Abstract
Master transcriptional regulators of development often function through dispersed cis elements at endogenous target genes. While cis-elements are routinely studied in transfection and transgenic reporter assays, it is challenging to ascertain how they function in vivo. To address this problem in the context of the locus encoding the critical hematopoietic transcription factor Gata2, we engineered mice lacking a cluster of GATA motifs 2.8 kb upstream of the Gata2 transcriptional start site. We demonstrate that the −2.8 kb site confers maximal Gata2 expression in hematopoietic stem cells and specific hematopoietic progenitors. By contrast to our previous demonstration that a palindromic GATA motif at the neighboring −1.8 kb site maintains Gata2 repression in terminally differentiating erythroid cells, the −2.8 kb site was not required to initiate or maintain repression. These analyses reveal qualitatively distinct functions of 2 GATA motif-containing regions in vivo.
Introduction
GATA factors are often expressed in overlapping and reciprocal patterns in hematopoiesis. Accrued evidence supports a model in which GATA factors cross-regulate transcription of their respective genes through a “GATA switch.”1,2 This hypothesis has been investigated at the Gata2 locus, where several conserved GATA motif-containing regions span ∼ 100 kb. GATA-2 has a critical role in the emergence and maintenance of hematopoietic stem/progenitor cells3-10 and appears to positively regulate its own transcription. During erythroid cell differentiation, GATA-1 mediates repression of Gata2 expression. A cis-element located −2.8 kb upstream from the 1S Gata2 promoter has been proposed to mediate GATA-2-dependent positive feedback before GATA-1-instigated repression.11-13 Removal of this site would thus be predicted to abrogate positive feedback, reducing Gata2 expression in hematopoietic stem/progenitor cells. We previously demonstrated that a distinct cis-element −1.8 kb upstream of the Gata2 promoter maintains repression of Gata2 in late-stage erythroblasts, though it is not required to initiate repression.14 As the patterns of GATA-factor binding at the −1.8 kb and −2.8 kb sites are similar,11-13 we addressed whether the −2.8 kb site is also required for GATA-1-mediated Gata2 repression.
We demonstrate that the −2.8 kb site is not required for initiation or maintenance of Gata2 repression during erythroid differentiation. By contrast, the −2.8 kb site confers maximal Gata2 expression in HSCs and progenitors in specific contexts. Comparison of the −2.8 kb and −1.8 kb sites illustrates how GATA motifs function uniquely and collectively to achieve complex and dynamic transcriptional control at the Gata2 locus during hematopoietic development.
Methods
Generation of Gata2 Δ-2.8 knockin mice
We replaced a 400 bp region containing 6 GATA motifs with a LoxP-PGKneo-LoxP cassette (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Details of the cloning strategy are available on request. Gene targeted ES cell clones were identified by Southern blotting (supplemental Figure 1B-C). Confirmation of Cre-mediated neo excision was performed with PCR before blastocyst injection (supplemental Figure 1D). All mouse studies were conducted under guidelines associated with protocols approved by Harvard Medical School.
Gene expression and chromatin occupancy
Sampling of fetal liver and bone marrow cells, flow cytometric analysis and cell sorting, quantitative ChIP assay, and real-time reverse-transcriptase PCR were performed as published.14
Results and discussion
Previous studies demonstrated a critical role for Gata2 in the emergence and maintenance of hematopoietic stem cells (HSCs).3,4 The −2.8 kb region drives exogenous expression of a reporter gene in HSCs in a manner dependent on GATA15 and E-box16 motifs, and has been proposed to be the primary cis-element conferring Gata2 expression in these contexts.
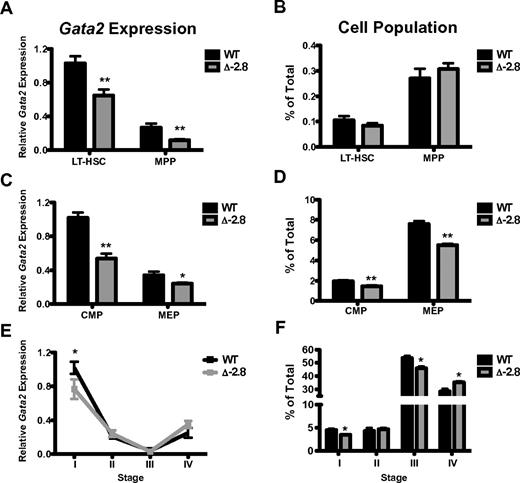
Mice homozygous for deletion of a 400 bp region located −2.8 kb upstream of the Gata2 1S promoter (supplemental Figure 1) were born at expected Mendelian ratios (data not shown). We quantified Gata2 expression in sorted long-term HSCs (LT-HSCs) and multipotent progenitors (MPPs) from embryonic day (E) 12.5 fetal livers. Δ-2.8 LT-HSCs and MPPs exhibited reduced Gata2 expression versus control cells (Figure 1A), but this reduction did not alter the frequency of the populations (Figure 1B). Gata2 serves critical roles in the maintenance and specification of multipotential progenitors.5-10 Expression of Gata2 was reduced in common myeloid progenitors (CMPs) and megakaryocyte-erythroid progenitors (MEPs) of Δ-2.8 fetal livers (Figure 1C), and was associated with a modest reduction in the frequency of these populations (Figure 1D). Quantitative ChIP analysis with enriched progenitor cells from Δ-2.8 and wild-type E14.5 fetal livers revealed a selective reduction of GATA-2 occupancy at the −2.78 kb site (surrogate for measuring occupancy at the deleted −2.8 kb site; supplemental Figure 2). These results indicate that the −2.8 kb site contributes to Gata2 expression in fetal liver-derived multipotent and committed erythroid progenitors in part through GATA-2 binding.
−2.8 kb GATA switch site regulates Gata2 expression in hematopoietic stem and progenitor cells, but is dispensible in erythroid cells. Gata2 expression within LT-HSC and MPP populations (LT-HSC defined as Lindimckit+Sca1+CD150+ and MPP defined as Lindimckit+Sca1+CD34+ or Lindimckit+Sca1+CD150−) from E12.5 fetal livers normalized to β-actin (A). Frequencies of LT-HSCs and MPPs in wild-type and Δ-2.8 fetal livers (B). Gata2 expression within CMP and MEP populations (with CMP defined as LindimSca1−,c-kit+CD34+, FcγR− and MEP defined as Lindim Sca1−c-kit+CD34−, FcγR−) from E12.5 fetal livers normalized to β-actin (C). Frequencies of CMPs and MEPs from wild-type and Δ-2.8 fetal livers (D). Gata2 expression was assessed by qPCR in Stage I through Stage IV sorted erythroid cells, corresponding to CD71loTer119− (committed erythroid progenitors, Stage I), CD71hiTer119− (proerythroblasts, Stage II), CD71hiTer119+ (basophilic erythroblasts, Stage III), and CD71loTer119+ (late erythroblasts, Stage IV) from wild-type and Δ-2.8 embryos. (E). Relative number of cells in Stage I-IV was determined in wild-type and Δ-2.8 E13.5 fetal liver cells, based on CD71 and Ter119 expression (F). Mean ± SEM. Statistical significance was assessed by 2-sided Student t test. *P ≤ .05 and **P ≤ .01.
−2.8 kb GATA switch site regulates Gata2 expression in hematopoietic stem and progenitor cells, but is dispensible in erythroid cells. Gata2 expression within LT-HSC and MPP populations (LT-HSC defined as Lindimckit+Sca1+CD150+ and MPP defined as Lindimckit+Sca1+CD34+ or Lindimckit+Sca1+CD150−) from E12.5 fetal livers normalized to β-actin (A). Frequencies of LT-HSCs and MPPs in wild-type and Δ-2.8 fetal livers (B). Gata2 expression within CMP and MEP populations (with CMP defined as LindimSca1−,c-kit+CD34+, FcγR− and MEP defined as Lindim Sca1−c-kit+CD34−, FcγR−) from E12.5 fetal livers normalized to β-actin (C). Frequencies of CMPs and MEPs from wild-type and Δ-2.8 fetal livers (D). Gata2 expression was assessed by qPCR in Stage I through Stage IV sorted erythroid cells, corresponding to CD71loTer119− (committed erythroid progenitors, Stage I), CD71hiTer119− (proerythroblasts, Stage II), CD71hiTer119+ (basophilic erythroblasts, Stage III), and CD71loTer119+ (late erythroblasts, Stage IV) from wild-type and Δ-2.8 embryos. (E). Relative number of cells in Stage I-IV was determined in wild-type and Δ-2.8 E13.5 fetal liver cells, based on CD71 and Ter119 expression (F). Mean ± SEM. Statistical significance was assessed by 2-sided Student t test. *P ≤ .05 and **P ≤ .01.
To examine the function of this site at a different developmental stage, we isolated LT-HSC and MPP populations from adult Δ-2.8 and wild-type bone marrow. Gata2 expression was reduced in adult MPPs, but not LT-HSC (supplemental Figure 3A). We observed a similar reduction in Gata2 in MPP from Δ-1.8 mice (supplemental Figure 3A), implying some functional overlap. Loss of either site did not alter LT-HSC, MPP, CMP, GMP, or MEP numbers in adult mice (supplemental Figure 3B-C). No differences in hematopoietic colony formation or peripheral blood parameters were apparent in the Δ-2.8 or −1.8 adult mice (supplemental Table 1 and supplemental Figure 3D). Competitive primary and secondary transplant experiments revealed no statistically significant differences in the ability of Δ-2.8 or Δ-1.8 HSCs to contribute to hematopoiesis in these assays (supplemental Figure 3). Thus, while both sites confer maximal Gata2 expression in certain contexts, they are not essential for hematopoiesis in the mouse.
Our prior work demonstrated that the −1.8 kb site maintains GATA-1-mediated repression of Gata2 during erythroid differentiation.14 As the 2 sites have similar patterns of GATA-factor binding,11-13 we investigated Gata2 repression during erythroid differentiation in Δ-2.8 mice. Gata2 expression was reduced in Stage I of erythroid differentiation in the Δ-2.8 mice (Figure 1E). Stage I contains MEP activity,17 consistent with of the findings of Figure 1C. Despite this reduction, Gata2 expression was normal in Stages II-IV in the Δ-2.8 fetal livers, indicating that the −2.8 kb site is not required for initiation or maintenance of GATA-1-mediated repression (Figure 1E). Only modest differences in the relative abundance of cells from each stage were apparent in Δ-2.8 versus wild-type mice (Figure 1F).
Based on the unexpected observation that the −1.8 kb, but not the −2.8 kb, site mediates repression, we reasoned that comparing their capacities to regulate nucleoprotein architecture would yield mechanistic insights. Quantitative ChIP analysis with Δ-2.8 and wild-type E14.5 fetal liver cells revealed low GATA-1 occupancy at the −2.78 kb site (Figure 2A). ChIP analysis of Pol II occupancy demonstrated no change on loss of the −2.8 kb site (data not shown), contrasting with our demonstration that loss of the −1.8 kb site results in Pol II accumulation.14 We quantified dimeH3K4 and trimeH3K27 marks associated with Gata2 repression.11,13,18 Results from the Δ-1.8 mice demonstrated locus-wide reduction in dimethylH3K4 with no significant reductions in trimethylH3K27 (supplemental Figure 5A-B and Snow et al14 ). By contrast, loss of GATA-1 occupancy on deletion of the −2.8 kb site results in locus-wide reduction in one of the marks associated with repression, trimeH3K27 (Figure 2C), while having minimal effects on dimeH3K4 (Figure 2B), and no influence on repression. Although these data implicate a function of the polycomb repressive complex 2 (PRC2)18 at the −2.8 kb site, reduction of H3K27 trimethylation does not reactivate Gata2 transcription (Figure 1).
The −2.8 kb GATA switch site establishes trimeH3K27 marks at the Gata2 locus. Quantitative ChIP analysis of the Gata2 locus in E14.5 fetal liver cells using antibodies to GATA-1 (A), dimeH3K4 (B), trimeH3K27 (C), and Total H3 (D). Updated model of the molecular regulation of Gata2 repression (E). Mean ± SEM. Statistical significance was assessed by 2-sided Student t test and *P ≤ .05 and **P ≤ .01.
The −2.8 kb GATA switch site establishes trimeH3K27 marks at the Gata2 locus. Quantitative ChIP analysis of the Gata2 locus in E14.5 fetal liver cells using antibodies to GATA-1 (A), dimeH3K4 (B), trimeH3K27 (C), and Total H3 (D). Updated model of the molecular regulation of Gata2 repression (E). Mean ± SEM. Statistical significance was assessed by 2-sided Student t test and *P ≤ .05 and **P ≤ .01.
In summary, our analyses of GATA motif function in vivo establish fundamentally different requirements for the −2.8 and −1.8 kb GATA switch elements. As GATA motifs within these sites conferred unique chromatin states and exerted different activities, these results support a model in which initiation/maintenance of Gata2 transcriptional control requires the collective actions of dispersed GATA factor complexes with qualitatively distinct contributions (Figure 2E). As GATA-1 occupies different permutations of GATA motifs, including canonical, palindromic, composite, and noncanonical,19 our results suggest that these permutations may differ considerably in their functionality in vivo. Individual HSs of the murine β-globin locus function additively to confer maximal expression of the β-like globin genes,20 differing from the differential −1.8 kb and −2.8 kb site activities at Gata2. Qualitatively distinct functions of GATA complexes were apparent at Gata2, highlighting the critical need to expand efforts to elucidate GATA motif function at a spectrum of endogenous loci.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge the technical assistance of Yuko Fujiwara, Minh Nguyen, and Mayumi Kaku, Grigoriy Losyev at the Brigham and Women's Flow Cytometry Core, John Daly and Susan Lazo-Kallanian at the Dana-Farber Cancer Institute HemNeo Flow Facility, and the staff of the Animal Resource Children's Hospital Boston. The authors thank Stephen Chan and Marc Kerenyi for critical comments.
This work was funded by National Institutes of Health grants DK068634 (E.H.B.) and HL32259 (S.H.O.) and by grant P30 CA014520 to the University of Wisconsin Paul Carbone Cancer Center from the National Cancer Institute. J.W.S. and J.J.T. were funded in part by Fellowships from the Leukemia & Lymphoma Society, and S.H.O. is an Investigator of the Howard Hughes Medical Institute.
National Institutes of Health
Authorship
Contribution: E.H.B. and S.H.O. conceived of the project; J.A.G., N.E.E, and J.W.S. built initial constructs, screened clones, and generated mice; J.W.S., T.F., K.D.J., and J.J.T performed experiments; J.W.S. and J.J.T. analyzed data; and J.W.S., J.J.T., S.H.O., and E.H.B wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for J.W.S. is Biology Department, Williams College, Williamstown, MA.
Correspondence: Emery H. Bresnick, Department of Cell and Regenative Biology, University of Wisconsin School of Medicine and Public Health, Wisconsin Institutes for Medical Research, Rm 4009, 1111 Highland Ave, Madison, WI 53705; e-mail: ehbresni@wisc.edu; and Jonathan W. Snow, Biology Department, Williams College, Williamstown, MA 01267, MA; e-mail: jonathan.snow@williams.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal