Abstract
Mutated CEBPA defines a subgroup of acute myeloid leukemia (AML). We have previously shown that C/EBPα or its AML mutants synergize with NF-κB p50 to activate antiapoptotic genes, including BCL2 and FLIP. Furthermore, p50 binds and activates the CEBPA gene in myeloid cells. We now report that C/EBPα or C/EBPα leucine zipper AML mutants bind in vivo to the nfkb1 (p50) promoter and induce its expression even in the presence of cycloheximide. Induction of p50 by C/EBPα depends on 2 conserved κB sites in the nfkb1 promoter. C/EBPα did not induce p65 expression. Thus, C/EBPα and p50 reciprocally regulate each other's expression, establishing a positive feedback relationship. Although p50 homodimers inhibit transcription, C/EBPα and p50 synergistically activate antiapoptotic genes. ChIP analysis showed that C/EBPα diminishes the occupation of histone deacetylase 1 (HDAC1) or HDAC3 on the endogenous FLIP promoter but not in mice lacking p50. Coimmunoprecipitation confirmed that C/EBPα, its AML variants, or C/EBPβ disrupt interaction between p50 and HDACs dependent on the C/EBP basic region. These findings suggest that C/EBPs displace HDACs from p50 homodimers bound to antiapoptotic genes, contributing to NF-κB dysregulation in leukemia, and that the C/EBPα:p50 complex is a potential therapeutic target.
Introduction
The transcription factor C/EBPα is essential for early myelopoiesis.1,2 The C/EBP family members have a characteristic structure, consisting of a leucine-rich dimerization domain (leucine zipper; LZ), a basic amino acid–rich DNA binding domain (basic region; BR) and 2 N-terminal trans-activation domains (TADs). All family members share the highly homologous C-terminal bZIP domain. The α-helical LZ mediates homodimerization or heterodimerization,3-5 a prerequisite for DNA binding through the BR.6 The N-terminus contains the less-conserved TADs. Two isoforms of C/EBPα are translated from the same mRNA by ribosomal scanning. The full-length protein is 42 kDa, and initiation of translation from internal AUG results in a truncated C/EBPαp30, lacking one TAD.7-9 Similarly, normal cells express 3 isoforms of the intronless C/EBPβ: LAP1, LAP2, and the truncated liver-inhibitory protein (LIP) isoform that is overexpressed in Hodgkin lymphoma and breast cancer.10,11
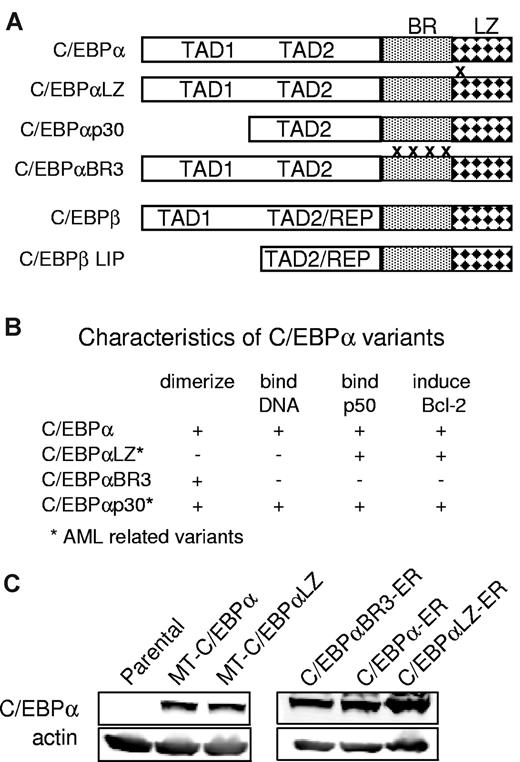
C/EBPα is mutated in ∼ 10% of patients with AML,7-9 defining a distinct clinical subgroup with a unique gene expression profile.12,13 Mutations occur in 2 clusters: changes in the N-terminal region result in the truncated C/EBPαp30 that binds DNA and has a dominant negative effect.9 Mutations in the LZ region are in-frame insertions or deletions that prevent dimerization and thus DNA binding and are designated herein C/EBPαLZ. Typically, both CEBPA alleles are affected, and the human disease can be recapitulated in knockin mice.14,15 Interestingly, C/EBPα null mutations are rare, and the mutated proteins are expressed by the leukemic blasts, suggesting a selective pressure and an active role in leukemogenesis for the mutated C/EBPα proteins. Accordingly, we showed that C/EBPα or its oncoproteins, including C/EBPαLZ that cannot bind DNA directly, protect hematopoietic cells from apoptosis.16,17 Through tethering to NF-κB p50, C/EBPα or its AML mutants activate several antiapoptotic genes, including BCL2 and FLIP, thereby inhibiting both the extrinsic and intrinsic apoptotic pathways.16,17 These effects do not depend on DNA binding but rather on integrity of the BR, explaining its conservation in C/EBPα AMLs. Interaction with p50 is necessary for induction of bcl-2 and FLIP but not of myeloid differentiation genes, such as the gene encoding neutrophil elastase.17 We have also shown that NF-κB p50 regulates C/EBPα expression by directly binding the CEBPA promoter and by synergizing with C/EBPα to activate its expression. Consequently, p50-null mice display a defect in granulopoiesis, both in vitro and in vivo.18
NF-κB is a group of closely related transcription factors that are a key regulator of a rapid-response system enabling cells to adjust their transcriptional program in the face of external stress.19,20 NF-κB regulates numerous genes involved in inflammation, immune response, proliferation, and apoptosis. Five members were described in mammalian cells: Rel A or p65, c-Rel, Rel B, NF-κB1 (p50/p105), and NF-κB2 (p52/p100), all characterized by a 300-aa Rel homology domain that mediates dimerization and DNA binding. Only p65, Rel B, and c-Rel contain a TAD, and dimers lacking an activation domain, such as p50 homodimers, mediate transcriptional repression.21 The p50:p65 heterodimer is the most common form of NF-κB in most cells and is retained in the cytosol by IκB. Canonical activation of NF-κB depends on phosphorylation of IκB followed by its ubiquitination and rapid degradation by the 26S proteasome to release NF-κB dimers, which in turn translocate to the nucleus and bind their target (κB) DNA sequences.22 In unstimulated cells p50 homodimers are present as the primary NF-κB species in the nucleus.21 p50 homodimers bind to DNA and recruit histone deacetylase 1 (HDAC1) or HDAC3 to repress NF-κB–dependent gene expression.23,24 Appropriate stimulation causes nuclear localization of NF-κB complexes containing phosphorylated p65 that displaces the p50-HDAC complexes and associates with CREB binding protein.23,24
Because of the importance of p50 to the antiapoptotic effect of C/EBPα and the regulation of C/EBPα by p50, we hypothesized that conversely C/EBPα regulates the expression of nfkb1, the gene encoding NF-κB p50. The studies described here show that expression of the mRNA encoding p50, but not p65, is directly induced by C/EBPα or its LZ leukemic mutants. This induction is primarily mediated through 2 evolutionary conserved κB sites in the nfkb1 promoter that bind p50 but not C/EBPα. In addition, we provide a mechanism for C/EBP:p50 transcriptional synergy, showing that C/EBPα, its AML mutants, or C/EBPβ displace HDAC1 or HDAC3 from p50 bound to κB sites to induce expression of NF-κB–regulated genes. Because NF-κB p50 homodimers exist in the nucleus of unstimulated cells, these findings identify an alternative, C/EBP-dependent means to activate NF-κB target genes.
Methods
Cell lines
Ba/F3 cells were cultured in RPMI with 10% heat inactivated fetal bovine serum (HI-FBS) and 1 ng/mL IL-3 (PeproTech).25 Clones expressing C/EBPα or the human AML-derived C/EBPαLZ mutant under the regulation of the zinc inducible metallothionein (MT) promoter were described as were Ba/F3 cells expressing fusion of C/EBPα, C/EBPαLZ, or C/EBPαBR3 to the ligand binding domain of the estrogen receptor (ER).16,17 Schematic representation of the relevant C/EBPα variants and their characteristics are presented in Figure 1A and B. Expression from the MT promoter was induced by culturing cells with 100μM zinc chloride, and the ER fusion proteins were activated by estradiol (E2) at 1μM, using ethanol as a vehicle control. U937 cells were cultured in RPMI with 10% HI-FBS, 293T cells were maintained in DMEM with 10% HI-FBS, and NIH-3T3 cells were grown in DMEM with 10% heat inactivated calf serum. The translation inhibitor cycloheximide was added 30 minutes before E2 to a final concentration of 50 μg/mL. C/EBPα expression was knocked down with the use of a human pLKO.1 lentiviral shRNA target gene set containing 4 lentiviral CEBPA shRNA constructs (RHS4533; Open Biosystems). Lentiviruses were generated by cotransfection with packaging plasmid in 293T cells according to the manufacturer's protocol. Cells were transduced in 12-well dishes in the presence of polybrene (4 μg/mL). Selection with puromycin (2 μg/mL) was started 48 hours after lentiviral transduction.
Chromatin immunoprecipitation
Seven to 10 million Ba/F3 or mouse spleen cells were used in each ChIP reaction as previously described,17,18 using antisera against C/EBPα, C/EBPβ, NF-κB p50, rabbit IgG (Santa Cruz Biotechnology), HDAC1, or HDAC3 (Abcam). Precipitation of DNA fragments corresponding to the promoters of interest was detected by PCR. Enrichment of each promoter of interest was analyzed with 2 sets of primers for the relevant (proximal) C/EBP binding site and an upstream negative control primer. The primer sequences are presented in Table 1.
Primers used for ChiP and RNA analysis
| Gene . | Species . | Forward sequence . | Reverse sequence . |
|---|---|---|---|
| ChIP | |||
| NFKB1 proximal | Human | GACGTCAGTGGGAATTTCC | GTGGCGAAACCTCCTCTTC |
| NFKB1 upstream | Human | TGCTGCATGGAATTACATCC | CATCTTAACTTGATAACATTTGC |
| Nfkb1 proximal 1 | Mouse | GTCCGTCCGTCTGTCTGCTC | CTCCTCTTCCTGCCCGGC |
| Nfkb1 proximal 2 | Mouse | CTCGACGTCAGTGGGAATTT | GCGAAACCTCCTCTTCCTG |
| Nfkb1 upstream | Mouse | CTTCATAAGTGGAACGCTGAG | GGTCTCACTACTGAGTTCAAG |
| Flip proximal 1 | Mouse | ATGGCCTCGATTCAGTTTTG | GATAAGGACGTCCCCAGAGT |
| Flip proximal 2 | Mouse | CTGTAACTTTCCTCCGCCCT | GAGAAGGGCTCCGAGAAGAT |
| Flip upstream | Mouse | CAAGGAGGTCGCTACGAGTC | AGCCAGCAAAGCCAGTAAAG |
| Reverse transcriptase PCR | |||
| Nfkb1 | Mouse | GGTATGCACCGTAACAGCAG | GCTTCCCTCTGTCATCCGT |
| RelA | Mouse | CTCATCCACATGAACTTGTGG | GCTTCTTCACACACTGGATC |
| mS16 | Mouse | CTTGGAGGCTTCATCCACAT | ATATTCGGGTCCGTGTGAAG |
| NFKB1 | Human | CTGTGGTTGAAATACTTCTGG | CAGAATGGCAGAAGATGATC |
| FLIP | Human | GGATAAATCTGATGTGTCCTC | ACTCAACCACAAGGTCCAAG |
| CEBPA | Human | AAGAAGTCGGTGGACAAGAACAG | GCGGTCATTGTCACTGGTCA |
| ABL | Human | AACACCCTAACCTGGTGCAG | CAAGTGGTTCTCCCCTACCA |
| GAPDH | Human | CCACCCATGGCAAATTCC | GATGGGATTTCCATTGATGACA |
| Actin | Human | TGCGTGACATTAAGGAGAAG | GCTCGTAGCTCTTCTCCA |
| Gene . | Species . | Forward sequence . | Reverse sequence . |
|---|---|---|---|
| ChIP | |||
| NFKB1 proximal | Human | GACGTCAGTGGGAATTTCC | GTGGCGAAACCTCCTCTTC |
| NFKB1 upstream | Human | TGCTGCATGGAATTACATCC | CATCTTAACTTGATAACATTTGC |
| Nfkb1 proximal 1 | Mouse | GTCCGTCCGTCTGTCTGCTC | CTCCTCTTCCTGCCCGGC |
| Nfkb1 proximal 2 | Mouse | CTCGACGTCAGTGGGAATTT | GCGAAACCTCCTCTTCCTG |
| Nfkb1 upstream | Mouse | CTTCATAAGTGGAACGCTGAG | GGTCTCACTACTGAGTTCAAG |
| Flip proximal 1 | Mouse | ATGGCCTCGATTCAGTTTTG | GATAAGGACGTCCCCAGAGT |
| Flip proximal 2 | Mouse | CTGTAACTTTCCTCCGCCCT | GAGAAGGGCTCCGAGAAGAT |
| Flip upstream | Mouse | CAAGGAGGTCGCTACGAGTC | AGCCAGCAAAGCCAGTAAAG |
| Reverse transcriptase PCR | |||
| Nfkb1 | Mouse | GGTATGCACCGTAACAGCAG | GCTTCCCTCTGTCATCCGT |
| RelA | Mouse | CTCATCCACATGAACTTGTGG | GCTTCTTCACACACTGGATC |
| mS16 | Mouse | CTTGGAGGCTTCATCCACAT | ATATTCGGGTCCGTGTGAAG |
| NFKB1 | Human | CTGTGGTTGAAATACTTCTGG | CAGAATGGCAGAAGATGATC |
| FLIP | Human | GGATAAATCTGATGTGTCCTC | ACTCAACCACAAGGTCCAAG |
| CEBPA | Human | AAGAAGTCGGTGGACAAGAACAG | GCGGTCATTGTCACTGGTCA |
| ABL | Human | AACACCCTAACCTGGTGCAG | CAAGTGGTTCTCCCCTACCA |
| GAPDH | Human | CCACCCATGGCAAATTCC | GATGGGATTTCCATTGATGACA |
| Actin | Human | TGCGTGACATTAAGGAGAAG | GCTCGTAGCTCTTCTCCA |
Reverse transcriptase and quantitative PCR
Total cellular RNAs were extracted (Nucleospin RNAII, Macherey-Nagel; Clontech Laboratories), and first-strand cDNA was synthesized from 1 μg of total RNA (ImProm II Reverse Transcriptase System; Promega) with the use of random hexamers as primers. Quantitative real-time PCR was performed with the use of the iQ SYBR Green Supermix and the iCycler iQ Real Time PCR Detection System (Bio-Rad Laboratories). Each sample was assayed in triplicate, and each experiment was repeated ≥ 3 times. Amplification of the endogenous mouse large ribosomal subunit (mS16), or human GAPDH gene transcripts was used as a reference to standardize between samples. Standard curves were constructed to ensure high amplification efficiencies and comparability across experiments. The comparative Ct method was used for quantification. Triplicate Ct values were generated for all assays, and 2−ΔΔCt values were then calculated. Fold expression was calculated as the ratio of normalized level of each target gene in the experimental and the control samples. The primer sequences are presented in Table 1.
Gel shift assay
Nuclear extracts were obtained from transiently transfected 293T cells and subjected to gel shift analysis as previously described.16 The sequences of the sense oligonucleotide probes with 4-bp overhang and mutations underlined were neutrophil elastase, 5′-TCGAGGCCAGGATGGGGCAATACAACCCG; κB1, 5′-GTACCGACGTCAGTGGGAATTTCCAGCCAGGAAG; mut-κB1, 5′-GTACCGACGTCAGTGCTAGCTTCCAGCCAGGAAG; κB2, 5′-GTACTCGGGGCGCGCGGGCTTCCCCCACCCCCGGA; and mut-κB2: 5′-GTACTCGGGGCGCGCGCTAGCCCCCACCCCCGGA.
Luciferase reporter assays
With the use of PCR, a DNA fragment from −1750 to +37 bp relative to the transcription initiation site (Ensembl database) of the murine nfkb1 gene was cloned as an MluI/XhoI fragment into the pGL3-luciferase reporter vector (Promega), to generate nfkb1-1750-Luc. PCR subfragment cloning was performed to create the following deletions of the nfkb1 promoter: nfkb1–912-Luc, –528-Luc, and –257-Luc, starting at −912, −528, and −257 bp, respectively. PCR mutagenesis was used to alter κB1 and κB2 sites to the sequences indicated above. All plasmid sequences were confirmed by DNA sequencing. The κBx2-Luc plasmid, containing 2 adjacent κB sites next to a minimal mouse α-actin core promoter linked to a luciferase reporter, was previously described,26 as was (C/EBP)2TK-Luc.27 NIH-3T3 cells seeded in 60-mm dishes were transfected with Lipofectamine 2000 (Invitrogen). Each plate was cotransfected with 1 μg of the reporter plasmid, 5 ng of CMV-β-Gal internal control, and 100 ng of CMV, CMV-C/EBP, or CMV-NF-κB plasmids. HeLa cells were seeded in 12-well dish and were cotransfected with FuGENE 6 (Promega) with 125 ng of reporter plasmid, 2 ng of CMV-β-Gal, and 5 ng of CMV, CMV-C/EBPβ, or CMV-C/EBPβ LIP. Cells were assayed for luciferase and β-galactosidase activity 2 days after transfection, and fold activation relative to the empty CMV vector after correction for β-galactosidase activity was determined. Trichostatin A (TSA) was kept as a 5-mM stock solution in DMSO and was added 24 hours after transduction to a final concentration of 250nM.
Mice
H2K-C/EBPα-Eμ transgenic (αTG) mice were previously described16,17 and were bred with nfkb1−/−28 (lacking p50) or wild-type (WT) B6;129PF1 strain-matched control mice (The Jackson Laboratory) to generate WT; WT;αTG, p50−/−, and p50−/−;αTG mice. Mice genotype was determined by PCR of tail DNA as described.17 Single-cell suspensions of BM or splenocytes were obtained with the use of a 70-μm cell strainer, and red cells were lysed with NH4Cl buffer. All animal experiments were approved by the Johns Hopkins University Institutional Animal Care and Use Committee.
Coimmunoprecipitation and Western blotting
293T cells were seeded in 10-cm dishes and cotransfected with Lipofectamine 2000 with combinations of 1.5 μg of CMV–FLAG-tagged HDAC1, 750 ng of CMV–FLAG-tagged HDAC3, 1 μg of CMV–NF-κB p50, and 3 μg of CMV-C/EBP. Two days after transfection cells were harvested in RIPA buffer (150mM NaCl, 50mM Tris, pH 7.4, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1mM EDTA) with 1mM PMSF and Protease Inhibitor Cocktail (Sigma), briefly sonicated, and spun down for 10 minutes at 13 000g. Supernatants were precleared with protein A/G-Sepharose beads and subsequently incubated with 30 μL of anti-FLAG M2 beads (Sigma-Aldrich) for 3 hours at 4°C. The beads were washed 4 times in RIPA buffer, and proteins were eluted with 1× Laemmli sample buffer at 95°C for 5 minutes. For endogenous coimmunoprecipitation, 20 million Ba/F3 cells were harvested in RIPA buffer. After preclearing, the lysates were equally divided and incubated for 3 hours with primary antibody. Immune complexes were collected by incubation with protein A/G-Sepharose beads for 1 hour, and samples were eluted in 1× Laemmli sample buffer. In all coimmunoprecipitation experiments, a 5% input sample was obtained, and 20% of this was loaded for Western blot analysis. Immunoprecipitated proteins from 30% of the total cell lysates are loaded in the coimmunoprecipitation lanes.
Protein samples were subjected to Western blotting as described,16 using the following antibodies: C/EBPα (sc9314), C/EBPβ (sc150), p50 (sc8414) (Santa Cruz Biotechnology), β-actin (AC15), FLAG (F7425) (Sigma-Aldrich), HDAC1 (ab7028), HDAC3 (ab7030) (Abcam), acetylated histone H3 (06–599), and acetylated histone H4(06–866) (Millipore). The membranes were visualized with the Odyssey infrared imaging system (LI-COR Biosciences).
Statistical analysis
Quantitative data are presented as mean ± SEM from ≥ 3 independent repetitions. Statistical comparisons between groups were carried out with the use of 2-tailed Student t test. P values of < .05 were considered significant.
Results
C/EBPα or C/EBPα myeloid oncoproteins induce NF-κB p50 expression
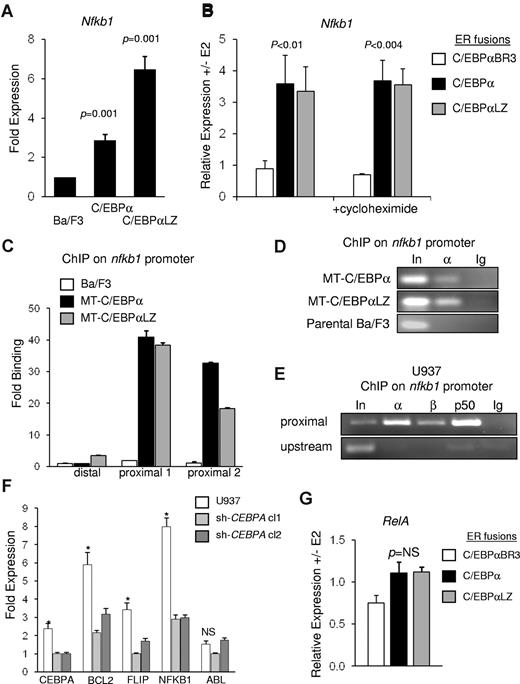
We previously demonstrated that direct interaction with NF-κB p50 is required for induction of bcl-2 and FLIP by C/EBPα.17 In addition, NF-κB p50 directly binds and induces the CEBPA gene.18 To study induction of NF-κB p50 by C/EBPα we used Ba/F3 cell lines expressing C/EBPα or a patient-derived C/EBPαLZ leukemic variant under control of the zinc-inducible MT promoter. C/EBPα is not expressed in parental Ba/F3 cells, and activation of MT-C/EBPα or MT-C/EBPαLZ resulted in similar expression (Figure 1C). Parental Ba/F3 cells and MT-C/EBPα or MT-C/EBPαLZ subclones were cultured with zinc chloride for 16 hours followed by extraction of total cellular RNA. Normalized expression of NF-κB p50 was analyzed with quantitative reverse transcriptase PCR. Compared with parental cells, C/EBPα or C/EBPαLZ significantly induced p50 expression by 2.8- or 6.4-fold, respectively (Figure 2A), on average from 3 independent repetitions. To validate direct induction by C/EBPα, we used Ba/F3 cells expressing equal amounts (Figure 1C) of C/EBPα-ER, C/EBPαLZ-ER, or the C/EBPαBR3-ER BR mutant that cannot bind DNA, interact with NF-κB p50, or induce antiapoptotic genes. Activation with E2 allows translocation of the C/EBPα isoforms into the nucleus and binding to DNA. Total cellular RNA was extracted 7 hours after adding E2 or ethanol vehicle, and expression of p50 was assessed by quantitative reverse transcriptase PCR. Compared with vehicle-treated cells, C/EBPα or C/EBPαLZ induced p50 RNA expression 3.5-fold, whereas C/EBPαBR3 was not effective (Figure 2B). Addition of the translation inhibitor cycloheximide before adding E2 did not change p50 induction (Figure 2B), suggesting direct activation of the nfkb1 gene by C/EBPα or its C/EBPαLZ leukemic variant.
Diagram properties and expression of C/EBPα proteins. (A) Diagram depicting C/EBPα (p42), the location of C/EBPαLZ in-frame duplications and deletions, truncated C/EBPαp30, the basic region mutant C/EBPαBR3, and C/EBPβ or its LIP isoform. BR indicates basic region; LZ, leucine zipper; TAD, trans-activation domain; REP, repression domain. (B) Properties of the C/EBP variants used. (C) Total cellular proteins extracted from parental Ba/F3 cells or clones expressing the indicated C/EBPα isoforms were subjected to Western blotting with the use of C/EBPα or actin antisera.
Diagram properties and expression of C/EBPα proteins. (A) Diagram depicting C/EBPα (p42), the location of C/EBPαLZ in-frame duplications and deletions, truncated C/EBPαp30, the basic region mutant C/EBPαBR3, and C/EBPβ or its LIP isoform. BR indicates basic region; LZ, leucine zipper; TAD, trans-activation domain; REP, repression domain. (B) Properties of the C/EBP variants used. (C) Total cellular proteins extracted from parental Ba/F3 cells or clones expressing the indicated C/EBPα isoforms were subjected to Western blotting with the use of C/EBPα or actin antisera.
C/EBPα directly regulates nfkb1 gene expression. (A) Parental Ba/F3 cells and clones expressing MT-C/EBPα or MT-C/EBPαLZ were cultured with zinc chloride, and RNA was extracted after 16 hours. p50 transcripts were measured and normalized to mS16 levels with the use of quantitative reverse transcriptase PCR. Mean and SE from 3 independent experiments are shown. (B) Ba/F3 cells carrying the indicated C/EBPα-ER variant were cultured with (+) or without (-) estradiol (E2) for 7 hours, and normalized p50 mRNA levels were assessed by quantitative reverse transcriptase PCR. p50 transcripts levels were similarly assessed in the presence of cycloheximide. The average of RNA ratios with or without E2 from 3 independent experiments is shown. (C) Parental Ba/F3 cells and clones expressing MT-C/EBPα or MT-C/EBPαLZ were cultured with zinc chloride for 16 hours and subjected to ChIP analysis. After immunoprecipitation with C/EBPα antiserum, enrichment of DNA fragments corresponding to the proximal or the distal regions of the nfkb1 promoter were determined with quantitative real-time PCR relative to input. Averages from 3 repetitions are presented. (D) A representative gel of ChIP of C/EBPα of C/EBPαLZ on the nfkb1 promoter is shown. (E) U937 lysates were subjected to ChIP analysis with the use of antiserum against C/EBPα (α) C/EBPβ (β), NF-κB p50 (p50), or normal rabbit IgG (Ig). PCR was used to amplify a DNA fragment centered at 20 bp or an upstream fragment starting at −2284 bp of the NFKB1 gene. (F) U937 cells were transduced with vectors expressing shRNA against C/EBPα. Total cellular RNA was extracted from parental cells and from 2 clones expressing different shRNAs that resulted in effective knockdown of CEBPA. Expression of the indicated genes was measured with quantitative real-time PCR. *P ≤ .001 for the comparison between parental cells and either of the clones expressing an shRNA targeting CEBPA. (G) mRNA levels of NF-κB p65 were evaluated in Ba/F3 cells expressing ER fusions of C/EBPα or its indicated variants, 7 hours after induction with E2. The average normalized transcript level ratios with or without E2 from 3 independent repetitions are shown.
C/EBPα directly regulates nfkb1 gene expression. (A) Parental Ba/F3 cells and clones expressing MT-C/EBPα or MT-C/EBPαLZ were cultured with zinc chloride, and RNA was extracted after 16 hours. p50 transcripts were measured and normalized to mS16 levels with the use of quantitative reverse transcriptase PCR. Mean and SE from 3 independent experiments are shown. (B) Ba/F3 cells carrying the indicated C/EBPα-ER variant were cultured with (+) or without (-) estradiol (E2) for 7 hours, and normalized p50 mRNA levels were assessed by quantitative reverse transcriptase PCR. p50 transcripts levels were similarly assessed in the presence of cycloheximide. The average of RNA ratios with or without E2 from 3 independent experiments is shown. (C) Parental Ba/F3 cells and clones expressing MT-C/EBPα or MT-C/EBPαLZ were cultured with zinc chloride for 16 hours and subjected to ChIP analysis. After immunoprecipitation with C/EBPα antiserum, enrichment of DNA fragments corresponding to the proximal or the distal regions of the nfkb1 promoter were determined with quantitative real-time PCR relative to input. Averages from 3 repetitions are presented. (D) A representative gel of ChIP of C/EBPα of C/EBPαLZ on the nfkb1 promoter is shown. (E) U937 lysates were subjected to ChIP analysis with the use of antiserum against C/EBPα (α) C/EBPβ (β), NF-κB p50 (p50), or normal rabbit IgG (Ig). PCR was used to amplify a DNA fragment centered at 20 bp or an upstream fragment starting at −2284 bp of the NFKB1 gene. (F) U937 cells were transduced with vectors expressing shRNA against C/EBPα. Total cellular RNA was extracted from parental cells and from 2 clones expressing different shRNAs that resulted in effective knockdown of CEBPA. Expression of the indicated genes was measured with quantitative real-time PCR. *P ≤ .001 for the comparison between parental cells and either of the clones expressing an shRNA targeting CEBPA. (G) mRNA levels of NF-κB p65 were evaluated in Ba/F3 cells expressing ER fusions of C/EBPα or its indicated variants, 7 hours after induction with E2. The average normalized transcript level ratios with or without E2 from 3 independent repetitions are shown.
To assess binding of these proteins to the endogenous nfkb1 promoter while avoiding interference by the ER fragment, parental Ba/F3, MT-C/EBPα, or MT-C/EBPαLZ cells were cultured with zinc for 16 hours and subjected to ChIP analysis with the use of C/EBPα antisera or rabbit IgG. Precipitated DNA was subjected to PCR amplifying a fragment spanning from −4 to 120 bp, using 2 primer sets, or an upstream fragment located at −810 bp. Both C/EBPα and the C/EBPαLZ variant bound the proximal promoter region of nfkb1 but not to the distal region (Figure 2C-D). Of note, C/EBPαLZ interacts with the promoter despite its inability to bind DNA directly.
U937 is a human AML-derived cell line expressing C/EBPα, C/EBPβ, and p50.16 U937 cell lysates were subjected to ChIP with the use of antibodies against C/EBPα, C/EBPβ, p50, or normal rabbit IgG, followed by PCR amplification of a DNA fragment centered at 21 bp of the human NFKB1 promoter or an upstream fragment centered at −2284 bp (Figure 2E). Endogenous C/EBPα, C/EBPβ, and p50 bound the proximal segment of the NFKB1 promoter but not to the upstream element. U937 cells were transduced with lentiviral vectors expressing 2 different shRNAs targeting CEBPA, resulting in reduced expression by ∼ 60%. As a consequence, the endogenous expression of NFKB1 and the previously described C/EBPα antiapoptotic targets FLIP or BCL2 were significantly diminished. In contrast, expression of the mRNA encoding the ABL tyrosine kinase was not affected by knockdown of CEBPA (Figure 2F).
The most common species of NF-κB is the p50:p65 heterodimer. To assess p65 induction, C/EBPα-ER or its variants were induced with E2, and total cellular RNA was extracted after 7 hours. Remarkably, in contrast to p50, NF-κB p65 RNA was not significantly induced by C/EBPα (Figure 2G). Together, these findings indicate that C/EBPα or the C/EBPαLZ variant bind the nfkb1 promoter and directly induce its expression and that C/EBPα regulates NFKB1 in human AML cells.
C/EBPα or C/EBPαLZ oncoproteins activate the nfkb1 promoter dependent on 2 NF-κB sites
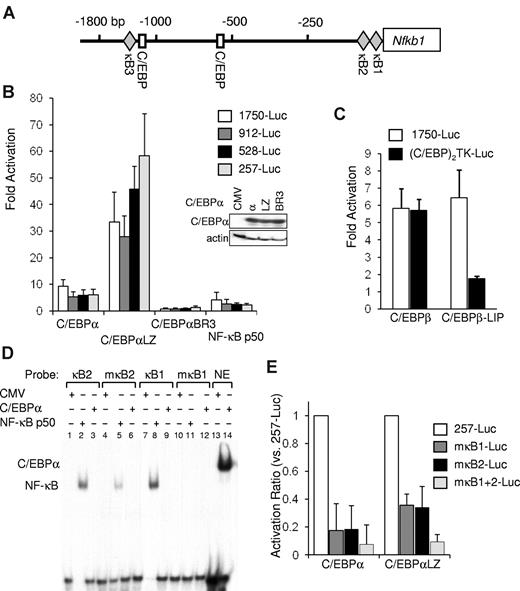
The murine nfkb1 promoter contains 2 proximal κB consensus sites, a C/EBP consensus site at −553 bp and adjacent C/EBP and κB consensus sites at −1347 and −1375 bp, respectively (Figure 3A). The sequences of the 2 proximal κB sites are 100% conserved between mouse and human. In contrast, sequence comparison shows that the 85-bp region between these sites is only 85% conserved, and the identity of the flanking sequences is < 70% (not shown). Nfkb1–1750-Luc, containing −1735/37 bp of the murine nfkb1 promoter linked to a luciferase reporter, was cotransfected with empty CMV vector or with CMV expression plasmids encoding C/EBPα, C/EBPαLZ, C/EBPαBR3, or NF-κB p50, as well as CMV–β-Gal as an internal control. Equal expression of the C/EBPα variants was confirmed by Western blotting (Figure 3B). Relative to the empty CMV vector, nfkb1–1750-Luc was activated 9-fold by C/EBPα and 32-fold by the C/EBPαLZ mutant but not by the C/EBPαBR3 BR variant (Figure 3B). NF-κB p50 minimally activated its own promoter. We next generated a truncation series of this construct designated nfkb1–912-Luc, –528-Luc, or –257-Luc. C/EBPα or its LZ mutant similarly activated each of these constructs, including the nfkb1–257-Luc promoter fragment that does not contain a C/EBP consensus site (Figure 3B).
C/EBPα regulates nfkb1 gene expression via 2 conserved κB sites. (A) Diagram of the murine nfkb1 promoter, which lacks a TATAA box. Locations of the indicated transcription factor binding sites are marked relative to the transcription initiation site. (B) NIH-3T3 cells were transiently cotransfected with 1 μg of nfkb1–1750-Luc or its truncated variants nfkb1–912, –528-Luc, or –257-Luc, 5 ng of CMV-β-Gal, and 100 ng of CMV, CMV-C/EBPα variants, or CMV-NF-κB p50. Fold activation of the full-length or truncated promoters relative to empty CMV plasmid was determined after adjustment for β-galactosidase activity. The averages from 3 independent experiments are shown. Expression level of the various C/EBPα variants was determined by Western blotting. (C) Fold activation of the nfkb1-Luc or (C/EBP)2TK-Luc reporter by C/EBPβ or its LIP isoform, relative to empty CMV was determined by cotransfection in HeLa cells as described earlier. Luciferase and β-galactosidase activity was measured 48 hours after transfection, and the average corrected activity from 3 experiments is presented. (D) 293T cells were transiently transfected with 3 μg of CMV-C/EBPα, CMV-p50, or empty CMV vector as indicated. Nuclear extracts were subjected to gel shift analysis with the use of oligonucleotides corresponding to the κB sites from the nfkb1 promoter (κB1 or κB2), their corresponding mutants (mκB1 or mκB2), or a consensus C/EBP-binding site from the neutrophil elastase (NE) promoter. (E) NIH-3T3 cells were cotransfected with 1 μg of the nfkb1–257-Luc reporter or its variants carrying clustered point mutations of either one or both κB sites in the nfkb1 promoter (mκB1-, mκB2-, or mκB1+2-Luc, respectively) and with 100 ng of CMV, CMV-C/EBPα, or CMV-C/EBPαLZ and 5 ng of CMV-β-Gal. Corrected activation by the indicated C/EBPα isoform relative to empty CMV was determined, and the average activation ratio of each mutant relative to the WT reporter from 3 experiments is presented.
C/EBPα regulates nfkb1 gene expression via 2 conserved κB sites. (A) Diagram of the murine nfkb1 promoter, which lacks a TATAA box. Locations of the indicated transcription factor binding sites are marked relative to the transcription initiation site. (B) NIH-3T3 cells were transiently cotransfected with 1 μg of nfkb1–1750-Luc or its truncated variants nfkb1–912, –528-Luc, or –257-Luc, 5 ng of CMV-β-Gal, and 100 ng of CMV, CMV-C/EBPα variants, or CMV-NF-κB p50. Fold activation of the full-length or truncated promoters relative to empty CMV plasmid was determined after adjustment for β-galactosidase activity. The averages from 3 independent experiments are shown. Expression level of the various C/EBPα variants was determined by Western blotting. (C) Fold activation of the nfkb1-Luc or (C/EBP)2TK-Luc reporter by C/EBPβ or its LIP isoform, relative to empty CMV was determined by cotransfection in HeLa cells as described earlier. Luciferase and β-galactosidase activity was measured 48 hours after transfection, and the average corrected activity from 3 experiments is presented. (D) 293T cells were transiently transfected with 3 μg of CMV-C/EBPα, CMV-p50, or empty CMV vector as indicated. Nuclear extracts were subjected to gel shift analysis with the use of oligonucleotides corresponding to the κB sites from the nfkb1 promoter (κB1 or κB2), their corresponding mutants (mκB1 or mκB2), or a consensus C/EBP-binding site from the neutrophil elastase (NE) promoter. (E) NIH-3T3 cells were cotransfected with 1 μg of the nfkb1–257-Luc reporter or its variants carrying clustered point mutations of either one or both κB sites in the nfkb1 promoter (mκB1-, mκB2-, or mκB1+2-Luc, respectively) and with 100 ng of CMV, CMV-C/EBPα, or CMV-C/EBPαLZ and 5 ng of CMV-β-Gal. Corrected activation by the indicated C/EBPα isoform relative to empty CMV was determined, and the average activation ratio of each mutant relative to the WT reporter from 3 experiments is presented.
C/EBPβ is expressed as a full-length protein or as the truncated LIP isoform capable of having a dominant-negative effect by dimerizing with full-length C/EBPs. C/EBPβ or the LIP isoform both activated nfkb1–1750-Luc (Figure 3C). In contrast, the (C/EBP)2TK-Luc reporter, carrying 2 C/EBP binding sites, was activated by C/EBPβ but not by C/EBPβ LIP (Figure 3D). These data suggest indirect activation of the nfkb1 promoter independent of C/EBPβ or LIP DNA binding, but instead by interaction with DNA-bound NF-κB. Although LIP itself lacks a TAD, it can apparently cooperate with NF-κB to induce nfkb1 promoter activity.
We further defined the role of the 2 proximal κB sites in C/EBP activation of the nfkb1 promoter. 293T cells were transfected with an empty vector or with plasmids expressing CMV–NF-κB p50 or C/EBPα. Nuclear extracts were obtained, and gel shift analysis showed strong binding of NF-κB p50 but not C/EBPα to labeled oligonucleotides whose sequence corresponds to the κB1 or κB2 sites of the nfkb1 promoter (Figure 3D lanes 2, 3, 8, and 9). Introduction of clustered point mutations into the κB sites resulted in significantly diminished binding of p50 to oligonucleotides carrying mκB1 or mκB2 (Figure 3D lanes 5 and 11). C/EBPα avidly binds a C/EBP consensus oligonucleotide from the neutrophil elastase promoter (Figure 3D lane 14) but neither of the κB oligonucleotides. Using site-directed mutagenesis we generated variants of nfkb1–257-Luc carrying mutations in the κB1 or κB2 sites or in both sites. We studied activation of these reporter variants by C/EBPα or C/EBPαLZ relative to an empty CMV vector. Disruption of either κB site abrogated 65%-80% of reporter activation, and mutating both sites resulted in reduction of activation by either C/EBPα or C/EBPαLZ to < 10% (Figure 3E). Together, these data indicate that C/EBPα or its LZ AML variant that cannot bind DNA activate the nfkb1 promoter by interaction with NF-κB bound to 2 proximal, highly conserved κB sites.
C/EBPα or a C/EBPαLZ oncoprotein displaces endogenous HDAC from target promoters
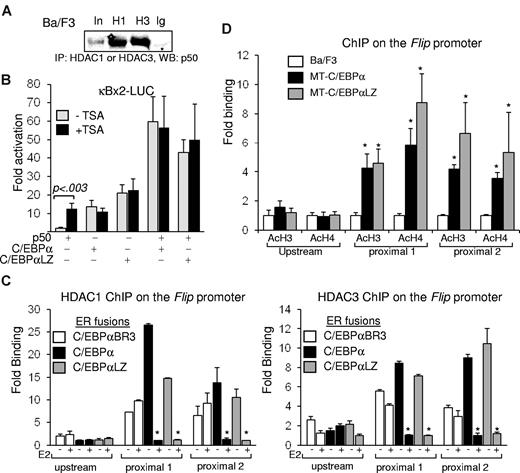
NF-κB p65 is held in the cytoplasm by IκBα and on stimulation enters the nucleus to bind DNA, predominantly as p65:p50 heterodimers. In contrast, p50:p50 homodimers are present in the nucleus bound to chromatin in resting cells and inhibit transcription because of lack of a TAD and recruitment of corepressors, including HDACs.21,23,24,29 Indeed, resting Ba/F3 lysates subjected to coimmunoprecipitation with the use of HDAC1 or HDAC3 antisera showed association between endogenous NF-κB p50 and HDAC1 or HDAC3 (Figure 4A). Therefore, induction of p50 transcription by C/EBPα would be expected to result in repression of target genes. However, our data indicate that C/EBPα and p50 synergistically activate antiapoptotic genes.16,17 We therefore hypothesized that C/EBPα modulates recruitment of corepressors by p50. NIH-3T3 cells were transiently transfected with κBx2-Luc, containing a luciferase reporter under the regulation of 2 κB binding sites, and combinations of vectors expressing p50, C/EBPα, C/EBPαLZ, or empty vector. Luciferase activity relative to the empty CMV vector, and after normalization to β-galactosidase expression, was determined in the presence or absence of the pan HDAC inhibitor TSA. Although p50 minimally activates κBx2-Luc, the addition of TSA allows p50 to activate this reporter > 12-fold. In contrast, TSA did not have a significant effect on activation by C/EBPα, its LZ mutant, or by p50 combined with these C/EBPα isoforms, suggesting that in the presence of C/EBPα, p50 does not recruit HDACs; therefore, TSA has no additional effect (Figure 4B).
C/EBP displaces HDACs from NF-κB p50. (A) Ba/F3 cell lysates were subjected to immunoprecipitation (IP) with the indicated antisera, and coprecipitated NF-κB p50 was detected by Western blotting (WB). (B) NIH-3T3 cells were transiently cotransfected in duplicate with 1 μg of κB×2-Luc, 5 ng of CMV–β-Gal, and the indicated combinations of CMV, CMV-p50, CMV-C/EBPα, or CMV-C/EBPαLZ. TSA or vehicle was added 24 hours after transfection to one set, and luciferase and β-galactosidase activities were determined after an additional 24 hours. Corrected fold activation relative to empty CMV was calculated, and an average from 3 experiments is presented. (C) Ba/F3 cells carrying the indicated ER fusions of C/EBPα variants were cultured without (-) or with (+) estradiol (E2) for 5 hours and subjected to ChIP analysis with the use of antisera for HDAC1 (left) or HDAC3 (right). Enrichment of a DNA fragment corresponding to the proximal or the distal regions of the Flip promoter, determined with quantitative real-time PCR relative to input DNA, in 3 independent ChIP experiments is shown. *P < .001 for the comparison between without (-) and with (+) E2. (D) Parental Ba/F3 cells or clones carrying MT-C/EBPα were cultured with zinc for 16 hours and subjected to ChIP analysis with the use of antisera for C/EBPα (α), HDAC1 (H1), HDAC3 (H3), or normal rabbit IgG (Ig). PCR was used to detect the precipitated FLIP promoter. Endogenous expression of HDAC1 or HDAC3 was determined by Western blotting in similarly treated cells. (E) The indicated Ba/F3 cell clones were cultured with zinc for 16 hours, and chromatin DNA was immunoprecipitated with antibodies against acetyl-histone H3 (AcH3) or acetyl-histone H4 (AcH4). The precipitation of DNA fragments corresponding to the proximal or distal regions of the Flip promoter was determined with the use of quantitative real-time PCR relative to input DNA, and the average from 3 experiments is presented. *P < .01 for the comparison between parental Ba/F3 and cells expressing MT-C/EBPα or MT-C/EBPαLZ.
C/EBP displaces HDACs from NF-κB p50. (A) Ba/F3 cell lysates were subjected to immunoprecipitation (IP) with the indicated antisera, and coprecipitated NF-κB p50 was detected by Western blotting (WB). (B) NIH-3T3 cells were transiently cotransfected in duplicate with 1 μg of κB×2-Luc, 5 ng of CMV–β-Gal, and the indicated combinations of CMV, CMV-p50, CMV-C/EBPα, or CMV-C/EBPαLZ. TSA or vehicle was added 24 hours after transfection to one set, and luciferase and β-galactosidase activities were determined after an additional 24 hours. Corrected fold activation relative to empty CMV was calculated, and an average from 3 experiments is presented. (C) Ba/F3 cells carrying the indicated ER fusions of C/EBPα variants were cultured without (-) or with (+) estradiol (E2) for 5 hours and subjected to ChIP analysis with the use of antisera for HDAC1 (left) or HDAC3 (right). Enrichment of a DNA fragment corresponding to the proximal or the distal regions of the Flip promoter, determined with quantitative real-time PCR relative to input DNA, in 3 independent ChIP experiments is shown. *P < .001 for the comparison between without (-) and with (+) E2. (D) Parental Ba/F3 cells or clones carrying MT-C/EBPα were cultured with zinc for 16 hours and subjected to ChIP analysis with the use of antisera for C/EBPα (α), HDAC1 (H1), HDAC3 (H3), or normal rabbit IgG (Ig). PCR was used to detect the precipitated FLIP promoter. Endogenous expression of HDAC1 or HDAC3 was determined by Western blotting in similarly treated cells. (E) The indicated Ba/F3 cell clones were cultured with zinc for 16 hours, and chromatin DNA was immunoprecipitated with antibodies against acetyl-histone H3 (AcH3) or acetyl-histone H4 (AcH4). The precipitation of DNA fragments corresponding to the proximal or distal regions of the Flip promoter was determined with the use of quantitative real-time PCR relative to input DNA, and the average from 3 experiments is presented. *P < .01 for the comparison between parental Ba/F3 and cells expressing MT-C/EBPα or MT-C/EBPαLZ.
To define the direct effect of C/EBPα on HDAC recruitment to the FLIP promoter, Ba/F3 cells carrying ER fusions of C/EBPα, C/EBPαLZ, or C/EBPαBR3 were cultured with or without E2 for 5 hours and subsequently subjected to ChIP analysis with the use of HDAC1 or HDAC3 antisera. Precipitation of a DNA fragment corresponding to the FLIP promoter, relative to input, was measured by quantitative real-time PCR. Each quantitative analysis included 2 separate primer sets amplifying DNA fragments within the region between −26 and 134 bp relative to the transcription initiation site and a primer set amplifying an upstream fragment, centered at −1544 bp. FLIP is a target of the C/EBPα:p50 complex17 and was used as a model in our ChIP experiments. On average, from ≥ 4 experiments, activation of C/EBPα or its LZ variant resulted in a significant 90% reduction in the association of HDAC1 or HDAC3 with the endogenous FLIP promoter. In contrast, activation of the BR mutant C/EBPαBR3 did not have a significant effect on HDAC occupancy (Figure 4C). Induction of C/EBPα expression did not have an effect on the cellular content of HDAC1 or HDAC3 (not shown). Bcl-3, an atypical IκB, may bind p50 homodimers and serve as a coactivator or corepressor in a gene-specific manner30,31 ; however, C/EBPα did not have an effect of bcl-3 occupancy on the FLIP promoter (not shown).
In correlation with the displacement of HDACs, ChIP analysis showed that, on the FLIP promoter, induction of MT-C/EBPα or C/EBPαLZ resulted in 3.5- to 5.8-fold or 4.6- to 8.7-fold increase in histone H3 or histone H4 acetylation, respectively (Figure 4D). Of note, increased histone acetylation was restricted to the proximal promoter region in which C/EBPα displaced HDAC1 or HDAC3.
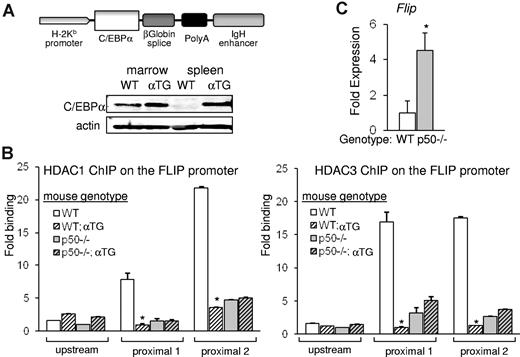
C/EBPα displaces endogenous HDAC from nuclear NF-κB p50
To study if the displacement of HDAC from the FLIP promoter reflects displacement of HDAC bound to p50, we used transgenic mice expressing C/EBPα from the H2Kb promoter and the lymphoid-specific Eμ enhancer (Figure 5A). C/EBPα is expressed in spleen cells of the αTG mice but not of control littermates, and expression of exogenous C/EBPα in splenocytes is at a level similar to its endogenous expression in nucleated WT marrow cells (Figure 5A). Nfkb1−/− mice (lacking p50) and strain-matched WT controls were bred with αTG mice to generate p50−/−;αTG and matched WT;αTG mice. Single-cell suspensions of splenocytes were subjected to ChIP analysis to assess occupancy of HDAC1 or HDAC3 on the FLIP promoter in cells carrying or lacking the C/EBPα transgene. On average, from 3 independent experiments, in WT mice the ectopic presence of C/EBPα resulted in a significant reduction of endogenous HDAC1 or HDAC3 on the FLIP promoter to 15% or 6%, respectively. In contrast, in p50−/− mice, the C/EBPα transgene did not have an effect on HDAC1 or HDAC3 occupancy on the FLIP promoter (Figure 5B). In correlation with the notably decreased HDAC occupancy on the FLIP promoter in p50−/− mice, expression of FLIP mRNA increased in these mice (Figure 5C). These findings suggest that C/EBPα displaces primarily p50-bound HDAC1 or HDAC3 from the FLIP promoter and that diminished presence of p50:HDAC complexes is associated with up-regulation of gene expression.
C/EBPα preferentially displaces HDAC from NF-κB p50. (A) A diagram depicting the H2K-Eμ–C/EBPα transgene. Total cellular extracts from BM-nucleated cells or splenocytes of WT or H2K-Eμ–C/EBPα transgenic (αTG) mice were subjected to Western blotting for C/EBPα or actin. (B) Single-cell suspensions of splenocytes from WT, WT;αTG, p50−/−, or p50−/−;αTG mice were subjected to ChIP analysis with the use of HDAC1 (left) or HDAC3 (right) antibodies, and enrichment of precipitated DNAs corresponding to the proximal or the distal regions of the Flip promoter were determined with quantitative real-time PCR relative to input. Averages from 3 repetitions are presented. (C) Flip transcripts were measured in splenocytes of WT or p50−/− mice and normalized to mS16 levels with the use of quantitative reverse transcriptase PCR. Mean and SE from 3 independent experiments are shown.
C/EBPα preferentially displaces HDAC from NF-κB p50. (A) A diagram depicting the H2K-Eμ–C/EBPα transgene. Total cellular extracts from BM-nucleated cells or splenocytes of WT or H2K-Eμ–C/EBPα transgenic (αTG) mice were subjected to Western blotting for C/EBPα or actin. (B) Single-cell suspensions of splenocytes from WT, WT;αTG, p50−/−, or p50−/−;αTG mice were subjected to ChIP analysis with the use of HDAC1 (left) or HDAC3 (right) antibodies, and enrichment of precipitated DNAs corresponding to the proximal or the distal regions of the Flip promoter were determined with quantitative real-time PCR relative to input. Averages from 3 repetitions are presented. (C) Flip transcripts were measured in splenocytes of WT or p50−/− mice and normalized to mS16 levels with the use of quantitative reverse transcriptase PCR. Mean and SE from 3 independent experiments are shown.
C/EBPs or C/EBPα myeloid oncoproteins disrupt the HDAC:NF-κB p50 complex
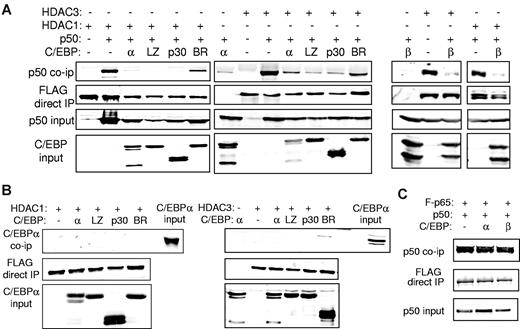
To evaluate the effect of C/EBPα or its variants on interaction between NF-κB p50 and HDAC1 or HDAC3, we expressed p50 and FLAG-tagged HDAC1 or HDAC3 in 293T cells with or without a C/EBPα variant. Cell lysates were immunoprecipitated with the use of anti-FLAG beads, and the precipitated proteins or input extracts were subjected to Western blotting. As expected, p50 interacted with HDAC1 or HDAC3. Coexpression of C/EBPα or its C/EBPαLZ or C/EBPαp30 AML oncoprotein mutants reproducibly interrupted interaction between p50 and HDAC1 and to a lesser extent HDAC3 (Figure 6A). Importantly, the C/EBPαBR3 BR mutant, which cannot interact with p50, induce p50, bcl-2, or FLIP or protect from apoptosis, also did not interfere with the p50:HDAC interaction (Figure 6A). Similar to C/EBPα, C/EBPβ also disrupted NF-κB p50 interaction with HDAC1 or HDAC3 (Figure 6A). As a control, we show that neither C/EBPα nor its leukemic variants interact directly with either HDAC1 or HDAC3 (Figure 6B). In addition, C/EBPα does not disrupt the association between NF-κB p50 and NF-κB p65 (Figure 6C).
C/EBP interrupts interaction between NF-κB p50 and HDAC. (A) 293T cells were cotransfected with the indicated combinations of 1 μg of CMV-p50, 3 μg of the indicated C/EBPα isoform (left) or C/EBPβ (right), and 1.5 μg of CMV-FLAG-HDAC1 or 750 ng of CMV-FLAG-HDAC3. Cell extracts were immunoprecipitated (IP) with the use of anti-FLAG beads, and the precipitated lysates or input extracts were Western blotted with the indicated antibodies. (B) 293T cells were transfected with similar quantities of C/EBPα, CMV-FLAG-HDAC1, or CMV-FLAG-HDAC3 as indicated, and cell extracts were immunoprecipitated with anti-FLAG beads followed by Western blotting with the use of the indicated antibodies. Representative blots from 3 experiments are shown. (C) 293T cells were cotransfected with 1 μg of CMV-p50, 1 μg of CMV-FLAG-p65, and 3 μg of the indicated CMV-C/EBP plasmid. Anti-FLAG beads were used to immunoprecipitate cell extracts, and the precipitated lysates or input extracts were subjected to Western blotting with the use of the indicated antibodies.
C/EBP interrupts interaction between NF-κB p50 and HDAC. (A) 293T cells were cotransfected with the indicated combinations of 1 μg of CMV-p50, 3 μg of the indicated C/EBPα isoform (left) or C/EBPβ (right), and 1.5 μg of CMV-FLAG-HDAC1 or 750 ng of CMV-FLAG-HDAC3. Cell extracts were immunoprecipitated (IP) with the use of anti-FLAG beads, and the precipitated lysates or input extracts were Western blotted with the indicated antibodies. (B) 293T cells were transfected with similar quantities of C/EBPα, CMV-FLAG-HDAC1, or CMV-FLAG-HDAC3 as indicated, and cell extracts were immunoprecipitated with anti-FLAG beads followed by Western blotting with the use of the indicated antibodies. Representative blots from 3 experiments are shown. (C) 293T cells were cotransfected with 1 μg of CMV-p50, 1 μg of CMV-FLAG-p65, and 3 μg of the indicated CMV-C/EBP plasmid. Anti-FLAG beads were used to immunoprecipitate cell extracts, and the precipitated lysates or input extracts were subjected to Western blotting with the use of the indicated antibodies.
Discussion
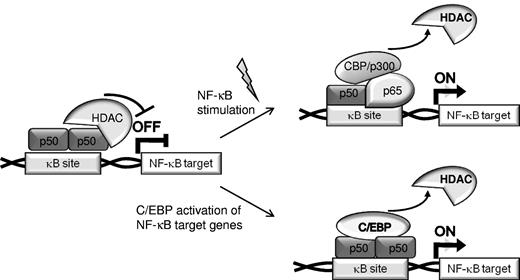
Transcription factors cooperate in gene regulation by binding to adjacent cis elements or by protein/protein interaction. C/EBPα synergizes with NF-κB p50, but not with p65, to activate antiapoptotic genes, including CFLAR (encoding for FLIP), BCL2, and MCL1,16,17,32 and the presence of p50 is important for the antiapoptotic effect of C/EBPα. In the absence of canonical NF-κB activation, p50 homodimers bind DNA and repress NF-κB–dependent gene transcription.21,23 The presence of p50 homodimers in the nucleus reflects in part the 30-fold greater affinity of IκBα for p65 compared with p50.33 Various cellular stimuli result in IκB phosphorylation by the IκB kinase complex and its subsequent ubiquitination and proteasomal degradation, leading to translocation of p50:p65 heterodimers into the nucleus where they displace p50 homodimers from κB sites, bind DNA, and activate NF-κB responsive genes. We now demonstrate that C/EBPα, its variants expressed in AML, or C/EBPβ have the capacity to displace HDACs from NF-κB p50:p50 homodimers in unstimulated cells to activate NF-κB target genes whose expression would otherwise be repressed (Figure 7). Thus, we define a mechanism underlying an alternative pathway for activation of NF-κB target genes. In addition, C/EBPs potentially also complex with p50:p65 heterodimers to augment transcription in stimulated cells.
Two pathways for activation of NF-κB target genes. In unstimulated cells, expression of NF-κB–regulated genes is often repressed by recruitment of HDACs by p50:p50 homodimers bound to nuclear κB sites (top). Canonical NF-κB stimulation results in displacement of p50 homodimers, binding of p65:p50 heterodimers, and recruitment of coactivators to chromatin (top). Alternatively, C/EBPs or C/EBPα AML mutants displace HDACs bound to p50 homodimers, resulting in activation of NF-κB–regulated gene independent of activation of the NF-κB system (bottom).
Two pathways for activation of NF-κB target genes. In unstimulated cells, expression of NF-κB–regulated genes is often repressed by recruitment of HDACs by p50:p50 homodimers bound to nuclear κB sites (top). Canonical NF-κB stimulation results in displacement of p50 homodimers, binding of p65:p50 heterodimers, and recruitment of coactivators to chromatin (top). Alternatively, C/EBPs or C/EBPα AML mutants displace HDACs bound to p50 homodimers, resulting in activation of NF-κB–regulated gene independent of activation of the NF-κB system (bottom).
Within a few hours of C/EBPα activation by E2, HDAC occupancy on the FLIP promoter was diminished, resulting in the expected increase of histone acetylation marks. Similar reduction in HDAC binding to the FLIP promoter was seen in cells expressing C/EBPα or C/EBPαLZ from the zinc-regulated MT promoter or in αTG mice, confirming that HDAC displacement is not because of the ER segment. Expression of the C/EBPα transgene in splenocytes resulted in a significant reduction of both HDAC1 and HDAC3 interaction with the FLIP promoter. In contrast, no significant effect was seen when C/EBPα is expressed in p50−/− mice. Interestingly, the HDAC occupancy on the Flip promoter in WT mice expressing the C/EBPα transgene is similar to the level seen in the p50−/− mice. Together, these data suggest that, on this promoter, C/EBPα specifically displaces HDACs recruited to NF-κB p50 rather than from other bound transcription factors.
NF-κB p50 binds the CEBPA promoter and regulates its expression in synergy with C/EBPα itself. Consequently, mice lacking p50 have impaired granulopoiesis explained in part by diminished C/EBPα levels.18 Our current finding that C/EBPα induces NF-κB p50 expression indicates that these proteins reciprocally induce each other's expression, forming a transcriptional positive feedback loop. Of note, this cooperation is specific, because C/EBPα had no effect on p65 transcription paralleling the functional transcriptional synergy between C/EBPα and p50 but not p65 for BCL2 gene induction and the apparent increased affinity of C/EBPα for p50 compared with p65.16
NF-κB activity is controlled primarily through posttranscriptional regulation. Less is known about the transcriptional regulation of members of this family.29 Function of the prototypic p50:p65 dimers depends on IκB kinase activation, whereas p50 homodimers are the most prevalent NF-κB species in the nuclei of resting cells.21 nfkb1 is translated as a p105 precursor that in part is constitutively cleaved to p50 by the 20S proteasome independent of ubiquitination, allowing translocation of active p50 homodimers into the nucleus.34 We now find that C/EBPα or a C/EBPαLZ AML mutant that cannot bind DNA directly can occupy the endogenous nfkb1 promoter and induce its expression. Activation of C/EBPα-ER results in rapid induction of nfkb1 mRNA that is not abrogated by cycloheximide, consistent with direct transcriptional induction. C/EBPα activation of the nfkb1 promoter depends on the integrity of 2 κB sites, which p50 binds but C/EBPα cannot bind directly. These sites are strictly conserved between mouse and human. Together these data indicate that nfkb1 is a member of a group of genes, including bcl-2 and FLIP,16,17 in which C/EBP or C/EBP oncoproteins contribute to trans-activation by tethering to NF-κB bound to DNA rather than by direct DNA binding. We previously demonstrated stronger activation of the bcl-2 and higher induction of Flip expression be C/EBPαLZ compared with C/EBPα.16,17 A potential explanation may be that the WT C/EBPα binds DNA both directly through C/EBP sites and indirectly by cooperation with p50. In contrast, the C/EBPαLZ mutant cannot bind DNA. Thus, although expressed at a similar level (Figures 1C, 3C), more C/EBPαLZ is available to interact with p50 and derepress target genes, as suggested by the increased histone acetylation marks (Figure 4D). C/EBPβ LIP was reported to increase the activity of the IL-6 promoter but only if a κB binding site is intact.35 Because LIP lacks a TAD, derepression of NF-κB–responsive genes is a potential mechanism. Indeed, LIP activated the nfkb1–1750-Luc reporter containing 2 κB sites but failed to activate the (C/EBP)2TK-Luc reporter. Similarly, a C/EBPαΔ3-8 variant that lacks all TADs effectively induces endogenous bcl-2.16 Derepression secondary to diminished HDAC recruitment to the promoter may also account for the higher Flip expression in p50−/− compared with WT mice.
Our data provide a mechanism whereby C/EBPα or its AML mutants can activate NF-κB target genes and contribute to their dysregulated expression. Current approaches to target NF-κB activation are centered around controlling the cytoplasmic regulation of NF-κB activation. Our findings lend support to the idea that disrupting the C/EBP:NF-κB p50 complex may offer a complementary approach for targeting NF-κB activity in the nucleus to favor apoptosis in AML or other malignancies in which C/EBP family members are expressed. Moreover, inhibiting C/EBP:p50 interaction may synergize with cytoplasmic inhibitors of NF-κB and may be specifically useful in cancers that lack canonical NF-κB activation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
We thank Dr R. Dikstein (Weitzman Institute, Israel) for providing the κBx2-Luc plasmid.
This work was supported by the St Baldrick's Foundation and the Elsa Pardee Foundation (I.P.-P.), the National Institutes of Health (grants R01 HL082948, R01 HL089176, U01 HL099775, and U01 HL100397; A.D.F.), and the Samuel Waxman Cancer Research Foundation (A.D.F.).
National Institutes of Health
Authorship
Contribution: I.P.-P. designed and performed the research, analyzed the data, and wrote the manuscript; S.H. and J.D. performed the research; and A.D.F. designed the research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ido Paz-Priel, Bunting-Blaustein Cancer Research Bldg I, Rm 208, 1650 Orleans St, Baltimore, MD 21231; e-mail address: ipazpri1@jhmi.edu.