Abstract
Erythropoietin (EPO), the key hormone in red blood cell renewal, is mainly produced in the adult kidney. Anemia and hypoxia substantially enhance EPO expression to increase erythropoiesis. Investigations of the cellular physiology of renal EPO production have been hampered by the lack of an adequate human cell line. In the present study, we present the human kidney cell line REPC (for renal Epo–producing cells), established from an explanted human kidney exhibiting EPO gene expression and release of the EPO protein in an oxygen-dependent manner. Hypoxic induction of EPO mRNA showed the typical transient increase and peak in expression after 36 hours under continuous conditions of hypoxia. Bioactive EPO protein accumulated in the culture supernatant. The induction of EPO gene expression in REPCs critically depended on the activation of hypoxia-inducible transcription factors (HIFs). SiRNA treatment revealed that the expression of EPO was largely dependent on the activation of the transcription factor complex HIF-2. In addition, hepatic nuclear factor 4α was shown to be critically involved in hypoxia-induced renal EPO expression. Using the human kidney cell line REPC, we provide for the first time a powerful tool with which to study the cellular and molecular regulation of renal EPO production.
Introduction
The glycoprotein hormone erythropoietin (EPO) regulates the proliferation and differentiation of erythroid progenitor cells, and therefore controls blood oxygen capacity and homeostasis. In adults, EPO is produced mainly in the kidney, whereas during fetal life the liver is the main source.1 Little constitutive EPO production is necessary to maintain the physiologic renewal of erythrocytes, but EPO expression increases dramatically during tissue hypoxia (eg, in high altitudes or with anemia).2,3 In 1993, interstitial peritubular fibroblasts localized in the deep cortex and superficial outer medulla were identified as the EPO-producing cells in the kidney by colocalization of the fibroblast-specific 5′-ectonucleotidase CD73 with EPO mRNA.4 During the last decades, great efforts were made to isolate these EPO-producing cells. However, until now, isolation either failed or resulted in the loss of the capability to produce EPO. In 1987, Goldberg et al identified 2 human hepatoma cell lines displaying oxygen-dependent EPO expression, HepG2 and Hep3B.5 Using these cells, the regulation of hepatic EPO production was found to depend upon hypoxia-inducible factors (HIFs).6 HIF transcription factor complexes consist of an oxygen-regulated α subunit and a constitutive β subunit, and were recognized as the key regulators of hypoxia-inducible gene expression in general.7 During hypoxia, the α subunit becomes stabilized and translocates to the nucleus, where it dimerizes with the β subunit and stimulates HIF target gene expression.8 The oxygen-dependent instability of HIF-α is attributed to the activity of a family of oxygen- and 2-oxo-glutarate–dependent dioxygenases (PHDs), which hydroxylate HIF-α at specific proline residues, initiating its ubiquitination and proteasomal degradation under normoxic conditions.9 To date, 3 different isoforms of HIF (HIF-1, HIF-2, and HIF-3) have been identified.10 Although HIF-1 is ubiquitously expressed, the expression pattern of HIF-1/2 in the kidney appears to be specific.11 Studies using transgenic mice with inducible knockouts of either HIF-1α or HIF-2α12,13 and PHD inhibition studies in rats14 have underscored the importance of HIF-2α for renal and hepatic EPO expression in rodents.15 In human hepatoma and neuroblastoma cells, a predominant role of HIF-2α in the regulation of EPO was proposed using an siRNA approach.16 In contrast, in human renal allograft biopsies and in the developing kidney, HIF-1α was predominantly detected by immunohistochemistry.17,18 Until now, there has been no reliable information about the contribution of HIF-1 and HIF-2 to EPO expression in the human adult kidney because of the lack of a suitable model.
In this study, we present a human kidney cell line, REPC (for renal Epo–producing cells), which is able to express EPO in an oxygen-dependent manner and will, therefore, become a powerful new model with which to study the molecular mechanisms of human renal EPO expression.
Methods
Cell culture
TK173 renal fibroblasts were originally isolated and described by G. Engler-Blum,19 and HepG2 cells, HK2 cells, and HEK293 cells were obtained from ATCC. REPCs were established from the tumor-free renal tissue of an anonymized male patient suffering from a kidney tumor. Approximately 1 g of kidney tissue was cut in small pieces (3 × 3 mm) and digested with collagenase type II for 30 minutes. The cell suspension was sieved through 150-mm and 50-μm meshes and resuspended in medium. All cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2mM l-glutamine, penicillin (100 U/mL), and streptomycin (100 μg/mL) in a humidified atmosphere of 5% CO2 in air at 37°C. REPCs grown in 75-cm2 culture flasks were split 1:5 after reaching confluence. Cell shape varied between large, hyaline cells of epithelial character when cells were freshly subcultured and smaller, more fibroblast-like cells when cells grew to confluence. When injected into nude mice, REPCs did not form tumors after observation periods of up to 12 weeks. For experiments, cells were seeded on 6- or 96-well polystyrol culture dishes. Experiments were performed with 80% confluent cells if not otherwise indicated. Cell proliferation was measured with the methylthiazolyldiphenyl-tetrazolium bromide (MTT) assay. Hypoxic conditions were achieved by placing the culture dishes in a Heraeus incubator at 3% or 1% O2, 5% CO2, and N2 as a balance for the indicated time periods. At the end of the experiments, cells were lysed in 4M guanidine thiocyanate, followed by total RNA extraction for the determination of specific mRNAs by RT-PCR. In addition, total cell lysates and nuclear extracts were prepared and submitted to immunoblotting.
Chromosomal analysis
Chromosomal analysis was performed by the Institute of Human Genetics at the University of Duisburg-Essen.
Profiling of REPCs by gene array
Total RNA was extracted with the RNeasy kit (QIAGEN) from confluent cultures of REPCs incubated for 24 hours under normoxic or hypoxic conditions. RNAs were processed using the Affymetrix standard protocol (Version 2.0) with 5 μg of total RNA. Hybridization, washing, and staining of the arrays were done according to the Affymetrix GeneChip protocol Version 2.0. Identification of genes was performed using Affymetrix comparison analysis software (Affymetrix GCOS 1.4 MAS 5). Changes in gene expression between normoxic and hypoxic cells were defined as significantly increased at P < .002 or decreased at P > .998 (Affymetrix comparison statistical algorithms).
RNA preparation and RT-PCR
Total RNA was isolated by the acid guanidinium thiocyanate-phenol-chloroform extraction method.20 One microgram of total RNA was reverse transcribed into cDNA with oligodT15 as the primer for reverse transcriptase (AMV reverse transcriptase; Promega). Qualitative PCRs were carried out for β-actin as a housekeeping gene to test the quality of cDNA preparation and for EPO, HIF-1α, HIF-2α, and PHD1-3 to estimate the amount of specific cDNA before real-time PCR. The resulting PCR fragments were visualized on ethidium bromide–stained 1.5% agarose gels. Primers for real-time PCR (Invitrogen) were designed with Primer Express software (Applied Biosystems Software v2.0) at an amplicon sizes of 150 bp, an annealing temperature of 60°C, and a CG content of approximately 60%. The primer sequences used for qualitative and quantitative PCR are listed in Table 1.
Primer sequences used for real-time PCR and ChIP assay
| Primer . | Use . | Sequence . | Accession no. . |
|---|---|---|---|
| EPO | Real-time PCR | 5′ctccgaacaatcactgct | X02158 |
| 3′ggtcatctgtcccctgtcct | |||
| Actin | Real-time PCR | 5′tcacccacaatgtgcccatctacga | X00351 |
| 3′cagcggaaccgctcattgccaatgg | |||
| HIF-1α | Real-time PCR | 5′gctggccccagccgctggag | AF208487 |
| 3′gagtgcagggtcagcactac | |||
| HIF-2α | Real-time PCR | 5′aggactacagcctgtcgtcagc | NM_001430 |
| 3′ccttgcaggagcgtggag | |||
| EPO 3′enhancer | ChIP assay | 5′ccaatatgaactcttggc | L16588 |
| 3′ctgcagccttgccctgggc | |||
| FSP-1 | qualitative PCR | 5′gagaaggccctggatgtgat | NM_002961 |
| 3′cctcgttgtccctgttgctg | |||
| CD73 | qualitative PCR | 5′cgcaacaatggcacaattat | NM_002526 |
| 3′ctcgacacttggtgcaaaga | |||
| PHD2 | Real-time PCR | 5′gcacgacaccgggaagtt | NM_022051 |
| 3′ccagcttcccgttacagt | |||
| PHD3 | Real-time PCR | 5′ggccatcagcttcctcctg | NM_022073.3 |
| 3′ggtgatgcagcgaccatca | |||
| Collagen IV | qualitative PCR | 5′gcgctggtgggtcagaaggc | NM_001845.4 |
| 3′ctgccttcaaggtggacggc |
| Primer . | Use . | Sequence . | Accession no. . |
|---|---|---|---|
| EPO | Real-time PCR | 5′ctccgaacaatcactgct | X02158 |
| 3′ggtcatctgtcccctgtcct | |||
| Actin | Real-time PCR | 5′tcacccacaatgtgcccatctacga | X00351 |
| 3′cagcggaaccgctcattgccaatgg | |||
| HIF-1α | Real-time PCR | 5′gctggccccagccgctggag | AF208487 |
| 3′gagtgcagggtcagcactac | |||
| HIF-2α | Real-time PCR | 5′aggactacagcctgtcgtcagc | NM_001430 |
| 3′ccttgcaggagcgtggag | |||
| EPO 3′enhancer | ChIP assay | 5′ccaatatgaactcttggc | L16588 |
| 3′ctgcagccttgccctgggc | |||
| FSP-1 | qualitative PCR | 5′gagaaggccctggatgtgat | NM_002961 |
| 3′cctcgttgtccctgttgctg | |||
| CD73 | qualitative PCR | 5′cgcaacaatggcacaattat | NM_002526 |
| 3′ctcgacacttggtgcaaaga | |||
| PHD2 | Real-time PCR | 5′gcacgacaccgggaagtt | NM_022051 |
| 3′ccagcttcccgttacagt | |||
| PHD3 | Real-time PCR | 5′ggccatcagcttcctcctg | NM_022073.3 |
| 3′ggtgatgcagcgaccatca | |||
| Collagen IV | qualitative PCR | 5′gcgctggtgggtcagaaggc | NM_001845.4 |
| 3′ctgccttcaaggtggacggc |
Real-time PCR was performed with SYBR Green as the fluorescent dye (Eurogentec) on the Gene Amp 5700 Sequence Detection System (Applied Biosystems). The cDNA standards for real-time PCR were prepared from the specific PCR products using a DNA-purification kit (Roche). The amount of standard cDNA was determined photometrically; the standard concentration ranged from 100-0.01 fg/μL. Quantification was with 2-step real-time PCR with a denaturation step at 95°C for 10 minutes, and then 35 cycles of 95°C for 15 seconds and 60°C for 1 minute. The amounts of specific cDNAs were normalized to the expression of the housekeeping gene β-actin.
Whole-cell and nuclear extract preparation
For preparing whole-cell extracts, cells were washed once with ice-cold PBS and lysed with 80 μL of lysis buffer (150mM NaCl, 10mM Tris pH 7.9, 1mM EDTA, 0.1% IGEPAL, and 1× protease inhibitor cocktail [Roche]) for 20 minutes on ice. The lysates were centrifuged at 7000g rpm at 4°C for 5 minutes in a microcentrifuge, and supernatants containing cellular proteins were collected and stored at −80°C. Protein concentrations of the supernatants were quantitated using a protein assay reagent (Bio-Rad).
Nuclear proteins were prepared using the method of Schreiber et al.21 All procedures were carried out at 4°C. After incubation, cells were lysed with 150 μL of buffer A (10mM HEPES pH 7.9, 1.5mM MgCl2, 10mM KCl, 0.5mM PMSF, 0.5mM DTT, 0.4% IGEPAL, and 1× protease inhibitor cocktail) and incubated for 20 minutes on ice. Cell lysates were centrifuged at 7000g rpm at 4°C for 5 minutes. Nuclear pellets were resolved in 80 μL of buffer B (20mM HEPES, pH 7.9, 420mM NaCl, 1.5mM MgCl2, 0.2mM EDTA, 0.5mM PMSF, 0.5mM DTT, and 1× protease inhibitor cocktail) and homogenized on a magnetic stirrer for 30 minutes. After centrifugation at 18 200g at 4°C for 15 minutes, supernatants containing the nuclear proteins were transferred to −80°C. Protein concentrations were determined using the Bio-Rad kit.
Transfer of siRNA
siRNAs against HIF-1α, HIF-2α, PHD1, PHD2, and PHD3 were obtained using siGENOME (Dharmacon). siRNA (final concentration 50μM) was transfected with Oligofectamin according to the manufacturer's instructions. Cells were allowed to recover from transfection for 6 hours before starting the experiments. For long-term knockdown of HIF-1α and HIF-2α, cells were transfected with siRNA every second day. siRNA sequences for HIF-1α and HIF-2α differed in more than 5 bp according to Dharmacon.
Western blot analysis
After the addition of one-quarter volume of sample buffer (50mM Tris pH 6.8, 2% SDS, 5% β-mercaptoethanol, 0.0125% Bromophenol blue, and 1% glycine), 70 μg of total cell lysate per lane was subjected to 7.5% SDS-PAGE and transferred onto a nitrocellulose membrane (0.2-μm pore size; Schleicher & Schuell). The efficiency of protein transfer and equal loading was confirmed by staining the nitrocellulose membrane with Ponceau S (Sigma-Aldrich). Filters were blocked overnight at 4°C with 5% nonfat dry milk powder in TBS-T (10mM Tris pH 7.5, 100mM NaCl, and 0.05% Tween 20). Membranes were washed in TBS-T and incubated with a mouse monoclonal antibody against HIF-1α (Transduction Laboratories), HIF-2α (Novus Biologicals), or PHD2 and PHD3 (both Novus Biologicals) at room temperature for 2 hours with gentle shaking. α-Tubulin and RNA polymerase II (both Sigma-Aldrich) served as loading and transfer controls. After washing with TBS-T, HRP-conjugated anti-mouse IgG antibody (Sigma-Aldrich) or HRP-linked anti-rabbit IgG antibody (New England Biolabs) was added. Immunoreactive proteins were visualized by chemiluminescence. Membranes were incubated in a detection solution for 1 minute (100mM Tris/Cl pH 8.5, 2.65mM H2O2, 0.45mM luminol, and 0.625mM p-coumaric acid), followed by exposure to X-ray films (Agfa-Gevaert).
Immunofluorescence
REPC and HK2 cells were stained for microtubule-associated protein 2 (MAP-2), neurofilament light peptide (NF-L), N-cadherin, and CD73. Cells were cultured on cover slips to 60% confluence. Medium was removed and cells were washed with PBS and fixed with 250 μL of cooled methanol/acetone (1:1) at −20°C for 10 minutes. The solution was removed and cells were allowed to dry for 10 minutes at room temperature. After blocking, cells were incubated with antibodies directed against MAP-2, NF-L, N-cadherin, or CD73 (Santa Cruz Biotechnology) 1:50 in PBS and 3% BSA for 2 hours. Detection was performed with Alexa 568–conjugated, species-specific secondary antibodies. After washing with PBS, samples were fixed with mounting medium (Dako) on slides and analyzed the following day with a fluorescence microscope (Eclipse E1000; Nikon Instruments) using objective lenses (CFI Nikon Plan Fluor 40× H, NA = 1.3 in air at 22°C), a Nikon camera, and NIS Elements software F. 3.00, SP7 (Nikon). Lectin staining to discriminate between epithelial cells and fibroblasts was carried out by incubation with Alexa 568–conjugated peptide nucleic acid (75 μg/mL in PBS; Molecular Probes, Invitrogen) for 15 minutes at room temperature in the dark.
ELISA for EPO
EPO protein in cell culture supernatants was measured by ELISA (R&D Systems) according to the manufacturer's recommendations.
EPO bioassay
The biologic activity of secreted EPO was measured using the BaF3 bioassay system.22 Proliferation of these murine leukemia cell line BaF3 cells stably transfected with the human EPO receptor depends on the amount of biologically active EPO. BaF3 cells were incubated with cell-culture supernatants of REPCs and standard concentrations of EPO (Jansen-Cilag). After 24 hours, MTT was added to the cells and the formation of formazan was used as an indicator for cell proliferation. The amounts of EPO were calculated from the standard curve.
ChIP assay
At the end of the experiments, transcription factors bound to the respective DNA elements were cross-linked to DNA using ethylene glycol bis(succinimidylsuccinate) (2mM in PBS/1mM MgCl2, pH 8.0) for 45 minutes at room temperature. After fixation, cells were washed 3 times with PBS, 1% (vol/vol) formaldehyde was added, and cells were incubated once more for 15 minutes at room temperature. Cell were lysed in lysis buffer (50mM Tris-HCl, pH 8.0, 2mM EDTA, 0.1% IGEPAL, 10% glycerol; 1mM DTT, 1mM PMSF, and 1× protease inhibitor cocktail [Roche]) for 15 minutes on ice. Nuclei were collected by centrifugation at 1200g for 5 minutes, and resuspended in SDS lysis buffer (50mM Tris-HCl pH 8.0, 10mM EDTA, and 1% SDS) at room temperature. DNA was subjected to sonification (4 × 25% for 15 seconds, 1 × 30% for 10 seconds, 1 × 30% for 15 seconds, and 4 pulses × 25% for 15 seconds in a Branson sonifier), which resulted in DNA fragments of an average length of 300-500 bp as validated by agarose gel electrophoresis and ethidium bromide staining. DNA fragments were incubated with 4 μg of HIF-1α- or HIF-2α–specific antibodies in a spin column using the protein G immunoprecipitation kit (Sigma-Aldrich). After 1 hour of incubation, protein G agarose beads were added to the columns and incubation was continued overnight at 4°C with head-over-tail mixing. After washing, complexes were eluted from the agarose beads by adding elution buffer (1% SDS and 0.1M NaHCO3), heating for 1 hour at 65°C, and centrifugation at 12 000g for 30 seconds. After elution, DNA was extracted by phenol-chloroform-isoamyl alcohol and ethanol precipitation. DNA (2 μL) was subjected to PCR using primers specific for the EPO 3′ enhancer region (Table 1).
Statistics
Experiments were performed in triplicate. Values of cDNA quantification are given as means ± SD. The Dunnett test and the Tukey-Kramer test were used to calculate whether differences in the means between the treated and the control groups were significant.
Results
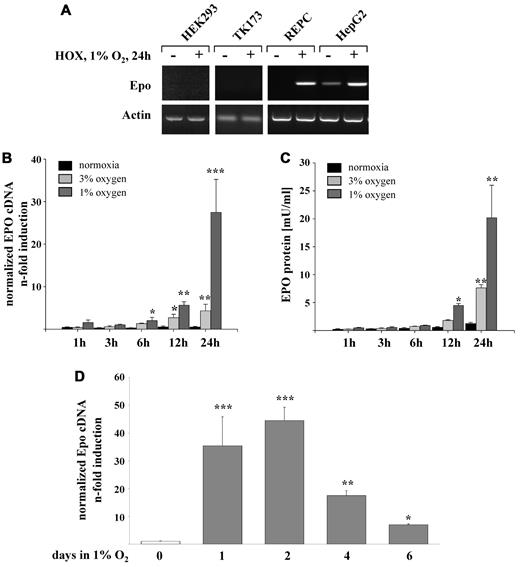
We screened different human kidney cell lines for their ability to express EPO in an oxygen-dependent manner. Only REPCs displayed a significant increase in EPO mRNA, which was comparable with the induction seen in the well-established EPO-producing human hepatoma cell line HepG2. Neither HEK293 nor the human renal fibroblast cell line TK173 expressed the EPO gene (Figure 1A). To analyze the time course of EPO gene expression and protein release, REPCs were incubated under normoxic and different hypoxic conditions. The magnitude of EPO mRNA expression and EPO protein release increased with the degree of hypoxia. A significant up-regulation of EPO gene expression was detected first after 6 hours of hypoxic incubation and increased further up to 24 hours, resulting in a 25-fold increase in cells incubated in 1% oxygen and a 5-fold increase in cells incubated in 3% oxygen (Figure 1B). EPO protein release followed the induction of EPO mRNA with a delay of 6 hours (Figure 1C). The maximum EPO expression was reached at day 2 after incubation at 1% O2, followed by a significant decrease during the following days. Nevertheless, EPO mRNA levels remained elevated 8-fold over the normoxic control after 6 days of hypoxic incubation (Figure 1D), and bioactive EPO protein showed a second increase at day 6 (Table 2).
Oxygen-regulated expression of EPO in REPCs. (A) Two established human renal cell lines (HEK 293 and TK173), REPCs, and HepG2 as a positive control were tested for hypoxia (24 hours at 1% O2)–induced expression of EPO. Shown are representative agarose gel electrophoreses stained with ethidium bromide. (B) Time course of EPO mRNA expression measured by real-time PCR and normalized to β-actin after up to 24 hours under conditions of 3% and 1% oxygen. Shown are the means ± SD; n = 6; *P < .05; **P < .01; ***P < .001. (C) Time course of EPO protein secretion after hypoxic stimulation of REPCs as measured by ELISA (R&D Systems). Shown are the means ± SD; n = 4; *P < .05; **P < .01; ***P < .001. (D) Time course of EPO mRNA expression measured by real-time PCR and normalized to β-actin after up to 6 days under 1% oxygen conditions. Shown are the means ± SD; n = 6; *P < .05; **P < .01; ***P < .001.
Oxygen-regulated expression of EPO in REPCs. (A) Two established human renal cell lines (HEK 293 and TK173), REPCs, and HepG2 as a positive control were tested for hypoxia (24 hours at 1% O2)–induced expression of EPO. Shown are representative agarose gel electrophoreses stained with ethidium bromide. (B) Time course of EPO mRNA expression measured by real-time PCR and normalized to β-actin after up to 24 hours under conditions of 3% and 1% oxygen. Shown are the means ± SD; n = 6; *P < .05; **P < .01; ***P < .001. (C) Time course of EPO protein secretion after hypoxic stimulation of REPCs as measured by ELISA (R&D Systems). Shown are the means ± SD; n = 4; *P < .05; **P < .01; ***P < .001. (D) Time course of EPO mRNA expression measured by real-time PCR and normalized to β-actin after up to 6 days under 1% oxygen conditions. Shown are the means ± SD; n = 6; *P < .05; **P < .01; ***P < .001.
Twenty-four-hour production rate of EPO during chronic hypoxia as measured with the Ba/F3 bioassay (n = 8)
| Incubation/day . | EPO protein, mU/mL/24 h . |
|---|---|
| Normoxia | Not detectable |
| Hypoxia (1% O2) | |
| 1 | 9.85 ± 0.436 |
| 2 | 20.14 ± 1.58 |
| 3 | 34.71 ± 1.08 |
| 4 | 5.10 ± 1.10 |
| 5 | 7.86 ± 0.28 |
| 6 | 17.98 ± 3.46 |
| Incubation/day . | EPO protein, mU/mL/24 h . |
|---|---|
| Normoxia | Not detectable |
| Hypoxia (1% O2) | |
| 1 | 9.85 ± 0.436 |
| 2 | 20.14 ± 1.58 |
| 3 | 34.71 ± 1.08 |
| 4 | 5.10 ± 1.10 |
| 5 | 7.86 ± 0.28 |
| 6 | 17.98 ± 3.46 |
For further characterization, we screened REPCs cultured for 24 hours under normoxic and hypoxic conditions by gene array analysis (Affymetrix). In total, 1800 genes were expressed in REPCs, the majority of which (71%) were not affected by hypoxia; approximately 2% of those genes were increased more than 2-fold by hypoxic incubation. Among these, enolase 2 (30.9-fold), EPO (11.1-fold), and carboanhydrase IX (10.9-fold) showed the strongest induction. In addition, we identified clusters of fibroblast markers, clusters of transcription factors possibly involved in hypoxia-regulated gene expression, and clusters of genes expressed in neuronal cells (Table 3).
Changes in gene expression induced by hypoxia at 1% O2 for 24 hours
| Cluster/gene . | Induction, -fold . |
|---|---|
| HIF target genes | |
| EPO | 11.1 |
| Adrenomedullin | 3.9 |
| Vascular endothelial growth factor | 2.8 |
| Lysyl oxidase | 3.1 |
| Carbonic anhydrase IX | 10.9 |
| Fibroblast markers | |
| Ecto-5′-nucleotidase CD73 | 0.4 |
| Procollagen-proline-4-hydroxylase | 1.2 |
| Procollagen-lysine-2-hydroxylase | 3.5 |
| Collagen type V | 1.4 |
| Collagen type III | 1.2 |
| FGFR1 | 0.6 |
| FGFR2 | 0.7 |
| Transcription factors | |
| HIF-1α | 0.7 |
| HIF-2α | 2.5 |
| ARNT | 0 |
| HNF4α | 1.8 |
| Neuronal markers | |
| Enolase 2 | 30.9 |
| Neural precursor cell protein (↓E9) | 2.7 |
| Brain-expressed X-linked 1 | 0.5 |
| GABA-receptor–associated protein 2 | 1.5 |
| vMAP-2 | 0.6 |
| NF-L (68 kDa) | 0.5 |
| Neurogenic differentiation 4 | 0.3 |
| Cluster/gene . | Induction, -fold . |
|---|---|
| HIF target genes | |
| EPO | 11.1 |
| Adrenomedullin | 3.9 |
| Vascular endothelial growth factor | 2.8 |
| Lysyl oxidase | 3.1 |
| Carbonic anhydrase IX | 10.9 |
| Fibroblast markers | |
| Ecto-5′-nucleotidase CD73 | 0.4 |
| Procollagen-proline-4-hydroxylase | 1.2 |
| Procollagen-lysine-2-hydroxylase | 3.5 |
| Collagen type V | 1.4 |
| Collagen type III | 1.2 |
| FGFR1 | 0.6 |
| FGFR2 | 0.7 |
| Transcription factors | |
| HIF-1α | 0.7 |
| HIF-2α | 2.5 |
| ARNT | 0 |
| HNF4α | 1.8 |
| Neuronal markers | |
| Enolase 2 | 30.9 |
| Neural precursor cell protein (↓E9) | 2.7 |
| Brain-expressed X-linked 1 | 0.5 |
| GABA-receptor–associated protein 2 | 1.5 |
| vMAP-2 | 0.6 |
| NF-L (68 kDa) | 0.5 |
| Neurogenic differentiation 4 | 0.3 |
REPCs were originally isolated and brought into culture from the tumor-free renal tissue of an anonymized male patient suffering from a kidney carcinoma. The morphology of the cells was similar to fibroblasts and astrocytes, having pronounced pseudopodia and cell processes (Figure 2A). Cells grow in culture to confluence and detach from the surface of the culture plate after reaching cell-to-cell contacts. Because malignant transformation of cells often causes abnormalities in chromosome content, we analyzed the chromosome set. Most REPCs display a diploid or hyperdiploid chromosome set. Trisomy and tetraploidy of several chromosomes occurred, but no chromosome aberration or translocation was detected in any of the analyzed karyograms (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). To obtain further information about the origin of these cells, we tested for the expression of markers specific for renal interstitial fibroblasts and for fibroblasts derived from epithelial cells after the epithelial-mesenchymal transition (EMT). Collagen IV and 5′-ectonucleotidase were constitutively expressed in REPCs, whereas fibroblast-stimulating protein-1 (FSP-1), which is specifically expressed in cells that undergo EMT, was not detectable (Figure 2B). Lectin staining characteristic of epithelial cells was only detectable on the established renal epithelial cell line HK2 (Figure 2C top panel); likewise, N-cadherin was notably present only on HK2 cells. Conversely, the neuronal markers MAP-2 and NF-L were expressed on REPCs, but not at all or rarely on HK2 cells. In addition, the distinct expression of CD73 in REPCs was confirmed at the protein level by immunofluorescence (Figure 2C bottom panels). The potential neuronal origin of REPCs prompted us to investigate whether REPCs respond to specific fibroblast or neuronal growth factors. Cells were cultured in the presence of basic fibroblast growth factor (bFGF) or brain-derived neurotrophic factor (BDNF). The proliferation of REPCs was significantly increased by bFGF and BNDF (Table 4), but EPO expression was not significantly influenced (supplemental Figure 4).
Characteristics of REPCs. (A) Morphology of REPCs at days 1 and 7 in culture. Shown are representative microscopic images at a final magnification of 400×. (B) Expression of kidney fibroblast–specific markers on REPCs compared with human kidney tissue. The EMT marker FSP-1 was not expressed in REPCs. Shown are representative agarose gels stained with ethidium bromide. (C) Differential expression of epithelial and neuronal markers on REPC and HK2 cells. REPCs were negative for the epithelial marker N-cadherin and lectin staining, but positive for MAP-2, NF-L, and CD73. (D) Proliferation of REPCs under normoxic and hypoxic conditions. REPCs were cultured for up to 7 days under normoxic and hypoxic conditions. Cells were counted every day, and cell proliferation was not influenced by hypoxia. Shown are the means ± SD; n = 6.
Characteristics of REPCs. (A) Morphology of REPCs at days 1 and 7 in culture. Shown are representative microscopic images at a final magnification of 400×. (B) Expression of kidney fibroblast–specific markers on REPCs compared with human kidney tissue. The EMT marker FSP-1 was not expressed in REPCs. Shown are representative agarose gels stained with ethidium bromide. (C) Differential expression of epithelial and neuronal markers on REPC and HK2 cells. REPCs were negative for the epithelial marker N-cadherin and lectin staining, but positive for MAP-2, NF-L, and CD73. (D) Proliferation of REPCs under normoxic and hypoxic conditions. REPCs were cultured for up to 7 days under normoxic and hypoxic conditions. Cells were counted every day, and cell proliferation was not influenced by hypoxia. Shown are the means ± SD; n = 6.
Proliferation of REPCs after 24 hours of incubation with bFGF or BDNF (n = 8)
| Incubation . | Proliferation, % . |
|---|---|
| Control | 100 ± 6.3 |
| BDNF | |
| 50 ng/mL | 134 ± 4.1 |
| 100 ng/mL | 132 ± 5.6 |
| 500 ng/mL | 137 ± 5.6 |
| 1 μg/mL | 143 ± 5.2 |
| bFGF | |
| 50 pg/mL | 143 ± 6.3 |
| 100 pg/mL | 150 ± 6.1 |
| 500 pg/mL | 143 ± 4.5 |
| 1 ng/mL | 149 ± 8.2 |
| Incubation . | Proliferation, % . |
|---|---|
| Control | 100 ± 6.3 |
| BDNF | |
| 50 ng/mL | 134 ± 4.1 |
| 100 ng/mL | 132 ± 5.6 |
| 500 ng/mL | 137 ± 5.6 |
| 1 μg/mL | 143 ± 5.2 |
| bFGF | |
| 50 pg/mL | 143 ± 6.3 |
| 100 pg/mL | 150 ± 6.1 |
| 500 pg/mL | 143 ± 4.5 |
| 1 ng/mL | 149 ± 8.2 |
Data are reported as means ± SD. P < .01 versus control for all values of BDNF and bFGF.
Because renal interstitial fibroblasts normally have to adapt to a hypoxic microenvironment, we investigated whether REPCs are able to cope with conditions of chronic hypoxia. The proliferation of REPCs did not vary between normoxic and hypoxic culture conditions (Figure 2C). Similar results were obtained by measuring cell viability using the MTT assay (data not shown).
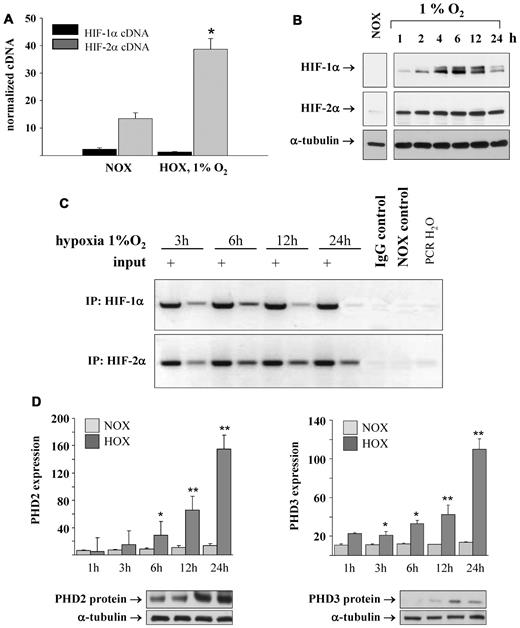
The transcription factor complex HIF was originally identified as the key regulator of hypoxia-inducible EPO gene expression in the human hepatoma cell lines HepG2 and Hep3B. Currently, 2 distinct HIFα isoforms are thought to be involved in hypoxia-induced gene expression, and there is debate regarding which HIF isoform is responsible for EPO gene expression in different tissues. We analyzed the expression of HIF-1α and HIF-2α mRNA in REPCs under normoxic and hypoxic conditions. Whereas HIF-1α mRNA was slightly down-regulated under hypoxia, HIF-2α mRNA was significantly increased, which confirmed our gene array data (Figure 3A). Moreover, the absolute normoxic expression level of HIF-2α mRNA was almost 10-fold higher than the expression of HIF-1α. A striking difference in the time course of protein accumulation between HIF-1α and HIF-2α was observed: HIF-1α accumulation was maximal after 6 hours of hypoxia whereas the maximum in HIF-2α accumulation was reached between 12 and 24 hours of hypoxic incubation (Figure 3B).
Expression and regulation of HIF-1α and HIF-2α. (A) HIF-1α and HIF-2α mRNA were expressed in REPCs. HIF-1α mRNA was constitutively expressed, whereas HIF-2α was significantly up-regulated by hypoxia (1% O2 for 6 hours). Shown are the means ± SD; n = 6; *P < .05. (B) Accumulation of HIF proteins during hypoxic incubation. HIF-1α protein accumulation peaked after 6 hours of hypoxic incubation. The HIF-2α protein became detectable after 1 hour of hypoxic incubation and remained elevated for up to 24 hours. Shown are representative immunoblots for HIF-1α and HIF-2α. α-Tubulin served as the loading control. Normoxic controls have been separated to indicate a repositioned lane from the same gel. (C) ChIP analysis of HIF-1 and HIF-2 DNA binding to the HRE of the EPO enhancer. Maximal HIF-1 binding was detected after 6 hours of hypoxia; after 24 hours, no HIF-1 binding to the EPO enhancer was observed. HIF-2 binding was detectable even after 24 hours under hypoxic conditions. (D) Regulation of the HIF-modifying prolyl hydroxylases and HIF target genes PHD2 and PHD3. Expression of both PHDs was continuously up-regulated by hypoxia. PHD mRNA expression was measured by real-time PCR and normalized to β-actin. PHD2 and PHD3 proteins were detected by immunoblot after 6 hours of hypoxic incubation. Shown are the means ± SD; n = 6; **P < .01.
Expression and regulation of HIF-1α and HIF-2α. (A) HIF-1α and HIF-2α mRNA were expressed in REPCs. HIF-1α mRNA was constitutively expressed, whereas HIF-2α was significantly up-regulated by hypoxia (1% O2 for 6 hours). Shown are the means ± SD; n = 6; *P < .05. (B) Accumulation of HIF proteins during hypoxic incubation. HIF-1α protein accumulation peaked after 6 hours of hypoxic incubation. The HIF-2α protein became detectable after 1 hour of hypoxic incubation and remained elevated for up to 24 hours. Shown are representative immunoblots for HIF-1α and HIF-2α. α-Tubulin served as the loading control. Normoxic controls have been separated to indicate a repositioned lane from the same gel. (C) ChIP analysis of HIF-1 and HIF-2 DNA binding to the HRE of the EPO enhancer. Maximal HIF-1 binding was detected after 6 hours of hypoxia; after 24 hours, no HIF-1 binding to the EPO enhancer was observed. HIF-2 binding was detectable even after 24 hours under hypoxic conditions. (D) Regulation of the HIF-modifying prolyl hydroxylases and HIF target genes PHD2 and PHD3. Expression of both PHDs was continuously up-regulated by hypoxia. PHD mRNA expression was measured by real-time PCR and normalized to β-actin. PHD2 and PHD3 proteins were detected by immunoblot after 6 hours of hypoxic incubation. Shown are the means ± SD; n = 6; **P < .01.
ChIP assays were performed to investigate whether HIF-1α or HIF-2α directly binds to the EPO enhancer under hypoxic conditions. A substantial increase in HIF-1α DNA binding was observed after 6 hours, which was less prominent but still present after 24 hours (Figure 3C). Quantitative analysis of the ChIP PCR revealed a 5-fold increase in HIF-1α DNA binding after 6 hours, which decreased to 2.2-fold after 24 hours. Binding of HIF-2α to the hypoxia-responsive element (HRE) of the EPO enhancer was likewise induced by hypoxia, but in contrast to HIF-1α, binding was pronounced even after 24 hours (Figure 3C). This indicates that HIF-1α may be responsible for the onset of hypoxia-induced EPO expression, whereas HIF-2α serves as a transcription factor during longer-lasting hypoxia.
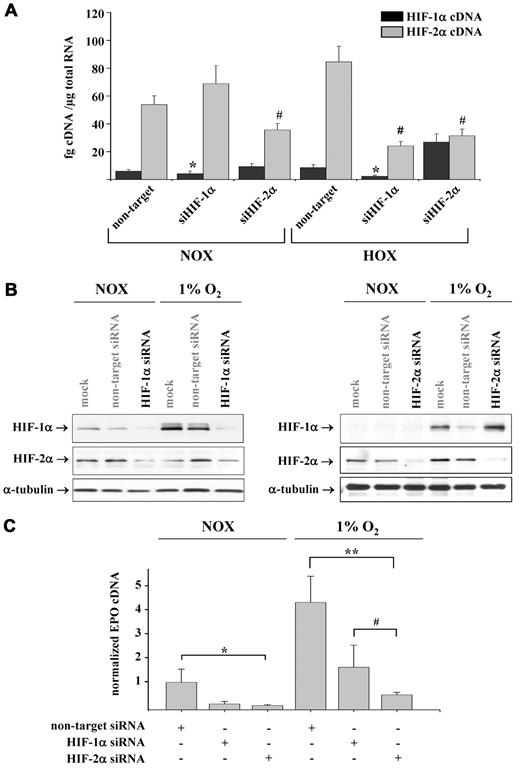
Because the regulation of HIF abundance and activity is regulated by PHDs, we analyzed the expression pattern of PHD1, PHD2, and PHD3. Whereas PHD2 and PHD3 were up-regulated under hypoxic conditions at both the mRNA and protein level (Figure 3D), the expression of PHD1 was not influenced by hypoxia (data not shown). To gain further insights into the role of the different PHD isoforms in the regulation of HIF-1α and HIF-2α stability, we knocked down PHDs by siRNA. Knockdown of PHD1 and PHD2 increased HIF-1α and HIF-2α stability and consequently EPO expression. Knockdown of PHD3 affected neither HIF stability nor EPO expression (supplemental Figure 1A-B). Because the stability of both HIF isoforms seemed to be regulated by PHD1 and PHD2, we knocked down HIF-1α and HIF-2α by siRNA to define their specific contribution to renal EPO expression. Efficiency of knockdown was assessed at the mRNA and protein levels. Specific knockdown efficiency was approximately 50% at the mRNA level; surprisingly, siRNA-mediated knockdown of HIF-1α significantly reduced hypoxia-induced HIF-2α expression (Figure 4A). At the protein level, the knockdown of HIF-1α and HIF-2α was almost complete (Figure 4B). Both HIF-1α and HIF-2α knockdown reduced the constitutive expression of EPO (Figure 4C). When cells were incubated under hypoxic conditions, the knockdown of HIF-2α led to a much more pronounced reduction in EPO mRNA expression than the knockdown of HIF-1α. These results were confirmed by analyzing the activity of an EPO Pro-312/EPO enhancer reporter construct after HIF siRNA treatment (supplemental Figure 2).
Importance of HIF-1 and HIF-2 for hypoxia-induced EPO expression in REPCs. (A) HIF-1α and HIF-2α proteins were knocked down by siRNA treatment, and the expression of the respective mRNAs was analyzed by real-time PCR and normalized to β-actin. Shown are the means ± SD; n = 6; *P < .05 with respect to the nontarget HIF-1α control; #P < .05 with respect to the nontarget HIF-2α control. (B) Effects of HIF-1α and HIF-2α siRNA treatment on the accumulation of HIF proteins under normoxic and hypoxic conditions. Shown are representative immunoblots for HIF-1α and HIF-2α. α-Tubulin served as the loading control. (C) Knockdown of HIF-1α and HIF-2α exerted different effects on the normoxic and hypoxia-induced EPO expression. Normoxic EPO expression was down-regulated by knockdown of HIF-1α and HIF-2α in a comparable manner. Hypoxia-induced EPO expression was more sensitive toward HIF-2α knockdown. EPO mRNA was quantitated by real-time PCR and normalized to β-actin. Shown are the means ± SD; n = 6, *P < .05; #P < .05; **P < .01.
Importance of HIF-1 and HIF-2 for hypoxia-induced EPO expression in REPCs. (A) HIF-1α and HIF-2α proteins were knocked down by siRNA treatment, and the expression of the respective mRNAs was analyzed by real-time PCR and normalized to β-actin. Shown are the means ± SD; n = 6; *P < .05 with respect to the nontarget HIF-1α control; #P < .05 with respect to the nontarget HIF-2α control. (B) Effects of HIF-1α and HIF-2α siRNA treatment on the accumulation of HIF proteins under normoxic and hypoxic conditions. Shown are representative immunoblots for HIF-1α and HIF-2α. α-Tubulin served as the loading control. (C) Knockdown of HIF-1α and HIF-2α exerted different effects on the normoxic and hypoxia-induced EPO expression. Normoxic EPO expression was down-regulated by knockdown of HIF-1α and HIF-2α in a comparable manner. Hypoxia-induced EPO expression was more sensitive toward HIF-2α knockdown. EPO mRNA was quantitated by real-time PCR and normalized to β-actin. Shown are the means ± SD; n = 6, *P < .05; #P < .05; **P < .01.
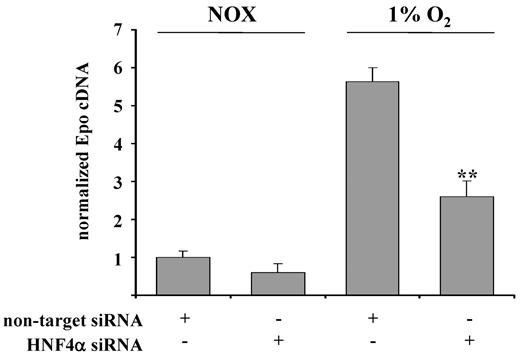
To search for other important transcription factors possibly involved in renal EPO expression, we investigated the role of hepatic nuclear factor 4 α (HNF4α), which was previously found to be involved in hypoxia-induced EPO expression.23 The requirement of HNF4α activation for EPO gene expression was demonstrated using an siRNA approach. Knockdown of HNF4α reduced hypoxia-induced EPO expression in REPCs by one-half (Figure 5).
Role of HNF4α in hypoxia-induced EPO expression in REPCs. SiRNA-mediated knockdown of HNF4α significantly inhibited hypoxia-induced EPO expression. EPO mRNA was quantitated by real-time PCR and normalized to β-actin. Shown are the means ± SD; n = 6; **P < .01.
Role of HNF4α in hypoxia-induced EPO expression in REPCs. SiRNA-mediated knockdown of HNF4α significantly inhibited hypoxia-induced EPO expression. EPO mRNA was quantitated by real-time PCR and normalized to β-actin. Shown are the means ± SD; n = 6; **P < .01.
Discussion
EPO is most important for stimulating the differentiation and proliferation of erythroid progenitors in the bone marrow. It is produced mainly in the adult kidney and the fetal liver,3 but EPO expression has also been demonstrated in several other tissues, where it may exert cytoprotective and antiapoptotic effects.24 Because the kidney was identified as the major source of circulating EPO in the adult,25 a lively debate about the cellular origin and molecular regulation of EPO arose. Colocalization of EPO expression and ecto-5-nucleotidase (CD73) in in situ hybridization studies provided evidence that peritubular interstitial fibroblasts, which are located in the juxtamedullary cortex, are the source of the EPO made in the kidneys.4 In rodents, it was shown that renal hypoxia caused by anemia, low inspiratory oxygen tension, or treatment with carbon monoxide (functional anemia) increased the number of EPO-producing cells in the kidney.26 During severe hypoxia, more and more EPO-producing cells were recruited and extended their localization to the subcapsular cortical tissue.27 In the adult human, more than 90% of the circulating EPO is produced by these cells in the kidney27 ; however, until the present study, the isolation and cultivation of EPO-producing cells from the kidney had failed, which was a major obstacle for studying the kidney-specific regulation of human EPO expression.
The first cellular model for oxygen-dependent regulation of EPO expression was introduced by Goldberg et al, who described the oxygen-regulated expression of EPO by the human hepatoma cell lines HepG2 and Hep3B.5 The use of these cell lines enabled the identification of HIF-1, the key transcription factor for oxygen-regulated EPO expression.28 Subsequently, HIF-1 was recognized as the “master regulator of oxygen homeostasis” and oxygen-regulated gene expression in general.29
We now introduce a human kidney cell line, REPC, which was isolated from the tumor-free tissue of a male patient suffering from renal carcinoma. Although REPCs proliferate under hypoxic conditions, they did not develop tumors when implanted into nude mice. EPO expression in REPCs displayed a dose-dependent increase when oxygen tension was reduced. Hypoxia-induced EPO production by REPCs showed the typical physiologic time course that was also observed in vivo.2 The maximum EPO mRNA expression was reached after 48 hours, and protein release was maximal after 3 days in hypoxia. We characterized REPCs as primary fibroblast-like cells with a potentially neuronal background. These cells are remarkably hypoxia tolerant, because incubation for up to 7 days in 1% oxygen did not inhibit cell proliferation or viability, which is consistent with the partially low oxygen tension found in the kidneys.30 The lack of FSP-1 and cadherin 9 expression, both markers for the EMT,31 makes it unlikely that REPCs are derived from EMT. 5-Ectonucleotidase (CD73), which was originally found to be strictly colocalized with renal EPO expression,4 was also found on REPC cells. In addition, REPCs lacked the expression of the epithelial marker N-cadherin and were negative for lectin staining. The assumption that REPCs do not derive from EMT was supported by the expression of the neuronal markers MAP-2 and NF-L, which were also found in EPO-expressing cells in a transgenic mouse model.32 In these mice, regulatory elements of the EPO gene drive GFP expression in vivo.32 We confirmed the distribution of these 2 markers on REPCs by immunofluorescence. Additional evidence for a neuronal origin of REPCs was provided by the detection of the Neural precursor cell protein that was down-regulated at embryonic day 9 (Table 2). Expression of this protein may be a clue to a physiologic migration of neuronal cells into the kidney during the early embryonic stages. The potential fibroblast-like/neuronal origin of REPCs is supported by the increase in their proliferation after stimulation with bFGF or BDNF.
Originally, HIF-1 was identified as the key transcription factor responsible for hypoxia-regulated EPO expression in human hepatoma cell lines by binding to the HRE located in the 3′-enhancer of the EPO.28,33 HIFs are heterodimeric transcription factors consisting of a constitutive β subunit, also known as aryl-hydrocarbon-nuclear-translocator (ARNT), and an oxygen-sensitive α subunit.34 Until now, 3 different HIFα isoforms were known: HIF-1α, which seems to be expressed in almost all cell types; HIF-2α, which displays a more tissue- and cell-specific expression pattern; and HIF-3α, which has a less clear role with respect to oxygen-regulated gene expression.34 Meanwhile, several studies using siRNA approaches35 and tissue-specific murine knockout models12 were performed to define the contribution of the different HIF isoforms to cell type–specific EPO expression. In rodents, it became obvious that renal EPO expression is linked to HIF-2 activation.15,36 Studies using different human tumor cell lines exhibited a less uniform correlation between distinct HIF isoforms and EPO expression. Whereas in the hepatoma cell line Hep3B and the neuroblastoma cell line Kelly, EPO expression seemed to be exclusively regulated by HIF-2,35 other neuronal cell lines displayed HIF-1–dependent EPO regulation.37 Both HIF isoforms were expressed and stabilized under hypoxic conditions in REPCs, but the time course and expression pattern differed between the isoforms (Figure 3). Whereas HIF-1α was only transiently stabilized, with a maximum after 6 hours of hypoxia, and HIF-1α mRNA expression decreased under these conditions, HIF-2α expression increased under hypoxic conditions and HIF-2α protein was detectable even after 24 hours of hypoxia. Similar results were observed when DNA binding of both isoforms to the human EPO enhancer was analyzed with the ChIP assay. A contribution of both isoforms to renal EPO expression was previously postulated by Yeo et al,38 who detected HIF-1 and HIF-2 binding to the EPO enhancer in the kidneys of mice exposed to mild hypoxia. However, in contrast to our findings, HIF-2 binding disappeared after 8 hours in that study. Because the abundance of HIFα proteins is regulated predominantly by posttranslational hydroxylation, resulting in proteasomal degradation,9,39 we investigated the expression of the HIF prolyl hydroxylases PHD1, PHD2, and PHD3 in REPCs under normoxic and hypoxic conditions. Whereas the oxygen sensors PHD2 and PHD3 were up-regulated during a 24-hour hypoxic incubation, PHD1 expression was not influenced. This agrees with previous work in which PHD2 and PHD3, but not PHD1 or FIH-1, were regulated by HIF.40-42 Knockdown of PHD1 and PHD2 resulted in an increased accumulation of HIF-1α and HIF-2α, as well as EPO expression, under normoxic conditions, whereas knockdown of PHD3 exerted no effects. Based on these findings, we assume that PHD3 is not critically involved in HIF regulation in REPCs. Together with the finding that HIF-2α mRNA expression was significantly induced by hypoxia in REPCs, one may hypothesize that HIF-2α escaped the PHD-initiated degradation by shifting the balance between new protein synthesis and degradation toward protein synthesis, thus stabilizing cellular protein levels. Therefore, under long-lasting hypoxic conditions, human renal EPO gene expression seems to be regulated predominantly by HIF-2.
To gain further insights into the contribution of HIF-1 and HIF-2 to EPO expression in REPCs, we knocked down both HIFα subunits by siRNA. Specific knockdown efficiency on the protein level was approximately 90% for both α subunit, but hypoxia-induced EPO expression was significantly reduced only in HIF-2α knocked-down REPCs. Interestingly, hypoxia-induced HIF-2α mRNA expression was reduced by HIF-1α siRNA treatment, which indicates that HIF-2α may be a gene that is directly or indirectly regulated by HIF-1. Computerized analysis of the HIF-2α promoter and the 5′UTR (−1859 to translation start site) revealed several possible HREs that could be accountable for the HIF dependency of HIF-2α expression. Therefore, the inhibition of hypoxia-induced EPO expression seen after HIF-1α knockdown could be partially the result of the suppression of HIF-2α by HIF-1α siRNA. This finding provides new insights into a possible intrinsic feedback loop of a tightly regulated sequence of HIF activation and deactivation during renal EPO expression. In addition, HNF4α was described as a coactivator of EPO gene expression,23 possibly by interacting with HIF-2. siRNA-mediated knockdown of HNF4α in REPCs significantly reduced hypoxia-induced EPO expression. Apart from HIFs and HNF4, other transcription factors such as GATA have been found to be involved in EPO gene expression.43,44 GATA-2 and GATA-3 are thought to be responsible for the repression of EPO gene expression.3 In this context, it is interesting that GATA-2 and GATA-3 expression was not detectable in REPCs. The importance of different promoter elements and the respective binding of transcription factors for cell type–specific EPO expression has not been fully elucidated and remains under investigation.
Based on the tissue from which REPCs were isolated, the oxygen-dependent regulation of EPO, and the expression of markers characterizing these cells as fibroblast-like with a potential neuronal origin, the human cell line REPC should provide a useful model with which to study the cellular and molecular mechanisms of renal-specific EPO expression.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the staff of the Institute of Human Genetics, University Duisburg-Essen, for their help with chromosome analysis.
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (GRK 1431 to J.F.).
Authorship
Contribution: S.F., P.F., L.G., and R.K. performed experiments; S.F. and J.F. analyzed results and made the figures; and S.F. and J.F. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joachim Fandrey, MD, Institut für Physiologie, Universität Duisburg-Essen, Hufelandstr 55, D-45122 Essen, Germany; e-mail: joachim.fandrey@uni-due.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal