IMiD compounds.
Overexpression of C/EBPβ induces resistance to IMiD compounds. (A) MM.1S cells (2 × 106) were transfected by electroporation with either empty vector pcDNA3.1 or WT-C/EBPβ plasmids. After selection, cells were treated with pomalidomide (10μM) for 2 days. Cells were then lysed, and whole cell lysates were analyzed for C/EBPβ expression by Western blotting. β-actin expression was probed for loading control. (B) MM.1S cells overexpressing WT-C/EBPβ were treated with pomalidomide (10μM) for 2 days, and DNA synthesis was measured by 3H-thymidine incorporation. The amount of DNA synthesis in treated cells relative to the amount in control cells (as a percentage) was graphed. Results are shown as triplicates of mean ± SD.
Overexpression of C/EBPβ induces resistance to IMiD compounds. (A) MM.1S cells (2 × 106) were transfected by electroporation with either empty vector pcDNA3.1 or WT-C/EBPβ plasmids. After selection, cells were treated with pomalidomide (10μM) for 2 days. Cells were then lysed, and whole cell lysates were analyzed for C/EBPβ expression by Western blotting. β-actin expression was probed for loading control. (B) MM.1S cells overexpressing WT-C/EBPβ were treated with pomalidomide (10μM) for 2 days, and DNA synthesis was measured by 3H-thymidine incorporation. The amount of DNA synthesis in treated cells relative to the amount in control cells (as a percentage) was graphed. Results are shown as triplicates of mean ± SD.
IMiD compounds down-regulate C/EBPβ by targeting the eIF4E
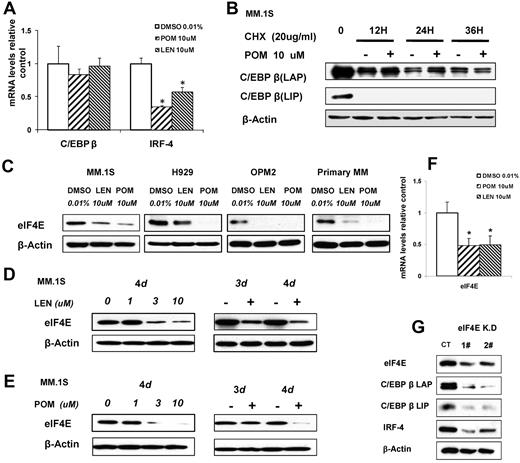
To further determine the mechanism of down-regulation of C/EBPβ, we examined the effects of pomalidomide and lenalidomide on the transcription and translation of C/EBPβ. Real-time PCR analysis showed that IMiD compounds did not down-regulate C/EBPβ mRNA, whereas the mRNA from the C/EBPβ-regulated gene IRF4 was significantly decreased (P ≤ .01; Figure 4A). This result suggested that IMiD compounds probably down-regulate C/EBPβ protein via either decreased protein translation or increased protein degradation, whereas IMiD compounds affect IRF4 protein levels by altering the amount of C/EBPβ-dependent IRF4 gene transcription. Therefore, we investigated whether IMiD compounds affect the half-life of C/EBPβ protein by treating MM cells with pomalidomide and cycloheximide, an inhibitor of protein biosynthesis, for 12, 24, and 36 hours. Even at a high dosage of 10μM, pomalidomide did not accelerate the decrease of C/EBPβ protein in the presence of cycloheximide (Figure 4B) compared with cycloheximide alone, demonstrating that pomalidomide does not enhance C/EBPβ protein degradation.
IMiD compounds down-regulate C/EBPβ by targeting its protein translation. (A) MM.1S cells were cultured with pomalidomide, lenalidomide, or 0.01% DMSO (control). Total RNA was extracted from 12-hour cultures, reverse transcribed to cDNA, and used for quantitative real-time PCR. Data were analyzed according to the ΔΔCt method. Results are shown as mRNA expression relative to control (DMSO). mRNA levels were normalized with β-actin mRNA expression as control. (B) MM.1S cells were incubated with pomalidomide (10μM) with and without cycloheximide (20 μg/mL) for 12, 24, or 36 hours. Cells were lysed, and whole cell lysates were analyzed by Western blotting for C/EBPβ. (C) MM.1S, H929, OPM2, or primary myeloma cells were cultured with lenalidomide, pomalidomide, or 0.01% DMSO as control for 3 days. Cells were lysed, and whole cell lysates were analyzed by Western blotting for eIF4E. β-Actin expression was probed for loading control. (D-E) MM.1S cells (2 × 106) were incubated with lenalidomide (D) or pomalidomide (E) at different concentrations with a fixed time period of 4 days, or for different time periods with fixed concentration of 10μM, or with DMSO 0.01% as control. Cells were lysed, and whole cell lysates were analyzed for eIF4E expression by Western blotting. β-Actin expression was probed for loading control. (F) MM.1S cells were incubated with lenalidomide or pomalidomide for 12 hours, and total RNA was extracted by Trizol and followed by real-time PCR. Data were analyzed according to the ΔΔCt method. Results are shown as mRNA fold compared with control (DMSO). (G) eIF4E knockdown cell lines were generated by lentiviral infection of MM.1S cells. 1# and 2# indicate different eIF4E shRNA sequences. Green fluorescence protein was used as control for eIF4E shRN-expressing cells. Cell lysates were analyzed by Western blotting to compare the levels of eIF4E, C/EBPβ, and IRF4. β-actin expression was probed for loading control.
IMiD compounds down-regulate C/EBPβ by targeting its protein translation. (A) MM.1S cells were cultured with pomalidomide, lenalidomide, or 0.01% DMSO (control). Total RNA was extracted from 12-hour cultures, reverse transcribed to cDNA, and used for quantitative real-time PCR. Data were analyzed according to the ΔΔCt method. Results are shown as mRNA expression relative to control (DMSO). mRNA levels were normalized with β-actin mRNA expression as control. (B) MM.1S cells were incubated with pomalidomide (10μM) with and without cycloheximide (20 μg/mL) for 12, 24, or 36 hours. Cells were lysed, and whole cell lysates were analyzed by Western blotting for C/EBPβ. (C) MM.1S, H929, OPM2, or primary myeloma cells were cultured with lenalidomide, pomalidomide, or 0.01% DMSO as control for 3 days. Cells were lysed, and whole cell lysates were analyzed by Western blotting for eIF4E. β-Actin expression was probed for loading control. (D-E) MM.1S cells (2 × 106) were incubated with lenalidomide (D) or pomalidomide (E) at different concentrations with a fixed time period of 4 days, or for different time periods with fixed concentration of 10μM, or with DMSO 0.01% as control. Cells were lysed, and whole cell lysates were analyzed for eIF4E expression by Western blotting. β-Actin expression was probed for loading control. (F) MM.1S cells were incubated with lenalidomide or pomalidomide for 12 hours, and total RNA was extracted by Trizol and followed by real-time PCR. Data were analyzed according to the ΔΔCt method. Results are shown as mRNA fold compared with control (DMSO). (G) eIF4E knockdown cell lines were generated by lentiviral infection of MM.1S cells. 1# and 2# indicate different eIF4E shRNA sequences. Green fluorescence protein was used as control for eIF4E shRN-expressing cells. Cell lysates were analyzed by Western blotting to compare the levels of eIF4E, C/EBPβ, and IRF4. β-actin expression was probed for loading control.
Next, we analyzed whether IMiDs exert their inhibitory effects by impairing C/EBPβ protein translation. Although eIF4E regulates the relative amount of C/EBPβ-LIP translated versus C/EBPβ-LAP isoforms, it has also been reported that changes in eIF4E levels selectively regulate the translation of tumorigenesis-related genes.32 Therefore, we analyzed the role of eIF4E in the translational regulation of C/EBPβ in MM cells treated with IMiD compounds. Both pomalidomide and lenalidomide down-regulated eIF4E protein in MM.1S, H929, OPM2, and primary MM CD138+ cells (Figure 4C), in a time- and a dose-dependent manner (Figure 4D-E). Furthermore, real-time PCR showed that eIF4E mRNA was down-regulated as early as 12 hours after the start of IMiD compound treatment (Figure 4F). To further address the role of eIF4E in C/EBPβ translational regulation in MM cells, we knocked down eIF4E in MM.1S cells by lentiviral shRNA transduction. Decreased eIF4E led to down-regulation of both C/EBPβ isoforms (LAP, LAP*, and LIP) and, as hypothesized, decreased downstream IRF4 expression (Figure 4G). These data indicate that eIF4E is critical for the translational regulation of C/EBPβ in MM cells and that down-regulation of eIF4E by IMiD compounds blocks the biosynthesis of C/EBPβ protein through impairment of its translation.
The eIF4E-C/EBPβ pathway is not affected in IMiD-resistant MM cell lines
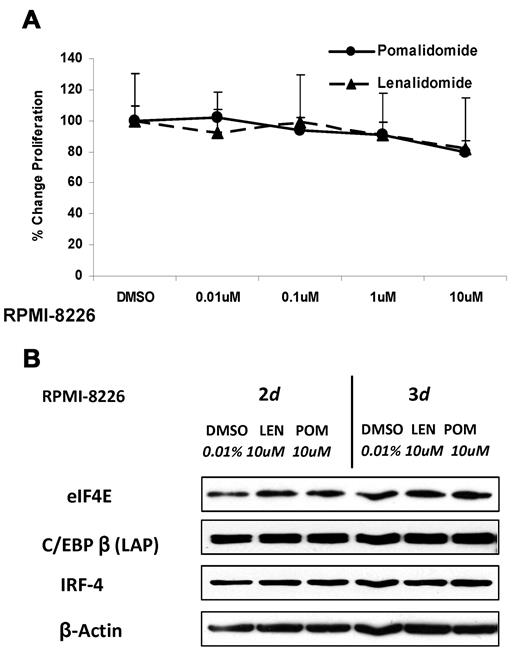
RPMI 8226 is a myeloma cell line, which is inherently resistant to the antiproliferative effects of IMiD compounds. As shown in Figure 5A, neither pomalidomide nor lenalidomide significantly impaired RPMI 8226 cell proliferation. Interestingly, in this cell line, we found that neither compound was effective in down-regulating eIF4E, C/EBPβ, or its downstream target IRF4 (Figure 5B). This indicates that the eIF4E-C/EBPβ pathway is critical for mediating the antiproliferative effects of IMiD compounds in MM cells and that other agents that affect this axis might enhance the sensitivity of MM cells to IMiD compounds.
Protein translation is not affected in IMiD-resistant MM cells. (A) RPMI 8226 cells (3 × 104) were cultured with different concentrations of lenalidomide, pomalidomide, or DMSO as control for 2 days. DNA synthesis was measured by 3H-thymidine incorporation. Results are shown as triplicates of mean ± SD. (B) RPMI 8226 cells were cultured with lenalidomide, pomalidomide, or DMSO for 2 or 3 days. Cells were lysed, and whole cell lysates were analyzed for eIF4E, C/EBPβ, and IRF4 by Western blotting. β-actin expression was probed for a loading control.
Protein translation is not affected in IMiD-resistant MM cells. (A) RPMI 8226 cells (3 × 104) were cultured with different concentrations of lenalidomide, pomalidomide, or DMSO as control for 2 days. DNA synthesis was measured by 3H-thymidine incorporation. Results are shown as triplicates of mean ± SD. (B) RPMI 8226 cells were cultured with lenalidomide, pomalidomide, or DMSO for 2 or 3 days. Cells were lysed, and whole cell lysates were analyzed for eIF4E, C/EBPβ, and IRF4 by Western blotting. β-actin expression was probed for a loading control.
Targeting mTOR as a strategy to overcome drug resistance of MM cells to IMiD compounds
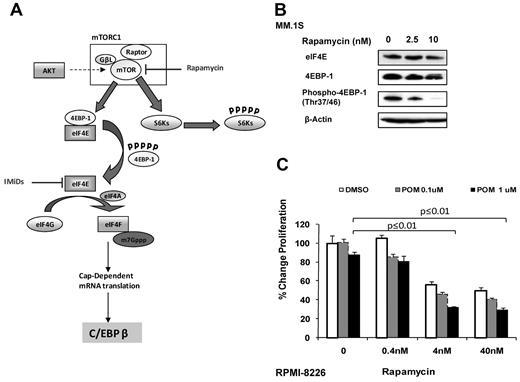
It has been reported that the activity of eIF4E is regulated by its associated protein 4EBP1.23 eIF4E typically is sequestered by hypophosphorylated 4EBP1, which blocks the assembly of the translational complex and results in restricted translation rates. The phosphorylation of 4EBP1 by mTOR, a central regulator of protein synthesis, leads to decrease in eIF4E binding capacity, thereby releasing eIF4E for translation initiation (Figure 6A). In contrast to IMiDs, the mTOR inhibitor rapamycin inhibited the phosphorylation of 4EBP1 but showed no effects on the eIF4E protein level (Figure 6B). This indicates that mTOR inhibition might further decrease the translational activity of eIF4E by suppressing 4EBP1 phosphorylation, in an additive manner with IMiD compounds. Therefore, we investigated the antiproliferative effect of rapamycin in combination with pomalidomide on IMiD-resistant RPMI 8226 cell line. The concomitant treatment of RPMI 8226 with rapamycin and pomalidomide overcame resistance, shown by a decrease of proliferation of 56% when cells were treated with pomalidomide (1μM) and rapamycin (4nM) versus pomalidomide alone (P ≤.01; Figure 6C). Our data show that rapamycin inhibits the mTOR complex, hence impairs 4EBP1 phosphorylation, and further blocks the assembly of the translational complex. In contrast, IMiD compounds inhibit the complex by down-regulation of eIF4E protein, suggesting that targeting different key factors of the translational complex might overcome resistance to IMiD compounds.
Combination of mTOR inhibitors and IMiD compounds might overcome resistance to IMiD compounds. (A) IMiD compounds inhibit the translation of C/EBPβ by down-regulation of eIF4E protein. Rapamycin targets the mTOR complex by inhibiting 4EBP1 phosphorylation, resulting in sequestering of eIF4E. This blocks the assembly of the translational complex and leads to decreased C/EBPβ translation. (B) MM.1S cells were incubated with rapamycin or DMSO 0.01% as control for 2 days. Cells were lysed, and lysates were analyzed for both total-and phospho-4EBP1 as well as for eIF4E by Western blot. β-actin expression was probed for loading control. (C) RPMI 8226 cells (3 × 104) were cultured with rapamycin or pomalidomide alone or in combination for 2 days. DMSO 0.01% treatment was used as a control. DNA synthesis was measured by 3H-thymidine incorporation. Results are shown as triplicates of mean ± SD.
Combination of mTOR inhibitors and IMiD compounds might overcome resistance to IMiD compounds. (A) IMiD compounds inhibit the translation of C/EBPβ by down-regulation of eIF4E protein. Rapamycin targets the mTOR complex by inhibiting 4EBP1 phosphorylation, resulting in sequestering of eIF4E. This blocks the assembly of the translational complex and leads to decreased C/EBPβ translation. (B) MM.1S cells were incubated with rapamycin or DMSO 0.01% as control for 2 days. Cells were lysed, and lysates were analyzed for both total-and phospho-4EBP1 as well as for eIF4E by Western blot. β-actin expression was probed for loading control. (C) RPMI 8226 cells (3 × 104) were cultured with rapamycin or pomalidomide alone or in combination for 2 days. DMSO 0.01% treatment was used as a control. DNA synthesis was measured by 3H-thymidine incorporation. Results are shown as triplicates of mean ± SD.
Discussion
IMiD compounds belong to a novel class of anticancer drugs with numerous effects on the human immune system and antitumor activity in various malignant disorders,33 including MM,34-37 chronic lymphocytic leukemia,38 and non-Hodgkin lymphoma.39 Nevertheless, the exact mechanism by which IMiD compounds, such as pomalidomide and lenalidomide, directly induce their antitumor activity is still elusive. In general, these compounds inhibit proliferation and tumor growth of cell lines or primary tumor cells both in vitro and in vivo.1,40
C/EBPβ plays an important role in the regulation of growth and differentiation of B cells.13 Our previous work demonstrated that C/EBPβ is an important TF that controls growth and proliferation of myeloma cells and regulates IRF4 and an extensive network of IRF4 target TFs, such as XBP1 and BLIMP1. Overexpression of C/EBPβ in MM cells up-regulates IRF4, XBP1, and BLIMP1, whereas silencing of C/EBPβ leads to down-regulation of these TFs, accompanied by significant inhibition of cell proliferation.15 In this study, we found that pomalidomide and lenalidomide down-regulate the expression of all C/EBPβ isoforms (LAP, LAP*, and LIP), in both MM cell lines and primary MM cells. Overexpression of C/EBPβ in MM cells showed that, although pomalidomide significantly down-regulated endogenous C/EBPβ protein in the control cells, exogenous (overexpressed) C/EBPβ was resistant to these effects and rescued cells from pomalidomide-induced inhibition of cell proliferation, indicating that C/EBPβ mediates the inhibition of cell proliferation by IMiD compounds. In addition, lenalidomide and pomalidomide also decreased levels of the C/EBPβ downstream TF IRF4. Initially, IRF4 was identified as an oncogene associated with the chromosomal translocation t(6;14) (p25;q32) in MM,41 but it also is a well-defined factor for normal plasma cell differentiation.16,42 Recently, IRF4 has been reported as a critical factor controlling MM survival19 and as a prognostic marker in patients with MM associated with poor survival.43 The importance of IRF4 as a target of lenalidomide in MM is also supported by a study of Lopez-Girona et al (oral and written communication with Peter Schafer, Celgene Corporation, August 11, 2010). This group found that higher IRF4 expression in MM patients was associated with a significantly worse survival. The negative prognostic impact of IRF4 expression was overcome by treatment with lenalidomide confirming the central role of IRF4 in MM pathogenesis and as a target for lenalidomide treatment in MM.
Our results demonstrated that IMiD compounds decrease the mRNA level of IRF4, but not of C/EBPβ. This and the fact that there was no increase in C/EBPβ protein degradation suggested that IMiD compounds block C/EBPβ protein translation in MM cells. This is in accordance with previous reports that translation of the C/EBPβ LAP isoform relative to the LIP isoform is regulated at the translation initiation site by eIF4E.12,20 It is further known that an increase in eIF4E level or activity results in increased translation of mRNA with highly complex 5′-untranslated regions, including c-Myc, Cyclin D1, and VEGF, all associated with proliferation. Accordingly, eIF4E expression was found to be elevated in cancer of the breast, head and neck, bladder, lung, prostate, and acute myeloid leukemia compared with normal tissue,32 but has not been described so far for MM. Our studies are in accordance with this because the exogenous transfected C/EBPβ, which is lacking any untranslated sequences in the mRNA and free from translational control by eIF4E, was resistant to the down-regulation by IMiD compounds. Further, we show that knockdown of eIF4E in MM cells significantly down-regulated C/EBPβ and IRF4, indicating that C/EBPβ is under translational regulation of eIF4E. This is further supported by the fact that in IMiD-resistant RPMI 8226 MM cells, lenalidomide and pomalidomide, do not affect expression of eIF4E, C/EBPβ, and IRF4, suggesting that the insensitivity might be responsible for its drug resistance. In this set of studies, there were no significant differences between lenalidomide and pomalidomide. Therefore, the observation that pomalidomide can overcome resistance to lenalidomide in MM patients36 requires further evaluation. The critical role of protein translation in MM cells is further supported by the fact that lenalidomide blocks secretion of VEGF44,45 and c-Myc,46 ; these have been reported to be regulated by eIF4E control of translation initiation.24,25 Regulating eIF4E expression and controlling translation by IMiD compounds could be responsible for its multiple functions, including immune modulation, antiangiogenic, anti-inflammatory, and antiproliferative effects.3,47,48 Therefore, eIF4E as a regulator of cytokines critical for survival and proliferation might be a potential new target for treatment of MM.49
Our data showed that IMiD-resistant cell lines, such as RPMI 8226 cells, responded to IMiDs after treatment with rapamycin. Further, mTOR inhibitors, such as rapamycin, which decrease 4EBP1 phosphorylation in combination with IMiD compounds, might overcome drug resistance. Rapamycin blocks the phosphorylation of 4EBP1, which is the inhibitory binding partner of eIF4E. Blocking of phosphorylation allows 4EBP1 to bind to eIF4E, resulting in inhibition of eIF4E translation initiation. In contrast, IMiD compounds decreased eIF4E protein levels, indicating that IMiD compounds and mTOR inhibitors target different steps during translation that together impair translational activity through complementary mechanisms, resulting in a stronger anti-MM effect. This is in accordance with a study by Raje et al that reported that the combination of lenalidomide and rapamycin showed strong synergism in MM inhibition.50 In addition, blocking translation by rapamycin and IMiD compounds might help to overcome resistance to IMiD compounds or reinduce sensitivity.
In conclusion, our studies, for the first time, provide evidence that the IMiD compounds lenalidomide and pomalidomide down-regulate eIF4E, which inhibits the translation of C/EBPβ and, as a consequence, decreases the downstream transcription of IRF4. This in turn down-regulates the network of IRF4-driven TFs, resulting in inhibition of MM growth. The increased understanding of the molecular effects of IMiD compounds on myeloma cells will contribute to the development of improved therapeutic strategies and to overcome drug resistance in the treatment of MM.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Robert W. Sobol for providing lentivirus particles, Ms Rita Bhutta for excellent preparation of the manuscript, and the UPCI Writing Group for review of the manuscript.
This work was supported in part by the Leukemia & Lymphoma Society, the Multiple Myeloma Research Foundation, and the Celgene Corporation.
Authorship
Contribution: S. Li designed and performed experiments and wrote the manuscript; R.P. designed and performed experiments; S.A.M. performed IHC of bone marrow biopsy samples and analyzed IRF4 staining; P.S. and M.M. designed experiments and discussed the manuscript; H.O. designed and performed experiments and discussed the manuscript; and D.L.G. and S. Lentzsch designed experiments and discussed and wrote the manuscript.
Conflict-of-interest disclosure: P.S. is an employee of Celgene Corporation, the manufacturer of IMiD compounds. S. Lentzsch receives research funding from Celgene Corporation. The remaining authors declare no competing financial interests.
Correspondence: Suzanne Lentzsch, Division of Hematology/Oncology, University of Pittsburgh Cancer Institute, 5150 Centre Ave, #568, Pittsburgh, PA 15232; e-mail: lentzschs@upmc.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal