Abstract
CCAAT/enhancer binding protein-α (CEBPA) mutations in acute myeloid leukemia (AML) patients with a normal karyotype (NK) confer favorable prognosis, whereas NK-AML patients per se are of intermediate risk. This suggests that blocked CEBPA function characterizes NK-AML with favorable outcome. We determined the prognostic significance of CEBPA DNA binding function by enzyme-linked immunosorbent assay in 105 NK-AML patients. Suppressed CEBPA DNA binding was defined by 21 good-risk AML patients with inv(16) or t(8;21) (both abnormalities targeting CEBPA) and 8 NK-AML patients with dominant-negative CEBPA mutations. NK-AML patients with suppressed CEBPA function showed a better overall survival (P = .0231) and disease-free survival (P = .0069) than patients with conserved CEBPA function. Suppressed CEBPA DNA binding was an independent marker for better overall survival and disease-free survival in a multivariable analysis that included FLT3-ITD, NPM1 and CEBPA mutation status, white blood cell count, age and lactate dehydrogenase. These data indicate that suppressed CEBPA function is associated with favorable prognosis in NK-AML patients.
Introduction
Karyotype abnormalities detectable at diagnosis in approximately 50% of acute myeloid leukemia (AML) patients are considered the most important prognostic factor.1-3 The 5-year overall survival (OS) of AML patients with a normal karyotype (NK) is between 35% and 45%,1,3 but individual outcome of such patients may vary greatly. Therefore, additional molecular markers, such as mutations in the genes encoding CCAAT/enhancer binding protein-α (CEBPA), FLT3, NPM1, IDH1 and IDH2, C-KIT, RAS, or WT1, have been reported to identify subgroups among NK-AML patients.4-18
In the hematopoietic system, the transcription factor CEBPA is expressed in myelomonocytic cells and specifically up-regulated during granulocytic differentiation.19,20 Expression of CEBPA in myeloid precursor cells can trigger terminal differentiation.20-22 Remarkably, cebpa knockout mice exhibit a selective block in neutrophil differentiation at the myeloblast level, whereas other blood cells develop normally.23
Dominant-negative CEBPA mutations are observed in up to 15% of AML patients, preferentially in NK-AML patients.4,5 Leukemic cells from AML patients with CEBPA mutations express distinctive gene expression signatures at the RNA24-26 and the miRNA level.27 Favorable prognosis of CEBPA mutations in AML is restricted to patients with double CEBPA mutations.5-8,28,29 The expression of the AML1-ETO fusion protein suppresses CEBPA at the mRNA level,30 and CEBPA expression and function can be inhibited by fms-like tyrosine kinase 3 (FLT3) mutations.31,32 The CBFB-SMMHC fusion protein selectively blocks CEBPA translation through induction of the mRNA binding protein calreticulin.33 Finally, CEBPA can be silenced by promoter hypermethylation, and forced expression of TRIB2 is blocking CEBPA wild-type protein by physical interaction, ultimately inducing proteasomal-dependent degradation.26
As various karyotype abnormalities target CEBPA function, deficient CEBPA function can also be present in NK-AML, such as in patients with CEBPA mutations. We therefore hypothesized that various levels of CEBPA function in NK-AML patients might be of prognostic significance.
Methods
Patients
Ficoll separated mononucleated cells from bone marrow samples of untreated consecutive de novo AML patients from a single center (University Hospital, Berne, Switzerland) were collected at diagnosis. Clinical characteristics and course of all patients are given in supplemental Tables 1 and 2 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Informed consent was obtained according to the Declaration of Helsinki. A total of 20% of the patients received SAKK 30/95 chemotherapy, 56% were treated within SAKK 30/00, and 24% underwent SAKK 30/01 chemotherapy.34,35 Approval of all studies was obtained by decisions of the local ethics committee of Berne, Switzerland.
Stable cell lines
Assessment of CEBPA DNA binding activity
Nuclear extracts were prepared as previously reported.4,30,33 The DNA binding activity was determined using the ELISA-based assay for CEBP (TransAM, Active Motif).33 The 96-well plates were coated with the CEBP consensus binding site oligo 5′-CTTGCGCAATCTATA-3′. Recognition of CEBPA was ensured by a specific CEBPA antibody. The addition of a secondary antibody conjugated to horseradish peroxidase provided sensitive colorimetric quantization by spectrophotometric assessment. Specificity of the assay was further verified by the addition of an oligonucleotide in excess encoding a CEBP wild-type or mutated consensus binding sequence.
Quantitative real-time PCR
Real-time PCR was performed as described36 on an ABI Prism 7700 Sequence Detection System using TaqMan Universal PCR Master Mix. CEBPA expression was normalized to the reference gene ABL.
Immunoblotting
CEBPA was detected using a rabbit polyclonal antibody against CEBPA (1:250; Santa Cruz Biotechnology). Protein bands were quantitated on Bio-Rad Versadoc 3000 Imaging System using Quantity One software (Bio-Rad).
Statistical analysis
Standard deviations were applied. Differences between groups were tested using the log-rank χ2 test. For comparison of the groups, Mann-Whitney rank-sum test and Kruskal-Wallis tests were performed. The correlation coefficient was specified as Pearson correlation (r). For multivariate analysis, the Cox proportional hazards regression model was applied. All statistical analyses were performed using Statview Version 5.0.1 for Windows (SAS Institute).
Results and discussion
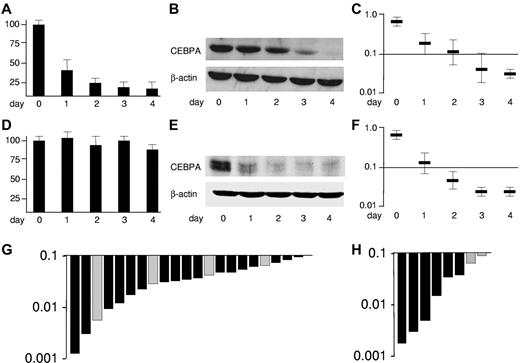
CEBPA is targeted by the leukemogenic fusion proteins CBFβ-SMMHC and AML1-ETO, encoded by inv(16) and t(8;21), respectively.30,33 Divergent mechanisms are involved as presented in Figure 1 depicting U937 cell lines allowing conditional expression of these fusion proteins using the tet-off system. Expression of AML1-ETO after withdrawal of tetracycline rapidly suppresses CEBPA mRNA (Figure 1A).30 Consequently, CEBPA protein is blocked (Figure 1B). An ELISA-based assay allowing spectrophotometric assessment (Figure 1C) depicts the decrease of CEBPA DNA binding activity.
CEBPA is suppressed in good-risk AML by the leukemic fusion proteins AML1-ETO and CBFβ-SMMHC. Conditional expression of AML1-ETO in U937-tet off-AML1-ETO cells blocks (A) CEBPA mRNA expression assessed by reverse-transcribed polymerase chain reaction, (B) CEBPA protein in Western blot analysis, and (C) CEBPA DNA binding function determined by an ELISA-based assay indicating the spectrophotometric result at 450-nm wavelength,30 whereas expression of CBFβ-SMMHC in U937-tet off-CBFβ-SMMHC (D) does not affect CEBPA mRNA but (E) blocks CEBPA translation by specific induction of the CEBPA mRNA binding protein calreticulin, (F) ultimately leading to blocked CEBPA DNA binding function.33 Results represent 3 independent experiments. Error bars represent SD. (G) Waterfall diagram presentations of CEBPA DNA binding function assessed by ELISA in 17 AML-M4eo patients with inv(16) (black bars) and in 4 AML-M2 patients with t(8;21) (gray bars) (H) as well as in NK-AML patients with double CEBPA mutations (n = 6) (black bars) and with single CEBPA mutations (n = 2) (gray bars).
CEBPA is suppressed in good-risk AML by the leukemic fusion proteins AML1-ETO and CBFβ-SMMHC. Conditional expression of AML1-ETO in U937-tet off-AML1-ETO cells blocks (A) CEBPA mRNA expression assessed by reverse-transcribed polymerase chain reaction, (B) CEBPA protein in Western blot analysis, and (C) CEBPA DNA binding function determined by an ELISA-based assay indicating the spectrophotometric result at 450-nm wavelength,30 whereas expression of CBFβ-SMMHC in U937-tet off-CBFβ-SMMHC (D) does not affect CEBPA mRNA but (E) blocks CEBPA translation by specific induction of the CEBPA mRNA binding protein calreticulin, (F) ultimately leading to blocked CEBPA DNA binding function.33 Results represent 3 independent experiments. Error bars represent SD. (G) Waterfall diagram presentations of CEBPA DNA binding function assessed by ELISA in 17 AML-M4eo patients with inv(16) (black bars) and in 4 AML-M2 patients with t(8;21) (gray bars) (H) as well as in NK-AML patients with double CEBPA mutations (n = 6) (black bars) and with single CEBPA mutations (n = 2) (gray bars).
Expression of CBFβ-SMMHC did not affect CEBPA mRNA expression (Figure 1D), whereas CEBPA translation was efficiently blocked (Figure 1E) by specific induction of the CEBPA mRNA binding protein calreticulin as previously reported.33 Again, the suppression of CEBPA DNA binding activity after CBFβ-SMMHC induction could be depicted using the ELISA assay (Figure 1F). In summary, the fusion proteins t(8;21) and inv(16) block CEBPA DNA binding activity, which can be assessed using an ELISA-based assay.
We determined the CEBPA DNA binding activity in nuclear extracts of leukemic cells from 17 patients with AML-M4Eo with inv(16) and 4 AML-M2 patients having the t(8;21) (Figure 1G). No differences between t(8;21) and inv(16) patients were observed. The range of CEBPA DNA binding activities in these 21 good-risk AML patients was selected to define AML patients with suppressed CEBPA DNA binding. The highest DNA binding activity scored at a value of 0.094. To validate this cut-off, we analyzed 8 NK-AML patients with dominant-negative CEBPA mutations. Again, they scored within the range observed in patients with inv(16) or with t(8;21) (Figure 1H). Notably, the 6 patients with double CEBPA mutations (with an N-terminal frame-shift and a C-terminal in-frame CEBPA mutation) had lower CEBPA DNA binding activities than the 2 patients with a single (N-terminal) CEBPA mutation. These observations are consistent with recent reports suggesting that AML patients with single versus double CEBPA mutations have divergent clinical characteristics.28,29
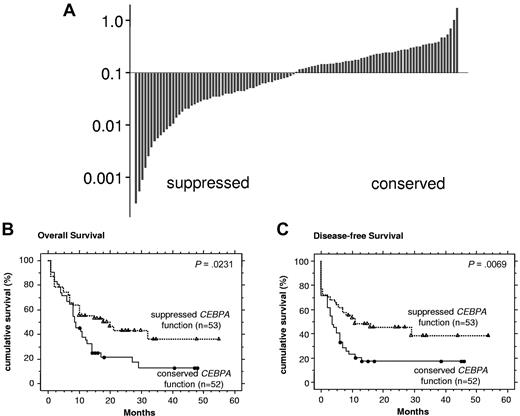
We then assessed CEBPA function in a cohort of 105 consecutive de novo NK-AML patients, including the 8 NK-AML patients with CEBPA mutations. Our cut-off of 0.094 separated NK-AML patients in 2 approximately equally sized groups (Figure 2A). We observed no significant clinical differences between patients with conserved versus suppressed CEBPA DNA binding activity (supplemental Table 3). In addition, no differences in the immunophenotype profile were detectable (data not shown).
Suppressed CEBPA DNA binding function is associated with favorable outcome in NK-AML patients. (A) Waterfall diagram presentations of CEBPA DNA binding function assessed by ELISA in 105 consecutive AML patients with an NK. The cut-off of 0.094 represents the highest value observed among inv(16) and t(8;21) AML patients in Figure 1G. (B) Kaplan-Meier curves are shown for OS and (C) DFS comparing NK-AML patients with suppressed versus normal CEBPA DNA binding function.
Suppressed CEBPA DNA binding function is associated with favorable outcome in NK-AML patients. (A) Waterfall diagram presentations of CEBPA DNA binding function assessed by ELISA in 105 consecutive AML patients with an NK. The cut-off of 0.094 represents the highest value observed among inv(16) and t(8;21) AML patients in Figure 1G. (B) Kaplan-Meier curves are shown for OS and (C) DFS comparing NK-AML patients with suppressed versus normal CEBPA DNA binding function.
Notably, NK-AML patients with suppressed CEBPA function showed a significantly more favorable course of their disease compared with patients with conserved CEBPA function: The OS was 18 months versus 9 months (P = .0231), and the disease-free survival (DFS) was 13 months versus 5 months (P = .0069) (Figure 2B-C). This difference was mainly the result of a higher relapse rate of the group with conserved CEBPA DNA binding function, whereas the rate of complete remissions (CR1) was not different (supplemental Table 4).
We intended to verify that NK-AML patients with suppressed CEBPA DNA binding activity also expressed lower levels of CEBPA protein. Sufficient protein for Western blot analyses was available from 51 of the 105 NK-AML patients (supplemental Figure 1A). As expected, there was a strong correlation between NK-AML patients with suppressed CEBPA DNA binding function and patients with suppressed 42-kDa CEBPA protein expression. Here, patients were stratified according to the median 42-kDa CEBPA protein value because the ELISA assay has produced 2 equally sized groups (supplemental Figure 1B). The clinical characteristics and the clinical course of these 2 groups are depicted in supplemental Tables 5 and 6. Patients with suppressed (versus conserved) 42-kDa CEBPA protein tended to have a more favorable OS and DFS (supplemental Figure 1C-D). The small number of evaluable patients in both groups led to insufficient statistical power to reach significance.
In a multivariable analysis, suppressed CEBPA DNA binding function turned out to be an independent prognostic factor for OS and DFS in NK-AML patients (supplemental Table 7). This analysis included CEBPA function, CEBPA mutation, FLT3-ITD mutation, NPM1 mutation, age, white blood cells, and lactate dehydrogenase. NPM1 mutations were reported to be associated with NK-AML10 and to predict favorable prognosis.11-14 Our results suggest that the impact on outcome of various levels of CEBPA function acts independently from the NPM1 mutation status. In addition, we found no evidence that the prognostic value of CEBPA function correlated with the FLT3 mutational status.
In conclusion, we identified frequent suppression of CEBPA DNA binding function in NK-AML patients using an ELISA-based assay, suggesting that block of CEBPA function in NK-AML is a more frequent event than commonly anticipated. Our data indicate that suppressed CEBPA function is associated with favorable outcome in NK-AML patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Daniel Helbling and Julian Schardt for technical support.
This work was supported by the Swiss National Science Foundation (SF 310000-113761) and Oncosuisse (OCS 01731082005 and KFS-02449-08-2009).
Authorship
Contribution: J.F. performed research and analyzed data; T.P. analyzed data and wrote the paper; V.P. performed research; D.R. analyzed data; and B.U.M. designed the study, analyzed data, and wrote the paper; and all authors read and approved the report in its final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Beatrice U. Mueller, Department of Internal Medicine, University Hospital, 3010 Berne, Switzerland; e-mail: beatrice.mueller@insel.ch.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal