To the editor:
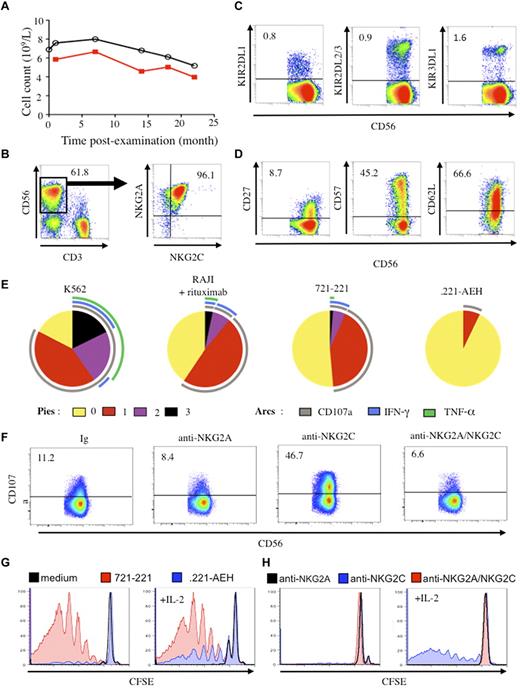
In April 2009, we saw a 64-year-old woman with persistent indolent proliferation of CD3−CD56+ large granular lymphocytes.1 She had no other clinical symptoms and no autoimmune events, and her clinical course has remained benign over 22 months. Neither clinical nor imaging studies have shown signs of lymphoma or acute infection. IgG serology is negative for HIV-1/2, human T cell lymphotrophic virus (HTLV)–1/2, hepatitis B virus, hepatitis C virus, and HHV8, and positive for EBV, CMV, HSV, and varicella zoster virus. Most notably, we have observed an elevated lymphocyte count, ranging from 5.2 to 8.0 × 109/L, extremely unbalanced with CD3−CD56dim natural killer (NK) cells accounting for 62.0%-83.2% of lymphocytes overall (Figure 1A). Intriguingly, more than 95% of the NK cells simultaneously coexpress inhibitory NKG2A and triggering NKG2C receptors (Figure 1B), although coexpression of these markers is generally observed in only 1%-2% of decidual and peripheral blood NK cells from healthy donors.2-4 However, half of NK cells express HLA-DR, an activation marker, and present an intermediate cell-differentiation phenotype with a high proportion of CD57+ cells and an insignificant quantity of killer cell immunoglobulin–like receptors (KIR) (Figure 1C-D),4 although her genome includes all KIR variants.
Phenotypic and functional features of NK cells. (A) Number of lymphocytes (black line) and CD3−CD56+ NK cells (red line) over the examination period. (B) Flow cytometric analysis of CD3−CD56+ NK cells among lymphocytes and coexpression of NKG2A and NKG2C on NK cells. (C) Expression of inhibitory KIR (KIR2DL1, KIR2DL2/3, and KIR3DL1) on CD3−CD56+ NK cells. (D) Expression of CD27, CD62L, and CD57 cell-differentiation markers on CD3−CD56+ NK cells. Numbers correspond to the proportion of positive cells. (E) Polyfunctionality assays of resting CD3−CD56+ NK cells tested against various targets including K562, 721.221, .221-AEH, and Raji cells in the presence of 1 μg/mL rituximab (E/T ratio of 1/1). Cells were stained with mAbs for CD107a degranulation, and intracellular production of IFN-γ and TNF-α. All data were analyzed with the Boolean gate algorithm of Flow Jo Version 8.8 (TreeStar). Pestle software Version 1.6 was used to remove the background, and pie charts, generated using the Spice software Version 5.2 (NIAI freeware), present the frequency of NK cells positive for 0, 1, 2, or 3 responses (to CD107a, IFN-γ, and TNF-α). Arcs depict the frequency of cells positive for CD107a, IFN-γ, and/or TNF-α, as described.10 (F) Redirect killing assays of CD3−CD56+ NK cells against the P815 cell line in the presence of 5 μg/mL mAbs specific for NKG2A and NKG2C, or the matched isotype control (Ig) (E/T ratio of 1/1). Numbers correspond to the proportion of positive cells. (G) Proliferation of NK cells after stimulation for 7 days with IL-2 and/or irradiated 721.221 or .221 AEH cells measured by cell dilution of CFSE. (H) Proliferation of NK cell after stimulation for 7 days with IL-2 and/or irradiated P815 cells plus anti-NKG2A, anti-NKG2C, or both mAbs, measured by cell dilution of CFSE.
Phenotypic and functional features of NK cells. (A) Number of lymphocytes (black line) and CD3−CD56+ NK cells (red line) over the examination period. (B) Flow cytometric analysis of CD3−CD56+ NK cells among lymphocytes and coexpression of NKG2A and NKG2C on NK cells. (C) Expression of inhibitory KIR (KIR2DL1, KIR2DL2/3, and KIR3DL1) on CD3−CD56+ NK cells. (D) Expression of CD27, CD62L, and CD57 cell-differentiation markers on CD3−CD56+ NK cells. Numbers correspond to the proportion of positive cells. (E) Polyfunctionality assays of resting CD3−CD56+ NK cells tested against various targets including K562, 721.221, .221-AEH, and Raji cells in the presence of 1 μg/mL rituximab (E/T ratio of 1/1). Cells were stained with mAbs for CD107a degranulation, and intracellular production of IFN-γ and TNF-α. All data were analyzed with the Boolean gate algorithm of Flow Jo Version 8.8 (TreeStar). Pestle software Version 1.6 was used to remove the background, and pie charts, generated using the Spice software Version 5.2 (NIAI freeware), present the frequency of NK cells positive for 0, 1, 2, or 3 responses (to CD107a, IFN-γ, and TNF-α). Arcs depict the frequency of cells positive for CD107a, IFN-γ, and/or TNF-α, as described.10 (F) Redirect killing assays of CD3−CD56+ NK cells against the P815 cell line in the presence of 5 μg/mL mAbs specific for NKG2A and NKG2C, or the matched isotype control (Ig) (E/T ratio of 1/1). Numbers correspond to the proportion of positive cells. (G) Proliferation of NK cells after stimulation for 7 days with IL-2 and/or irradiated 721.221 or .221 AEH cells measured by cell dilution of CFSE. (H) Proliferation of NK cell after stimulation for 7 days with IL-2 and/or irradiated P815 cells plus anti-NKG2A, anti-NKG2C, or both mAbs, measured by cell dilution of CFSE.
Consistent with recent studies exploring the relation between NK-cell responsiveness and receptors that engage self-MHC class I molecules,5 polyfunctional analysis showed that this patient's KIR−NKG2A+NKG2C+ NK cells were fully functional against MHC class I–deficient target cells (K562, and 721.221) and capable of antibody-dependent cellular cytotoxicity (Figure 1E).
Inhibitory NKG2A and activating NKG2C receptors are both specific for HLA-E, a nonclassical MHC class Ib molecule expressing peptides derived from signal sequences of other HLA class I molecules.6 Given their opposing functional effect, the coexpression of both markers raises the question of how NK cells can establish self-tolerance under this circumstance, as the lack of symptoms shows they do. A redirected killing assay against FcγR+ P815 targets in the presence of anti-NKG2C mAb showed that the NKG2C was functional, but more interestingly, costimulation of NKG2A and NKG2C with their respective mAbs demonstrated that the inhibitory function of NKG2A prevailed (Figure 1F). The NKG2A+NKG2C+ NK cells were clearly less polyfunctional against targets expressing HLA-E than against wild-type 721.221 cells (Figure 1E), and HLA-E+ target interaction inhibited NK-cell proliferation, except in the presence of IL-2 (Figure 1G), as described.7 A redirect killing assay confirmed that simultaneous engagement of NKG2C and IL-2 induced a marked proliferative response, abrogated by NKG2A engagement (Figure 1H).
We therefore report that the NKG2A inhibitory receptor plays a central inhibitory role, providing a regulatory feedback mechanism for controlling the activating NKG2C receptor, reminiscent of the activation-dependent expression of CTLA-4 on T lymphocytes that counteracts CD28-mediated costimulation.8 These findings are consistent with the higher affinity of NKG2A for HLA-E, which likely favors its competition with NKG2C for ligand engagement.9
To our knowledge, this case provides the first in vivo evidence that, in the absence of KIR, expression of NKG2A on NKG2C+ NK cells establishes an inhibitory threshold that prevents their potential autoreactivity against self-HLA-E+ cells and explains this woman's absence of autoimmune disorders or collateral damage.
In accordance with national ethics committee guidelines, the patient provided written informed consent before peripheral blood samples were collected for this study.
Authorship
Acknowledgments: This study was supported in part by Inserm and the Université Pierre et Marie Curie, Paris, France.
Contribution: V.B. and V.V. performed the research, analyzed the data, and drafted the manuscript; A.A. performed the research; and B.H., D.B., and A.M-K. conducted the clinical study and analyzed the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vincent Vieillard, Inserm UMR-S 945, Immunité et Infection, Hôpital Pitié-Salpêtrière, 83 boulevard de l'Hôpital, 75013 Paris, France; e-mail Vincent.vieillard@upmc.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal