Abstract
A subset of natural killer (NK) cells in normal mouse lymph node (LN) expresses CD127 (IL-7 receptor-α chain) and is thought to derive from the thymus. However, CD127+ NK cells are found in the LN of athymic mice. Therefore, the origin of CD127+ NK cells in the LN is unclear. Here, we have identified unique NK-cell progenitors (NKPs) in the LN that express the pan-NK cell marker CD49b and CD127 but lack CD122 and lineage markers. The LN NKPs develop in vitro into CD127+ NK cells that display natural cytotoxicity and cytokine production capacity. They also become CD127+ NK cells in lymphopenic mice that received a transplant. LN NKPs can be divided into stem cell antigen-1 (Sca-1)hi and Sca-1lo subsets. The latter comprise ∼ 60% of LN NKPs in normal mouse and < 10% of athymic mouse LN NKPs. Whereas both Sca-1hi and Sca-1lo NKPs develop into CD127+ NK cells in vitro, only those derived from Sca-1lo LN NKPs have rearranged TCRγ genes. Thus, CD127+ NK cells in the LN seem to be generated, at least in part, from both thymus-dependent Sca-1lo and thymus-independent Sca-1hi LN NKPs.
Introduction
Natural killer (NK) cells are lymphocytes of the innate immune system. They rapidly respond to tumors, viral infections, and allogeneic BM grafts by direct cytotoxicity and cytokine production.1-3 Mouse NK cells are identified by cell surface markers, including NK1.1 (NKRP1C), CD49b (Dx5), and NKp46.4 They also express multiple inhibitory receptors, including the Ly49 family and CD94-NKG2A heterodimer, which recognize classic and nonclassic MHC class I, respectively,5 as well as the stimulatory receptor NKG2D recognizing stress-induced ligands.6 NK cells acquire these receptors as they develop from hematopoietic stem cells in the BM.7 Committed NK cell progenitors (NKPs) in the BM have been identified by the lack of the mature NK-cell markers (NK1.1 and DX5) and the expression of the IL-2/IL-15 receptor β chain (CD122).8,9 They become immature NK cells expressing NKG2D, NK1.1, CD94-NKG2A, NKp46, and CD127 (IL-7Rα). Immature NK cells are neither cytotoxic nor do they produce IFNγ.8 As they further mature, they acquire the pan-NK cell marker CD49b (Dx5) and Ly49 as well as cytotoxicity and cytokine production capacities.10 Mature NK cells are heterogeneous and can be divided into subsets on the basis of their cell-surface receptors, including CD11b (Mac-1) and CD27.11 Although the relationship between different subsets of mature NK cells is still unclear, they are thought to indicate further maturation of NK cells.11,12
Most studies on mouse NK cells focus on spleen NK cells, and it is generally thought that they develop in the BM. However, NK cells are widely distributed, and those in various tissues differ from spleen NK cells in receptor expression patterns. Of particular interest are NK cells in the lymph node (LN). Approximately 20% of LN NK cells, whereas only ∼ 5% of splenic NK cells, express CD127 (α-chain of the IL-7 receptor). CD127+ LN NK cells have less Ly49 receptor expression, produce larger amounts of IFNγ, and are less cytolytic than CD127− NK cells.13 Most thymic NK cells are CD127+. Moreover, transplantation of newborn mouse thymus results in donor-derived CD127+ NK cells in the recipient mice, and athymic nude mice have significantly less CD127+ NK cells.13 Therefore, it has been suggested that CD127+ NK cells derive from the thymus.
We have previously reported that TCRγ genes are rearranged in some mouse NK cells.14 Although TCRγ gene rearrangement is rare (< 5%) among NK cells in most tissues, ∼ 20% of LN NK cells have rearranged TCRγ genes. Furthermore, those NK cells are undetectable in athymic mice, indicating that they are thymus dependent.15 Although CD127+ NK cells in the LN are thought to be thymus derived, TCRγ gene rearrangement is detectable at similar levels in both CD127+ and CD127− LN NK cells.15 Thus, the relationship between CD127+ LN NK cells and those with rearranged TCRγ genes has been unclear.
In this study, we reexamined CD127+ NK cells in the LN and found that athymic mouse LNs have normal percentages of CD127+ NK cells, indicating that most CD127+ LN NK cells are thymus independent. We then found a population of NKPs in the LN cells that does not express mature lymphoid and myeloid cell markers (lineage markers; Lin) and CD122 but express CD49b and CD127. They differentiate into CD127+ NK cells in vitro as well as in vivo. Our results suggest that at least some CD127+ LN NK cells develop from novel Lin−CD49b+CD127+ NKPs in the LN.
Methods
Mice
C57BL/6J (B6), B6-Ly5.1-Pep3b (B6.Pep3b), and NOD/SCID/IL-2Rγ-deficient (NSG) mice were bred and maintained in the Animal Resource Center of the British Columbia Cancer Research Center. Recombination activating gene 1 (RAG1)–deficient and Foxn1-deficient (nude) mice of the B6 background were obtained from The Jackson Laboratory. All mice were kept under specific pathogen-free conditions in the Animal Resource Center of the British Columbia Cancer Research Center. All animal use was approved by the animal care committee of the University of British Columbia, and animals were maintained and killed under humane conditions in accordance with the guidelines of the Canadian Council on Animal Care.
Flow cytometric analysis
Single-cell suspensions prepared from spleen, LN, BM, and blood; erythrocytes were analyzed by flow cytometer as described.15 The following FITC, PE, R-PE-cyanine 7 (Cy7), peridinin chlorophyll protein complex (PerCP)–Cy5.5, or allophycocyanin (APC)–conjugated monoclonal Abs (mAbs; BD Biosciences) were used (clone names given in parentheses): anti-CD3ϵ (145-2C11), anti-CD4 (GK1.5), anti-CD8α (53-6.7), anti-CD11b (M1/70), anti-CD11c (HL3), anti-CD25 (PC61), anti-CD45.2 (104), anti-CD49b (HMα2 and Dx5), anti-CD117 (2B8), anti-CD122 (TM-β1), anti-B220 (anti-RA3-6B2), TCRβ (H57-597), anti-TCRγδ (GL3), anti-Ly49D (4E5), Ly49G (4D11), anti–Ter-119 (Ly76), anti–Gr-1 (RB6-8C5), and anti-NK1.1 (PK136); from eBioscience: anti-CD24 (M1/69), anti-CD127 (A7R34), anti–stem cell antigen-1 (Sca-1; D7), and anti-NKp46 (29A1.4). Anti-Ly49H mAb (3D19) was a gift from Dr W. Yokoyama (Washington University, St Louis, MO).
TCRγ gene rearrangement
Genomic PCR to determine TCRγ rearrangement was carried out as described previously14 with modified primers. The following primers were used: forward 5′-CAGTTGGACATGGGAAGTTGGAG-3′ and reverse 5′-CAGAGGGAA TTACTATGAGCTTAGTT-3′.
Isolation and in vitro cultures of NK progenitors
BM and LN cells were incubated with 2.4G2 hybridoma supernatant to block Fc receptors. LN cells were stained with FITC-conjugated Lin antibodies (CD3ϵ, CD4, CD8α, CD11b, CD11c, CD19, CD122, B220, NK1.1, Gr-1, TCRβ, TCRγδ, and Ter-119), CD127-PE, CD49b-APC, and Sca-1–Pe–Cy7. BM cells were stained with a different set of FITC-conjugated antibodies for the isolation of BM NK progenitors (CD3ϵ, CD4, CD8α, CD11b, CD11c, CD19, B220, Gr-1, TCRβ, TCRγδ, and Ter-119), CD122-PE, and NK1.1-APC. Cells were sorted on a FACSAria II (BD Biosciences) for 2 rounds to a purity of > 99%. For bulk NK-cell differentiation cultures, the sorted cells were cultured in the presence of stem cell factor, Flt-3 ligand, IL-7, and IL-15 in 24-well plates containing a preformed stroma layer of OP9 cells as described previously.15 One-half of the media was replaced every 4-5 days, and cultures were analyzed after 12 days. For limiting dilution, the secondary sort was done in purity mode into 96-well plates at a low flow rate on preformed OP9 stroma (2 × 103 cells/well).
IFNγ production assay
NK cells were cultured in 150 μL of RPMI 1640 medium with 10% FCS, 2mM l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 × 10−5M β-mercaptoethanol in the presence of 1 ng/mL IL-12 and 0.5 ng/mL IL-18 or were left unstimulated for 18 hours at 37°C. IFN-γ production in culture supernatant was determined by the capture ELISA, following the manufacturer's instructions (R&D Systems). The detection limit was 62.5 pg/mL.
Cytotoxicity assay
The NK-sensitive cell lines RMA-S, YAC-1, and NK-resistant RMA cells were stained with CellTacker Green CMFDA (Invitrogen). The CFSE-labeled target cells (104) were mixed with effector NK cells at a 1:1, 1:2.5, and 1:5 ratio in 150 μL of RPMI 1640, 10% FCS on a 96-well plate. After 4 hours of incubation, cells were washed, stained with 1 μg/mL 7-aminoactinomycin D, and analyzed by FACS on a FACSCalibur. The level of cytotoxicity was determined as the percentage of 7-aminoactinomycin D–positive cells among CFSE+ target cells minus background.
Transplantation experiments
LN and BM NKPs from B6 mice were sorted to high purity (> 99%), and 1 × 103 cells were injected intravenously into NSG mice in 300 μL of PBS. Blood, spleen, and BM of injected mice were analyzed after 28 days.
Migration experiments
Splenocytes from B6.RAG1 knockout (KO) mice were either injected into B6.Pep3b mice or labeled with CellTacker Green CMFDA, following the manufacturer's instructions (Invitrogen) and injected into wild-type (WT) B6 mice. Approximately 18 hours later, single-cell suspensions were prepared from BM, spleen, blood, and LNs and analyzed by flow cytometry. For B6.Pep3b mice receiving unlabeled RAG1 KO spleen cells, cells were stained with anti–CD45.2-PerCP-Cy5.5, anti–NKp46-FITC, and anti–CD127-PE. For B6 mice receiving CFSE-labeled spleen cells, cells were stained with anti–NK1.1-PerCP-Cy5.5, anti–CD49b-APC, and anti–CD127-PE.
Results
LNs of athymic mice contain CD127+ NK cells
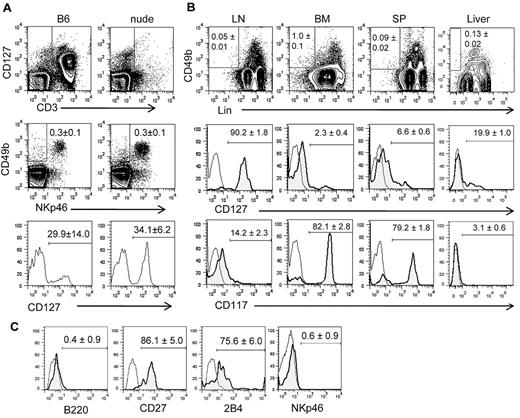
Vosshenrich et al13 have previously reported that the percentages of CD127+ cells among splenic and LN NK cells are much lower in athymic mice than in WT B6 mice. In the study, NK cells were identified by CD3−CD122+NK1.1+. Here, we reexamined the frequency of CD127+ NK cells with the use of the natural cytotoxicity receptor NKp46 to identify NK cells because it is thought to be a specific marker for mouse NK cells.16 WT and athymic nude (Foxn1−/−) B6 mice did not differ significantly in the frequencies of CD127+ cells among CD3−NKp46+ NK cells in the LN, spleen, and BM. Very few NK cells in the liver expressed CD127, and the frequency of CD127+ cells was slightly lower in nude liver than that of B6 liver (P < .05) (Figure 1A). As reported previously, CD127+ NK cells were relatively rich in the LN, whereas they comprise only a minor population in the spleen, BM, and liver. Analysis of young (4 weeks old), middle-aged (11 weeks old), and old (29 weeks old) WT and nude B6 mice (sex and age matched) showed that the frequency of LN CD127+ NK did not change with the age of the mice (data not shown). Individual analysis of axillary, inguinal, cervical, and popliteal LNs showed no significant difference among different LNs (data not shown). Therefore, LN cells were pooled for subsequent studies. Despite the lack of T cells, the LNs of nude mice contained significantly more lymphocytes than WT counterparts (63.0 ± 23.3 × 106 vs 27.5 ± 10.8 × 106; P < .01). NK cells were 2.7% of nude mouse LN cells and only 0.6% of B6 mouse LN cells. Thus, 2.1 ± 0.8 × 105 CD127+ NK cells are found in nude mouse LN compared with 2.1 ± 0.5 × 104 CD127+ NK cells in B6 mouse LN. Although most CD3−NKp46+ cells coexpressed high levels of NK1.1 or CD49b, some CD3−NKp46+ cells did not express NK1.1 or CD49b. Similarly, some CD3−NK1.1+ or CD3−CD49b+ did not express NKp46 (data not shown). Therefore, CD127 expression on LN NK cells defined by CD3−NK1.1+ or CD3−CD49b+ was also tested. Again, there was no significant difference between WT and athymic B6 mice in the frequency of CD127+ NK cells in the LN (Figure 1B). We further analyzed the phenotype of CD127+ and CD127− NK cells in the LN of WT and nude B6 mice and found no significant difference in the expression of Ly49s, CD122, KLRG1, CD11b, CD27, NK1.1, and DX5 with the only exception being CD117 (c-Kit), which was expressed at significantly (P < .05) higher levels in nude mouse LN NK cells (Figure 1C).
CD127+ NK cells are found in athymic mouse LNs. (A) NK cells in the LNs, spleen (SP), BM, and liver of WT B6 mice and Foxn1-KO (nude) B6 mice were identified by flow cytometry as CD3−NKp46+ (gated by rectangles in dot plots) The numbers show average (± SD) percentages of cells within the gates. The expression of CD127 on NK cells is shown by histograms. The numbers above and below the bars show the percentages of CD127+ NK cells in B6 and nude mice, respectively. (B) NK cells in the LNs of B6 and nude mice were identified by CD3−NK1.1+ (top) or CD3−CD49b+ (bottom) as shown by rectangles in dot plots, and CD127 expression on NK cells is shown in histograms. Histogram of B6 (thin line) and nude (bold line) are overlaid. The numbers above the bars in the histograms show the percentages of NK cells expressing CD127 (± SD) in B6 mice and below the bars are those in nude mice. (C) NK cells in the LNs of B6 and nude mice were gated as CD3−NKp46+, and the dot plots show the coexpression of CD127 and the indicated surface markers. The numbers above the bars in the dot plots show the percentages (± SD) of NK cells expressing both CD127 and the indicated marker, and the numbers below the bars show the percentages (± SD) of CD127− NK cells expressing the indicated marker. Results are representative of ≥ 3 individual experiments.
CD127+ NK cells are found in athymic mouse LNs. (A) NK cells in the LNs, spleen (SP), BM, and liver of WT B6 mice and Foxn1-KO (nude) B6 mice were identified by flow cytometry as CD3−NKp46+ (gated by rectangles in dot plots) The numbers show average (± SD) percentages of cells within the gates. The expression of CD127 on NK cells is shown by histograms. The numbers above and below the bars show the percentages of CD127+ NK cells in B6 and nude mice, respectively. (B) NK cells in the LNs of B6 and nude mice were identified by CD3−NK1.1+ (top) or CD3−CD49b+ (bottom) as shown by rectangles in dot plots, and CD127 expression on NK cells is shown in histograms. Histogram of B6 (thin line) and nude (bold line) are overlaid. The numbers above the bars in the histograms show the percentages of NK cells expressing CD127 (± SD) in B6 mice and below the bars are those in nude mice. (C) NK cells in the LNs of B6 and nude mice were gated as CD3−NKp46+, and the dot plots show the coexpression of CD127 and the indicated surface markers. The numbers above the bars in the dot plots show the percentages (± SD) of NK cells expressing both CD127 and the indicated marker, and the numbers below the bars show the percentages (± SD) of CD127− NK cells expressing the indicated marker. Results are representative of ≥ 3 individual experiments.
Normal B6 mouse LNs contain a unique population of Lin−CD49b+CD127+ cells
In the above-described analysis, we detected LN cells expressing CD49b but not NKp46. Some CD3−CD49b+NKp46− LN cells expressed CD127 (Figure 2A). Our initial characterization of this population showed that it contained CD11b+ cells, CD19+ cells, and other undefined cells. Therefore, we further characterized the population with the use of a cocktail of mAbs to mature hematopoietic cell markers (Lin), including CD122, NK1.1, CD3ϵ, CD4, CD8α, TCRβ, TCRγδ, CD11b, CD11c, CD19, B220, Gr-1, and Ter-119. Note that the Lin antibody cocktail in this study included anti-CD122, which would exclude conventional Lin−CD122+ NKPs. The frequency of Lin−CD49b+ cells was 0.05% ± 0.01% (n = 5) in the LN, 1.0% ± 0.1% (n = 3) in the BM, 0.14% ± 0.01% (n = 3) in the blood, 0.09% ± 0.02% (n = 3) in the spleen, and 0.13% ± 0.02% (n = 3) in the liver of normal B6 mice. Although most Lin−CD49b+ cells in the LN expressed CD127, only a small fraction of those in other tissues tested was CD127+ (Figure 2B). Conversely, most Lin−CD49b+ cells in the BM expressed CD117 (c-kit), but only a minor subset in the LN was CD117+. Lin−CD49b+ cells in the spleen were similar to those in the BM, although they had higher percentages of CD127+ and CD117− cells. Lin−CD49b+ cells in the LN are B220− but express CD27 and 2B4 (Figure 2C). To examine the relationship between Lin−CD49b+ LN cells and the conventional Lin−CD122+ NKPs, another Lin cocktail of mAbs without anti-CD122 was used to identify Lin−CD122+ cells in the LN. Those cells were mostly CD127+, and only ∼ 25% expressed CD49b. Similarly, only < 20% of Lin (minus CD122)−CD49b+ LN cells were CD122+ (supplemental Figure, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Therefore, Lin−CD122−CD49b+LN cells are different from conventional Lin−CD122+ NKPs.
A unique population of LN cells expresses CD49b and CD127 but not mature lymphocyte markers. (A) Freshly isolated LN cells were stained with antibodies to CD3 and CD127, and CD3− cells were gated as shown by rectangles (top). The gated cells were analyzed for the expression of the NK cell markers NKp46 and CD49b (center). CD49b+NKp46− cells were further gated as shown by rectangles in contour plots, and the expression of CD127 on CD3−NKp46−CD49b+ cells from wild-type B6 and athymic nude mice was analyzed (bottom). (B) Lin− (CD122, NK1.1, CD3ϵ, CD4, CD8α, TCRβ, TCRγδ, CD11b, CD11c, CD19, B220, Gr-1, Ter-119) CD49b+ cells from LN, BM, spleen (SP), and liver were gated, and the expression of CD127 and CD117 (c-Kit) was analyzed. (C) Lin−CD49b+CD127+ cells in the LN were gated and analyzed for the expression of B220, CD27, 2B4, and NKp46. All the FACS profiles are representative of 3 independent experiments, and the numbers show average (± SD) percentages of cells within the indicated gates calculated from ≥ 3 independent experiments.
A unique population of LN cells expresses CD49b and CD127 but not mature lymphocyte markers. (A) Freshly isolated LN cells were stained with antibodies to CD3 and CD127, and CD3− cells were gated as shown by rectangles (top). The gated cells were analyzed for the expression of the NK cell markers NKp46 and CD49b (center). CD49b+NKp46− cells were further gated as shown by rectangles in contour plots, and the expression of CD127 on CD3−NKp46−CD49b+ cells from wild-type B6 and athymic nude mice was analyzed (bottom). (B) Lin− (CD122, NK1.1, CD3ϵ, CD4, CD8α, TCRβ, TCRγδ, CD11b, CD11c, CD19, B220, Gr-1, Ter-119) CD49b+ cells from LN, BM, spleen (SP), and liver were gated, and the expression of CD127 and CD117 (c-Kit) was analyzed. (C) Lin−CD49b+CD127+ cells in the LN were gated and analyzed for the expression of B220, CD27, 2B4, and NKp46. All the FACS profiles are representative of 3 independent experiments, and the numbers show average (± SD) percentages of cells within the indicated gates calculated from ≥ 3 independent experiments.
Lin−CD49b+CD127+ LN cells differentiate into CD127+ NK cells in vitro and in vivo
Lin−CD49b+CD127+ LN cells were sorted from normal B6 mice to high purity (> 99%) and cultured on a confluent layer of OP9 stroma cells in the presence of standard NK differentiation cytokine cocktail.17 Conventional NKPs (Lin−CD122+)8 in the BM were also sorted (Lin antibody cocktail excluded anti-CD122) and cultured for NK cell differentiation for comparison. Both Lin−CD49b+CD127+ LN cells and conventional BM NKPs efficiently differentiated into NK cells expressing NKp46 and CD122 at the end of the 12 days of culture (Figure 3A). Further analyses of NK cells generated in vitro from Lin−CD49b+CD127+ LN cells (LN NKPs hereafter), and conventional BM NKPs showed most NK cells generated from the former expressed significantly higher levels of CD127 than those generated from the latter, whereas the expression of NKG2D, NKG2A, and Ly49 was the same. Limiting dilution assays showed that ∼ 1 in 36 Lin−CD49b+CD127+ LN cells and 1 in 20 BM NKPs differentiated into NK cells in vitro (Figure 3B).
B6 LN Lin−CD49b+CD127+ cells differentiate into mature NK cells in vitro and in vivo. (A) LN Lin−CD49b+CD127+ and BM Lin−CD122+ cells from B6 mice were purified by FACS and cultured with OP9 stroma cells and cytokines for differentiation into NK cells. Cells were harvested after 12 days in culture and analyzed by flow cytometry for the expression of indicated cell surface markers. Data are representative of 5 independent experiments. The numbers show the average (± SD) percentages of positive cells. (B) The frequency of NK-cell precursors among LN Lin−CD49b+CD127+ cells (LN NKP) and control conventional BM Lin−CD122+ NKPs (BM NKP) was determined by limiting dilutions analysis. Both cell types were sorted in pools of 3, 10, 30, and 90 cells (n = 40) onto OP9 stroma layers and cultured for 12 days. The frequency ± SEM of NK progenitors in each population was calculated by L-Calc software (StemCell Technologies). (C) The cytotoxicity of NK cells generated in vitro from LN NKPs and BM NKPs was tested with the use of YAC-1 as target. (D) The in vitro–generated NK cells were stimulated overnight with 1 ng/mL IL-12 alone or IL-12 and IL-18 (0.5 ng/mL). The level of IFN-γ secretion was measured by ELISA. Shown is 1 representative experiment of 3. (E) Nonirradiated NOD/Scid/IL-2Rγ KO (NSG) mice were intravenously injected with FACS-purified LN NKPs (103 cells per mouse). After 4 weeks, spleen cells were analyzed for the presence of CD45.2+ cells. CD45.2+ cells were gated for NKp46+ cells, and the expression of CD127 and CD49b was analyzed (top). CD45.2+ cells were analyzed for the expression of NK1.1 (NK cells), CD3 (T cells), and CD19 (B cells) (bottom).
B6 LN Lin−CD49b+CD127+ cells differentiate into mature NK cells in vitro and in vivo. (A) LN Lin−CD49b+CD127+ and BM Lin−CD122+ cells from B6 mice were purified by FACS and cultured with OP9 stroma cells and cytokines for differentiation into NK cells. Cells were harvested after 12 days in culture and analyzed by flow cytometry for the expression of indicated cell surface markers. Data are representative of 5 independent experiments. The numbers show the average (± SD) percentages of positive cells. (B) The frequency of NK-cell precursors among LN Lin−CD49b+CD127+ cells (LN NKP) and control conventional BM Lin−CD122+ NKPs (BM NKP) was determined by limiting dilutions analysis. Both cell types were sorted in pools of 3, 10, 30, and 90 cells (n = 40) onto OP9 stroma layers and cultured for 12 days. The frequency ± SEM of NK progenitors in each population was calculated by L-Calc software (StemCell Technologies). (C) The cytotoxicity of NK cells generated in vitro from LN NKPs and BM NKPs was tested with the use of YAC-1 as target. (D) The in vitro–generated NK cells were stimulated overnight with 1 ng/mL IL-12 alone or IL-12 and IL-18 (0.5 ng/mL). The level of IFN-γ secretion was measured by ELISA. Shown is 1 representative experiment of 3. (E) Nonirradiated NOD/Scid/IL-2Rγ KO (NSG) mice were intravenously injected with FACS-purified LN NKPs (103 cells per mouse). After 4 weeks, spleen cells were analyzed for the presence of CD45.2+ cells. CD45.2+ cells were gated for NKp46+ cells, and the expression of CD127 and CD49b was analyzed (top). CD45.2+ cells were analyzed for the expression of NK1.1 (NK cells), CD3 (T cells), and CD19 (B cells) (bottom).
The functional maturity of LN NKP-derived NK cells was also assessed. They efficiently killed the prototypic NK cell target YAC-1 (Figure 3D). They also killed MHC class I–deficient RMA-S cells but not control RMA cells expressing MHC class I, indicating that the cytotoxicity was inhibited by MHC class I (data not shown). On stimulation with IL-12 and IL-18, they produced a large amount of IFN-γ (Figure 3E). Taken together, LN NKP-derived NK cells had the functions of mature NK cells.
To assess the NK-cell potential of LN NKPs in vivo, we grafted 1 × 103 LN NKPs from B6 mice (CD45.2) into lymphopenic NSG mice (CD45.1). Four weeks later, we assessed the level of chimerism by staining for CD45.2+ cells in the recipient mice and characterized their maturation level. Because the recipient mice have no LNs, we recovered LN NKP-derived CD45.2+ cells from the BM, spleen, and blood. The percentages of NK cells among CD45.2+ cells varied, ranging from 12% to 38.8%, whereas no mature T and B cells could be detected (CD3 and CD19, respectively; Figure 3F). Remarkably, ∼ 40% of donor-derived NK (NKp46+) cells expressed CD127. These results show that LN NKPs differentiate into CD127+ NK cells in vitro as well as in vivo.
Sca-1lo LN NKPs are thymus dependent
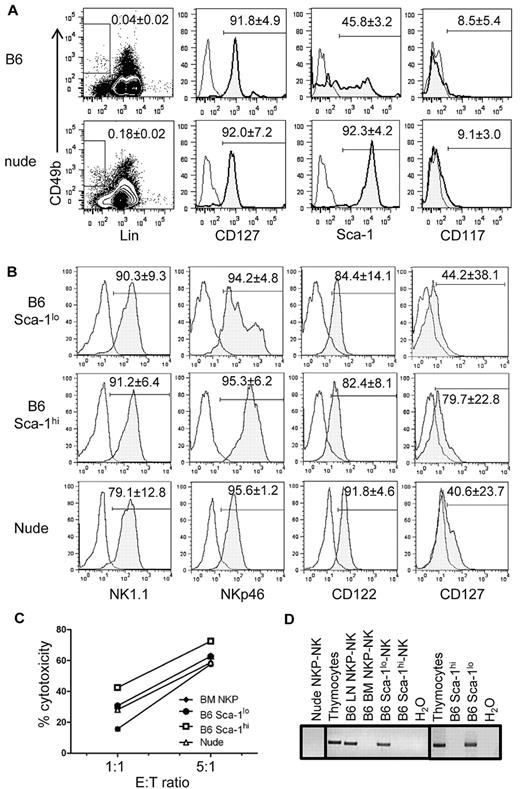
To determine whether Lin−CD49b+CD127+ LN NKPs derive from the thymus, we compared LN NKPs from WT and athymic nude (Foxn1−/−) B6 mice. Lin−CD49b+ cells were detected in nude mouse LNs at a higher frequency than in B6 mouse LNs (Figure 4A left). The total numbers of Lin−CD49b+ LN cells were ∼ 1.1 ± 0.3 × 105 in nude mouse and 1.1 ± 0.4 × 104 in WT B6 mouse. Both B6 and nude LN NKPs were CD127+CD117−. Further analysis showed a significant difference in the expression of Sca-1. Although approximately one-half (45.8% ± 3.2%; n = 7) of WT B6 LN NKPs expressed low levels of Sca-1, most nude mouse LN NKPs are Sca-1hi with few (7.0% ± 0.7%; n = 4) Sca-1lo cells. To test their NK-cell potential, nude mouse LN NKPs, as well as Sca-1lo and Sca-1hi LN NKPs, from WT mice were purified by cell sorting and cultured for NK-cell differentiation. They all differentiated into NK cells expressing NKp46, CD122, and variable levels of CD127 (Figure 4B). They were also functionally mature and efficiently killed YAC-1 cells (Figure 4C). Limiting dilution analysis of B6 LN NKP subsets showed that 1 in 21 Sca-1lo and 1 in 72 Sca-1hi LN NKPs differentiated into NK cells (data not shown).
Sca-1lo subset of LN NKPs is greatly reduced in athymic mouse LN. (A) WT (B6) and Foxn1−/− (nude) mouse LN Lin−CD49b+ cells were gated as shown by rectangles in dot plots, and CD127, Sca-1, and CD117 expressions were analyzed (histograms). (B) B6 LN Sca-1hi and Sca-1lo Lin−CD49b+CD127+ cells and nude LN Lin−CD49b+CD127+ cells were purified and cultured in vitro for NK-cell differentiation. NK cells thus generated were analyzed for the expression of indicated NK-cell markers. The numbers show average (± SD; n = 3) percentages of cells expressing the indicated markers. (C) The cytotoxicity of NK cells generated in vitro from Sca-1lo and Sca-1+ LN NKPs from B6 and LN NKPs from nude mice (B) were tested with the use of YAC-1 as target. (D) FACS-purified Sca-1lo or Sca-1hi LN NKPs from B6 and LN NKPs from nude mice were cultured for NK-cell differentiation as in panel B (left and middle). After 12 days, NKp46+ cells from the cultures were FACS-purified, and genomic DNA was extracted. Equivalent amounts of DNA were subjected to PCR for rearranged TCRγ genes and separated by agarose gel electrophoresis. DNA was also extracted from freshly isolated Sca-1lo and Sca-1hi LN NKPs from B6 mouse LN cells and analyzed for rearranged TCRγ genes as described before (right). Thymocytes served as a positive control, and NK cells generated from BM NKPs as a negative control.
Sca-1lo subset of LN NKPs is greatly reduced in athymic mouse LN. (A) WT (B6) and Foxn1−/− (nude) mouse LN Lin−CD49b+ cells were gated as shown by rectangles in dot plots, and CD127, Sca-1, and CD117 expressions were analyzed (histograms). (B) B6 LN Sca-1hi and Sca-1lo Lin−CD49b+CD127+ cells and nude LN Lin−CD49b+CD127+ cells were purified and cultured in vitro for NK-cell differentiation. NK cells thus generated were analyzed for the expression of indicated NK-cell markers. The numbers show average (± SD; n = 3) percentages of cells expressing the indicated markers. (C) The cytotoxicity of NK cells generated in vitro from Sca-1lo and Sca-1+ LN NKPs from B6 and LN NKPs from nude mice (B) were tested with the use of YAC-1 as target. (D) FACS-purified Sca-1lo or Sca-1hi LN NKPs from B6 and LN NKPs from nude mice were cultured for NK-cell differentiation as in panel B (left and middle). After 12 days, NKp46+ cells from the cultures were FACS-purified, and genomic DNA was extracted. Equivalent amounts of DNA were subjected to PCR for rearranged TCRγ genes and separated by agarose gel electrophoresis. DNA was also extracted from freshly isolated Sca-1lo and Sca-1hi LN NKPs from B6 mouse LN cells and analyzed for rearranged TCRγ genes as described before (right). Thymocytes served as a positive control, and NK cells generated from BM NKPs as a negative control.
We have previously shown that ∼ 20% of LN NK cells in WT B6 mice, but not nude mice, have rearranged TCRγ genes, indicating that TCRγ gene rearrangement is a unique marker for thymus-dependent NK cells.15 Therefore, we tested whether TCRγ genes are rearranged in NK cells generated in vitro from Sca-1lo and Sca-1hi subsets of LN NKPs. As expected from our previous report, NK cells generated from both LN NKPs of nude mice showed no TCRγ gene rearrangement. Interestingly, bulk B6 LN NKP-derived NK cells, but not BM NKP-derived NK cells, were positive for TCRγ rearrangement, and only those derived from the Sca-1lo subset of LN NKPs contained cells positive for TCRγ rearrangement (Figure 4D). B6 thymocytes were used as positive control for the genomic PCR analysis. To determine whether TCRγ genes are rearranged in NKPs or during the in vitro differentiation, Sca-1hi and Sca-1lo B6 LN NKPs were purified and tested for rearranged TCRγ genes. The results showed that freshly isolated Sca-1lo, but not Sca-1hi, LN NKPs have rearranged TCRγ genes. Those results collectively showed thymus-dependent Sca-1lo and thymus-independent Sca-1hi NKPs in the mouse LN differentiate into mature CD127+ NK cells.
CD127+ spleen NK cells do not preferentially migrate to the LN
The above-described results suggested that CD127+ NK cells in steady state LN may develop within the LN from the novel Lin−CD49b+CD127+ LN NKPs. Alternatively, CD127+ NK cells may develop elsewhere and preferentially migrate into the LN. To test the latter possibility, we intravenously injected CFSE-labeled spleen cells from RAG1-deficient mice into nonirradiated normal B6 mice and examined the migration of donor NK cells (CFSE+NK1.1+CD49b+) after ∼ 18 hours. The recovery of CFSE+ cells was the highest in the spleen (mean = 3.2 × 104/106 injected, SD = 1.6 × 104; n = 6) and lowest in the LN (Figure 5A left). When axillary, cervical, and inguinal LNs were individually isolated and analyzed, no preferential migration of CFSE+ cells into a specific LN was found. Therefore, all LNs of the recipients were pooled and further analyzed for donor NK cells. On average, 218 ± 50 (n = 6) CFSE+ cells were recovered in the LNs of recipients from 106 injected cells, and of those 68% ± 17% were NK cells (Figure 5A right). Approximately 33% of RAG1 KO spleen cells were NK cells. Thus, of ∼ 3.3 × 105 NK cells among 106 injected cells, only ∼ 148 NK cells (0.04%) were recovered in LNs. In this experiment, CD127 expression on CFSE+ NK cells in the LN was not reliably analyzed because of difficulty in compensating strong CFSE fluorescence. Therefore, we also injected unlabeled RAG1 KO spleen cells (CD45.2) into normal B6.Pep3b mice (CD45.1) and analyzed CD127 expression on CD45.2+NKp46+ NK cells in the injected mice. Because some RAG1 KO spleen cells were not strongly stained by anti-CD45.2, the total number of injected cells recovered in the recipients' lymphoid organs could not be determined in this analysis. However, the donor and host NK cells were readily identified by a combination of anti–CD45.2-PerCP-Cy5.5 and anti–NKp46-FITC staining (Figure 5B dot plots). The percentages of CD127+ cells among donor NK cells recovered in various lymphoid organs, including the LN of the recipients, were similar to that of the injected spleen cells (Figure 5B contour plots). These results showed that CD127+ NK cells do not preferentially migrate to the LN.
Splenic CD127+ NK cells do not preferentially migrate into the LN. (A) Spleen cells from RAG1 KO mice were labeled with CFSE and intravenously injected (3-3.5 × 106 per mouse) into nonirradiated B6 mice. Blood, spleen, LNs, and BM were isolated 18 hours after injection and analyzed by flow cytometry for the presence of CFSE+ cells. Left bar graph shows the total numbers of CFSE+ cells recovered after 18 hours. Right bar graph shows the percentages of NK cells (NK1.1+CD49b+) among CFSE+ cells recovered in the indicated lymphoid organs. (B) Unlabeled RAG1 KO spleen cells were intravenously injected into nonirradiated B6.Pep3b mice, and after 18 hours lymphoid organs were analyzed by flow cytometer for the expression of CD45.2, NKp4, and CD127. The donor NK cells (CD45.2+NKp46+) and the host NK cells (CD45.2−NKp46+) were gated as shown by rectangles in the dot plots, and the expression of CD127 is shown in contour plots. Data are representative of ≥ 3 mice. The numbers in contour plots show the percentages of CD127+ cells (mean ± SD).
Splenic CD127+ NK cells do not preferentially migrate into the LN. (A) Spleen cells from RAG1 KO mice were labeled with CFSE and intravenously injected (3-3.5 × 106 per mouse) into nonirradiated B6 mice. Blood, spleen, LNs, and BM were isolated 18 hours after injection and analyzed by flow cytometry for the presence of CFSE+ cells. Left bar graph shows the total numbers of CFSE+ cells recovered after 18 hours. Right bar graph shows the percentages of NK cells (NK1.1+CD49b+) among CFSE+ cells recovered in the indicated lymphoid organs. (B) Unlabeled RAG1 KO spleen cells were intravenously injected into nonirradiated B6.Pep3b mice, and after 18 hours lymphoid organs were analyzed by flow cytometer for the expression of CD45.2, NKp4, and CD127. The donor NK cells (CD45.2+NKp46+) and the host NK cells (CD45.2−NKp46+) were gated as shown by rectangles in the dot plots, and the expression of CD127 is shown in contour plots. Data are representative of ≥ 3 mice. The numbers in contour plots show the percentages of CD127+ cells (mean ± SD).
Discussion
Here, we report a novel population of NKPs in the mouse LN. Unlike conventional NKPs in the BM that are Lin−CD122+, the newly identified LN NKPs do not express CD122 but express CD127 and CD49b (pan-NK cell marker DX5/HMα2, very late antigen-2). NK cells generated in vitro from LN NKPs and those from BM NKPs differ as well. The former express CD127, whereas the latter are mostly CD127−. Although CD127 is expressed on immature NK cells, CD127+ NK cells generated from LN NKPs are functionally mature because they efficiently kill YAC-1 cells and produce IFN-γ on stimulation. On transplantation into lymphopenic NSG mice, LN NKPs differentiate into both CD127+ and CD127− NK cells, whereas most of the transplanted Lin−CD49b+CD127+ cells retain the expression of CD49b and CD127. Furthermore, LN NKPs can be divided into Sca-1hi and Sca-1lo subsets, both of which differentiate into CD127+ NK cells in vitro. Sca-1lo LN NKPs are greatly reduced in athymic Foxn1−/− mice, suggesting that most of them derive from the thymus.
Veinotte et al15 have previously detected thymus-dependent NK cells marked by TCRγ gene rearrangement, which are most abundant in the LN but undetectable in athymic mice. Interestingly, TCRγ gene rearrangement is not restricted to CD127+ NK cells because it is detected in both CD127+ and CD127− NK cells at similar levels. Consistent with those previous findings, in our current study TCRγ gene rearrangement is detectable in NK cells generated in vitro from B6 mouse LN NKPs but not athymic mouse LN NKPs. When B6 LN NKPs are divided into Sca-1hi and Sca-1lo subsets, TCRγ gene rearrangement is only detected in Sca-1lo NKPs and NK cells generated from them. Although most NK cells generated in vitro from LN NKPs are CD127+, both CD127+ and CD127− NK cells are generated from LN NKPs in mice that receive a transplant. Taken together, these results suggest that thymus-derived Sca-1lo LN NKPs, some of which have rearranged TCRγ genes, differentiate into CD127+ and CD127− NK cells. We have previously found that thymus-derived Lin−CD44+ NKPs in the LN resembling immature thymocytes develop into NK cells with rearranged TCRγ genes. CD127 was detected in only a minor subset of the Lin−CD44+ NKP population in this earlier study, and whether they develop into CD127+ NK cells was not tested. LN NKPs (Lin−CD127+CD49b+) in the current study are all CD44+ (data not shown) and seem to partially overlap with the Lin−CD44+ NKP population. It is possible that Sca-1lo LN NKPs derive from Lin−CD44+KitloCD127+Sca-1lo thymic progenitors reported by Terra et al18 However, the origin of Sca-1hi LN NKPs is currently unknown. They may directly migrate from the BM or alternatively develop from more immature lymphoid progenitors in the LN. Indeed, we have recently identified lymphoid progenitors that have T-, B-, and NK-cell potential in the LN (K.W., C.L., and F.T., manuscript in preparation), and some of the LN NKPs in this study may derive from those lymphoid progenitors.
Vosshenrich et al13 reported that most thymic NK cells are CD127+, athymic mice have greatly reduced CD127+ NK cells, and transplantation of thymus results in donor-derived CD127+ NK cells. On the basis of those findings they suggested that CD127+ NK cells are thymus derived. However, our current study did not find a reduction in CD127+ NK cells in athymic mice. The reason for the difference is still unclear. Whereas at least some CD127+ NK cells in WT B6 mice, particularly those with rearranged TCRγ genes, are thymus dependent, those in athymic mice are obviously thymus independent. It is possible that CD127+ NK cells in the athymic mouse LN may develop from LN NKPs. Alternatively, they may develop elsewhere and preferentially migrate into the LN. However, our migration study suggests that CD127+ NK cells do not preferentially migrate into the LN. Therefore, a relatively high percentage (∼ 20%) of CD127+ NK cells in the LN compared with spleen (∼ 5%) and peripheral blood (∼ 4%) cannot be explained by preferential migration of CD127+ NK cells from the circulating pool. Thus, it seems probable that CD127+ NK cells in the LN develop, at least in part, from thymus-dependent Sca-1lo and thymus-independent Sca-1hi LN NKPs.
Although some hematopoietic progenitors in the BM have been shown to express CD49b,19 the phenotype and lineage potential of LN NKPs is different from that of multipotent hematopoietic progenitors20 and the common lymphoid progenitor21,22 in the BM. LN NKPs do not express CD135 (fms-like tyrosine kinase receptor-3) and CD117 (c-Kit), and they only have NK potential. The developmental relationship between LN NKPs and conventional BM NKPs is still unclear. Although LN NKPs are CD122−, they acquire CD122 as they differentiate into mature NK cells while retaining CD127.
Our in vivo engraftment studies were performed in NSG mice that lack B, T, and NK cells and fail to develop lymph nodes during ontogeny. However, LN NKPs seem to have limited proliferative capacity in lymphopenic NSG mice compared with BM NKP. We assume that the poor engraftment potential was because of the absence of LNs in NSG. LN stroma cells are a source for IL-7,23 which seems to be required for the development of CD127+ NK cells.13 Remarkably, a large proportion of donor-derived cells that were recovered after 4 weeks shared a similar phenotype (CD49b+CD127+) with the injected LN NKPs, suggesting that they proliferate while maintaining their phenotype. Of note, the LN NKPs we described here are phenotypically different from mouse CD4+ lymphoid tissue inducer cells.24,25
Thus far, the physiologic relevance of LN NKP-derived NK cells is unknown. Under steady state conditions, NK cells in the LN localize to the interfollicular and cortex regions and stay in close proximity to dendritic cells, possibly allowing them to regulate initial immune responses in a timely manner.26 LN-resident NK cells expressing CD127 might also be involved in the maintenance of LN homeostasis by regulating the availability of IL-7 produced by stroma cells, a cytokine critical for B- and T-cell homeostasis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank C. Benz, D. Ko, and W. Xu for advice and excellent technical support.
This work was supported by grants from the Canadian Cancer Society Research Institute and the Deutsche Forschungsgemeinschaft (grant LU 1529/1-1; C.L.).
Authorship
Contribution: C.L. designed and performed the research, analyzed the data, and wrote the paper; K.W. designed and performed the research, analyzed the data, and wrote the paper; and F.T. designed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Fumio Takei, Terry Fox Laboratory, British Columbia Cancer Agency, 675 West 10th Ave, Vancouver, BC, Canada; e-mail: ftakei@bccrc.ca.
References
Author notes
C.L. and K.W. contributed equally to this study.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal